Abstract
The cell wall of fungal cells is important for cell integrity and cell morphogenesis and protects against harmful environmental conditions. The yeast cell wall is a complex structure consisting mainly of mannoproteins, glucan, and chitin. The molecular mechanisms by which the cell wall components are synthesized and transported to the cell surface are poorly understood. We have identified and characterized two homologous yeast proteins, Sbe2p and Sbe22p, through their suppression of a chs5 spa2 mutant strain defective in chitin synthesis and cell morphogenesis. Although sbe2 and sbe22 null mutants are viable, sbe2 sbe22 cells display several phenotypes indicative of defects in cell integrity and cell wall structure. First, sbe2 sbe22 cells display a sorbitol-remediable lysis defect at 37°C and are hypersensitive to SDS and calcofluor. Second, electron microscopic analysis reveals that sbe2 sbe22 cells have an aberrant cell wall structure with a reduced mannoprotein layer. Finally, immunofluorescence experiments reveal that in small-budded cells, sbe2 sbe22 mutants mislocalize Chs3p, a protein involved in chitin synthesis. In addition, sbe2 sbe22 diploids have a bud-site selection defect, displaying a random budding pattern. A Sbe2p–GFP fusion protein localizes to cytoplasmic patches, and Sbe2p cofractionates with Golgi proteins. Deletion of CHS5, which encodes a Golgi protein involved in the transport of Chs3p to the cell periphery, is lethal in combination with disruption of SBE2 and SBE22. Thus, we suggest a model in which Sbe2p and Sbe22p are involved in the transport of cell wall components from the Golgi apparatus to the cell surface periphery in a pathway independent of Chs5p.
INTRODUCTION
Yeast cell walls are essential for the maintenance of cell shape, prevention of lysis, and regulation of the uptake of substances from the environment. In spite of its apparent rigidity, the yeast cell wall is a dynamic structure that can be remodeled in response to different physiological states (e.g., budding, mating, and sporulation) or to morphological changes, such as in Candida albicans during the transition from yeast to hyphal growth (Cid et al., 1995). The Saccharomyces cerevisiae cell wall is a complex structure composed of mannoproteins, β-1,3-glucan, and β-1,6-glucan, all cross-linked to each other and to chitin, a N-acetylglucosamine polymer (Orlean, 1997). To mediate bud formation and growth, new plasma membrane and cell wall material must be directed properly to the site of growth, presumably through the secretory pathway. How cell wall components are transported to the cell surface and whether multiple pathways are involved remain poorly understood. The identification and characterization of components that participate in specific steps of this transport will be crucial for the understanding of how this process occurs at the molecular level.
Chitin is an essential structural component of the cell wall present at very low abundance (Shaw et al., 1991). Chitin deposition is spatially and temporally regulated throughout the yeast cell cycle and life cycle (for review, see Cid et al., 1995; Orlean, 1997). Three chitin synthase activities (CSI, CSII, and CSIII) have been described; each has a distinct function. CSIII activity is required for the formation of the chitin ring at the base of the bud and for chitin deposition in the lateral wall during vegetative growth, as well as for chitin synthesis during mating and sporulation (Shaw et al., 1991; Valdivieso et al., 1991). Several proteins, including Chs3p, Chs4p, Chs5p, Chs6p, and Chs7p, are required for the CSIII activity. Chs3p has significant homology with other chitin synthases and is the catalytic component of CSIII (Valdivieso et al., 1991). Chs3p is present at the cell periphery and in cytoplasmic patches and undergoes a dynamic localization during the cell cycle. Chs3p localizes at the incipient bud site in unbudded cells and at the bud neck in small-budded cells and cells undergoing cytokinesis (Chuang and Schekman, 1996; Santos and Snyder, 1997). Chs4p, Chs5p, Chs6p, Chs7p, and two members of the yeast SNAREs, Tlg1p and Tlg2p, are implicated in the highly regulated localization of Chs3p (DeMarini et al., 1997; Santos and Snyder, 1997; Holthuis et al., 1998; Ziman et al., 1998; Trilla et al., 1999). Chs4p is involved in the proper localization of Chs3p at the bud neck through an indirect interaction with the septins (DeMarini et al., 1997). Septins are highly conserved cytoskeletal proteins that assemble into filaments and are essential for cytokinesis (Longtine et al., 1996). Chs5p and Chs6p are required for the transport of Chs3p from an internal membrane compartment to the plasma membrane (Santos and Snyder, 1997; Ziman et al., 1998). Chs5p colocalizes with Kex2p in the trans-Golgi network (Santos and Snyder, 1997). Chs5p also has a chitin-independent role in cell fusion during mating (Santos et al., 1997). Chs7p is an endoplasmic reticulum (ER) protein required for the export of Chs3p from the ER (Trilla et al., 1999). Thus, these different proteins form a pathway for the transport of Chs3p, an important protein involved in cell wall synthesis. Because all of these proteins are not essential, it is possible that there are other as-yet-unidentified factors playing important functions for cell wall construction and transport of cell wall components.
Several proteins involved in cell wall synthesis are also implicated in cell polarity and morphogenesis (Cabib et al., 1998). Polarized cell growth in yeast is a complex process that requires the reorganization of the actin cytoskeleton, polarized secretion, and the function and regulation of signal transduction cascades (reviewed by Madden and Snyder, 1998). A number of proteins important for cell polarity in yeast have been identified. One such protein, Spa2p, is located at polarized growth sites, including the incipient bud site of unbudded cells, the bud tips of small-budded cells, the necks of cells undergoing cytokinesis, and the projection tips of mating cells (Snyder, 1989). spa2 mutants are defective in bud-site selection, apical bud growth, pseudohyphal growth, and mating projection formation (Gehrung and Snyder, 1990; Roemer et al., 1998). Spa2p interacts physically with other cell polarity proteins, such as Pea2p and Bud6p, and with components of two MAPK pathways, the mating signaling pathway and the Slt2p MAPK pathway (Sheu et al., 1998). This latter MAPK pathway functions downstream of PKC to maintain cellular integrity during polarized growth (for review, see Madden and Snyder, 1998).
chs5 and spa2 mutants share several common phenotypes affecting polarized growth processes: they are defective in mating projection formation and cell fusion during mating, and both exhibit altered bud-site selection in diploids during vegetative growth (Gehrung and Snyder, 1990; Santos et al., 1997; our unpublished results). Furthermore, the sequences of these two proteins display some common structural features: both show low-level similarity with filamentous proteins (Chs5p with neurofilaments and Spa2p with keratins and myosins), and both contain heptad repeats of unknown function (Gehrung and Snyder, 1990; Santos et al., 1997).
Here, we describe a genetic interaction between chs5 and spa2 mutants and show that the double mutant is inviable at high temperature. To identify additional components important for cell wall synthesis and cell morphogenesis, we have isolated high-copy-number suppressors of the growth defect of chs5 spa2 cells. We have characterized one of these suppressors, SBE2, and its highly related gene, SBE22. sbe2 sbe22 double mutants exhibit cell polarity and cell wall defects. We present evidence suggesting that Sbe2p and Sbe22p are novel Golgi proteins required for proper cell wall formation.
MATERIALS AND METHODS
Media and Microbiological Techniques
Genetic methods and growth media were as described by Guthrie and Fink (1991). Cell lysis was visualized on YPDA (rich medium supplemented with adenine) containing 0.001% methylene blue dye. YPDAS is YPDA containing 1 M sorbitol. Calcofluor sensitivity was analyzed on plates containing synthetic complete (SC) medium supplemented with 50 μg/ml fluorescent brightener 28 (calcofluor white; Sigma Chemical, St. Louis, MO). SDS sensitivity was analyzed on YPDA plates containing 0.0025% SDS. Yeast transformations were performed by the lithium acetate method of Ito et al. (1983).
Isolation of Multicopy Suppressors of chs5 spa2 Strain
A chs5Δ spa2Δ strain (Y1941) was transformed with a yeast genomic DNA library constructed in the multicopy plasmid YEp24 (Carlson and Botstein, 1982). After transformation, cells were incubated at 30°C for 24 h to allow recovery and then incubated at 37°C for 2 d. About 18,000 transformants were analyzed, and 18 transformants that reproducibly grew at 37°C were obtained. From each of these 18 transformants, plasmids were recovered for further analysis. Eight of them failed to suppress the growth defect of chs5 spa2 at 37°C when they were reintroduced into Y1941, indicating that the suppression event was not due to the plasmids. The genes present in the remaining 10 plasmids were identified by sequencing both ends of the insert.
Yeast Strains and Plasmids
Yeast strains used in this study are listed in Table 1. Construction of the chs5::ADE2 allele is described by Santos et al. (1997), 3Xmyc::CHS5 and CHS3::3XHA alleles are described by Santos and Snyder (1997), and spa2::URA3 and spa2::TRP1 alleles are described by Gehrung and Snyder (1990).
Table 1.
Yeast strains used in this study
| Strain | Genotypes |
|---|---|
| Y603 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 |
| Y601 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 spa2::URA3 |
| Y1935 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 |
| Y1936 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200chs5::ADE2 spa2::URA3 |
| Y1937 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 CDC3::3XHA |
| Y1938 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 CDC3::3XHA chs5::ADE2 |
| Y1939 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 CDC3::3XHA spa2::TRP1 |
| Y1940 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 CDC3::3XHA chs5::ADE2 spa2::TRP1 |
| Y1941 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3Δ-200 chs5::ADE2 spa2::TRP1 |
| Y270 | MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-901/trp1-901 his3-Δ200/his3-Δ200 |
| Y1942 | MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-901/trp1-901 his3-Δ200/his3-Δ200 sbe2::HIS3/sbe2::HIS3 |
| Y1943 | MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-901/trp1-901 his3-Δ200/his3-Δ200 sbe22: URA3/sbe22:URA3 |
| Y1944 | MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-901/trp1-901 his3-Δ200/his3-Δ200 sbe2::HIS3/sbe2::HIS3 sbe22:URA3/sbe22::URA3 |
| Y1310 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 3Xmyc::CHS5 CHS3::3XHA |
| Y1945 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 3Xmyc::CHS5 CHS3::3XHA sbe2::HIS3 |
| Y1946 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 3Xmyc::CHS5 CHS3::3XHA sbe2::HIS3 sbe22::TRP1 |
| Y1947 | MATα ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 3Xmyc::CHS5 CHS3::3XHA sbe2::HIS3 sbe22::TRP1 |
| Y1948 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 3Xmyc::CHS5 SBE2::3XHA |
| NY13a | MATa ura3-52 |
| NY26a | MATa ura3-52 sec2-59 |
| NY405a | MATa ura3-52 sec4-8 |
| NY17a | MATa ura3-52 sec6-4 |
| NY759a | MATa ura3-52 his4-619 sec7-1 |
| NY415a | MATa ura3-52 sec16-2 |
| Y604 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 |
| Y1949 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 sbe2::HIS3 sbe22::TRP1 |
| Y1950 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 chs5::ADE2 |
| Y1951 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 sbe2::HIS3 sbe22::TRP1 chs5::ADE2 |
| Y760 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 bck1:URA3 |
| Y1952 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 sbe2::HIS3 sbe22::TRP1 bck1::URA3 |
| Y602 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 spa2::URA3 |
| Y1953 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 sbe2::HIS3 sbe22::TRP1 spa2::URA3 |
| Y1954b | MATa ura3 leu2 ade2 lys2 his3 cdc24-4 |
| Y1955b | MATa ura3 lys2-801 ade2-101 trp1-901 his3-Δ200 sbe2::HIS3 sbe22::URA3 cdc24-4 |
| Y1956b | MATa cdc12-1 ura3 leu2 ade2 lys2 his3 |
| Y1957b | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 sbe2::HIS3 sbe22::URA3 cdc12-1 |
| Y1958 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 SBE2::3XHA |
| Y1959 | MATa ura3-52 lys2-801 ade2-101 trp1-901 his3-Δ200 SBE2::3Xmyc |
These strains are from P. Novick at Yale University.
cdc24-4 and cdc12-1 alleles originally come from J. Pringle's strains (University of North Carolina, Chapel Hill, NC), but they have been backcrossed into our background.
The complete ORFs of SBE2 (from −15 base pairs upstream of the ATG to the stop codon) and SBE22 (from the start codon to the stop codon) were deleted with the use of the PCR disruption procedure of Baudin et al. (1993). A sbe2::HIS3 null allele was generated using oligonucleotides 5′-CGGGCTTCCACCTTTGCTTCATTATTTTAC-TTCAGCTCTTTAGCTTTCTGTGACGCGCGTTTCGGTGATGACGGTG-3′ and 5′-GAACTTAAGAAGAGATAGTCTGGTCACCAA-ACTTTTAGTACGTGCCACATACACGGGGTGATGGTTCA-CGT-AGTGGGC-3′ to amplify the HIS3 gene from pRS313. Underlined portions of primers correspond to common sequences that flank each of the selectable markers within the pRS313–pRS316 series of plasmids (Sikorski and Hieter, 1989). Null alleles of SBE22 were created similarly. A sbe22::URA3 strain was generated with the use of oligonucleotides 5′-CAAATTTGTCCTTATCTTTAGTTAATAC-GGTCTAACTTGCCACGCTACTCAAGAAGCGCGTTTCGGTGATGACGGTG-3′ and 5′-CCAGTTTTTTTTTTCTTGTGCATGAGTGA-AATTACAGTTACAAAAAATAGGGTGATGGTTCACGTAG-TG-GGC-3′ to amplify the URA3 gene from pRS316. In addition, a sbe22::TRP1 null allele was generated with the use of the same oligonucleotides and pRS314 to amplify the TRP1 gene.
Strains containing the SBE2::3XHA or SBE2::3Xmyc alleles were constructed with the use of the PCR epitope-tagging method of Schneider et al. (1995). Both epitopes were integrated at the C terminus of the coding region (before the stop codon). Primers 5′-CCTCAATAATATTTTCACAAAGTGGTGGTTGCACTACTACCGAAAATTACGTAGGGAACAAAAGCTGG-3′ and 5′-CTTAAGAAGAGATAGTCTGGTCACCAAACTTTTAGTACGTGCCACATACA-CGCTACTATAGGGCGAATTGG-3′ were used. Proper formation of the hemagglutinin (HA)-tagged and myc-tagged alleles was confirmed by PCR and immunoblot analyses.
The green fluorescent protein (GFPS65T; Heim and Tsien, 1996) was fused to the N terminus of Sbe2p. A NotI site was created by PCR after the fourth codon of SBE2, creating the plasmid pBU55 (YEp24 vector). A fragment containing GFPS65T flanked by NotI sites and in frame with the SBE2 ORF was cloned in pBU55, creating pBU62. A BamHI/KpnI fragment from pBU62 containing SBE2::GFP was cloned in the same restriction sites of the centromeric vector pRS316, creating pBU65, the plasmid used in this study.
Mating Projection and Budding Pattern Analyses
For pheromone-treatment experiments, cells were grown to early log phase, α-factor (Sigma Chemical) was added to a final concentration of 5 μg/ml, and cells were incubated at 30°C with shaking for 45 min. Cultures were supplemented with a second addition of the same amount of α-factor and incubated for another 45 min. Microscopic examination of the cultures revealed that after 90 min most of the wild-type cells (90%) were unbudded and had formed shmoos.
Budding patterns were examined by staining cells with calcofluor. Mid log phase cells were fixed in 3.7% formaldehyde at 30°C with rotation, washed with water, and resuspended in 50 mM Tris, pH 7.5, containing 5 μg/ml calcofluor. Cells with two or more bud scars were scored and were classified into three categories: (1) bud scars only at one pole; (2) bud scars at both poles; and (3) random distribution of bud scars.
Thin Section Electron Microscopy
Cultures of wild-type (Y270), sbe2 (Y1942), sbe22 (Y1943), and sbe2 sbe22 (Y1944) cells were grown overnight at 30°C in YPDAS and then diluted into fresh YPDA medium and incubated for 6 h at 30°C (early log phase). Cells (15 ml) were fixed by adding 50% glutaraldehyde to a final concentration of 1%, incubated for 5 min on ice, and pelleted in a tabletop centrifuge at 4°C. Cell pellets were resuspended in 1 ml of PBS. Samples were subsequently processed according to the method described by Kaiser and Schekman (1990) but with 1% osmium tetroxide instead of permanganate for fixation.
Enzyme Assays
External invertase activity was measured as described by Goldstein and Lampen (1975). Internal invertase was determined by assaying spheroplast lysates prepared as described by Novick and Schekman (1979). The percentage of secreted invertase represents the level of external invertase divided by the total amount of the enzyme, external plus internal. For exoglucanase activity measurement, log phase cells grown in YPDA were diluted in fresh YPDA medium at OD600 = 0.25 and incubated at 25 or 37°C for 2 h. Cells were pelleted, and 200 μl of the culture supernatant was used for the assay, as described by Nebreda et al. (1986). The amount of exoglucanase liberated in the wild-type cells at 25°C was scored as 100% secretion, and all of the other values are relative to this one. Note that the sec6-4 strain used in these experiments has a different background than Y270 and Y1944.
Sucrose Density Gradient Centrifugation
Cell lysates were prepared from strain Y1948 and analyzed as described by Santos and Snyder (1997). Briefly, cells were grown in YPDA to mid log phase (2 × 107 cells/ml), and 2 g of cells (wet weight) was resuspended in 6 ml of 17% sucrose (wt/vol) in 50 mM Tris-HCl, pH 7.5, 1 mM EDTA containing protease inhibitors (Sigma Chemical) and 6 ml of glass beads. Cells were broken by vortexing, and the crude extract was centrifuged at 1500 × g for 10 min. The supernatant was layered on top of a 33-ml linear sucrose gradient (10–65%, wt/vol) in 50 mM Tris-HCl, 1 mM EDTA, pH 7.5. The tubes were centrifuged in a SW28 rotor at 25,000 rpm for 20 h at 4°C (Beckman Instruments, Fullerton, CA). One-milliliter fractions were collected from the bottom of the tube with the use of a peristaltic pump and analyzed by immunoblot analysis, as described by Santos and Snyder (1997). Monoclonal anti-HA (HA.11; BABCO, Richmond, CA) or anti-myc (9E10; BABCO) antibodies were used for detection of epitope-tagged Sbe2p and Chs5p, respectively. Rabbit polyclonal antibodies against Pma1p, Anp1p, and carboxypeptidase Y (CPY) (kindly provided by C. Slayman [Yale University, New Haven, CT], S. Munro [Laboratory of Molecular Biology, Cambridge, England], and P. Novick [Yale Univeristy], respectively) were used as markers of different cellular compartments. The reactive bands were detected with the use of anti-mouse or anti-rabbit alkaline phosphatase–conjugated antibodies (Jackson Immunoresearch, West Grove, PA) and CDP-Star (Boehringer Mannheim, Indianapolis, IN) detection reagent. Protein immunoblots were scanned, and the resulting data were analyzed with the use of the Multi-Analyst software from Bio-Rad (Richmond, CA).
Indirect Immunofluorescence
Indirect immunofluorescence was performed as outlined by Gehrung and Snyder (1990) and Pringle et al. (1991). Specific conditions for indirect immunofluorescence of epitope-tagged Chs3p and Chs5p are described by Santos and Snyder (1997).
RESULTS
CHS5 and SPA2 Genetically Interact
chs5 and spa2 mutants share several common phenotypes (see INTRODUCTION). In addition, we found that these mutants display common genetic interactions with mutations in genes involved in morphogenesis, suggesting that Chs5p and Spa2p might act in similar processes. It has been described that a spa2 disruption is lethal in combination with a septin mutant, cdc10-10 (Flescher et al., 1993), with mutations in genes of the Slt2p MAPK signaling cascade, such as BCK1 (SLK1) and SLT2 (Costigan et al., 1992), with mutations in SWI4, which encodes a transcription factor functioning downstream of the Slt2p MAPK pathway (Madden et al., 1997), and with mutations in genes required for bud emergence, such as BEM2 (Costigan et al., 1992). Similarly, we found that chs5 mutants are also lethal with mutations in septin genes, BCK1, SLT2, and BEM2 (our unpublished results).
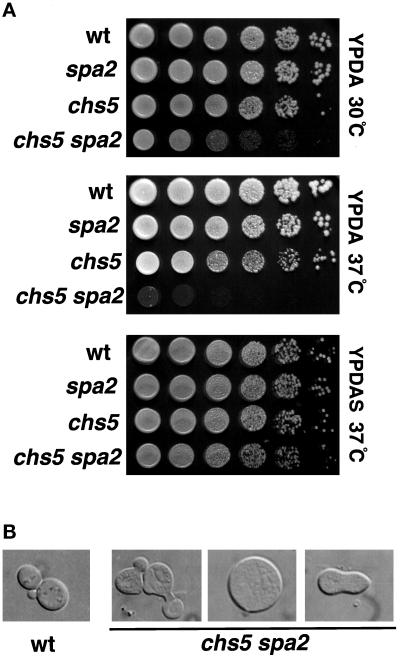
To test for a possible genetic interaction between CHS5 and SPA2, we constructed and analyzed chs5 spa2 strains. The chs5 spa2 double mutant displays a stronger cell fusion defect than either single mutant (our unpublished results). In addition, although chs5 and spa2 single mutants are indistinguishable from the wild type in growth rate at 25, 30, or 37°C, the chs5 spa2 double mutant exhibits a severe growth defect at 37°C that can be rescued by the addition of the osmotic stabilizer sorbitol (1 M) to the growth medium (Figure 1A). The double mutant also displays a growth defect at 30°C. Microscopic analysis of chs5 spa2 cells at 30 or 37°C reveals aberrant morphologies in many cells, including elongated bud necks, multiple projections or buds, large round cells, and a shmoo-like morphology (Figure 1B).
Figure 1.
chs5 spa2 mutant has a severe growth defect at 37°C that can be rescued by sorbitol. (A) Isogenic wild-type (Y603), spa2 (Y601), chs5 (Y1935), and chs5 spa2 (Y1936) strains were grown overnight to stationary phase at 30°C in YPDAS. Ten-fold serial dilutions starting with the saturated culture (∼5 × 107 cells/ml) were spotted in 5-μl drops onto YPDA plates (two upper panels) or YPDAS (lower panel). Plates were incubated at 30°C (upper panel) or 37°C (two lower panels). (B) Morphology of chs5 spa2 cells (Y1936) incubated at the restrictive temperature in YPDA for 6 h; elongated necks, large round cells, and shmoo-like cells are shown from left to right.
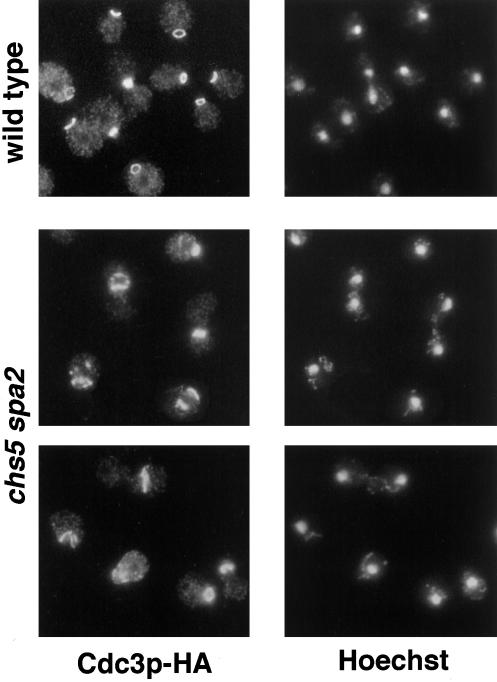
The existence of cells with elongated bud necks suggests a possible defect in neck organization in chs5 spa2 cells; therefore, we analyzed septin organization in this mutant by examining the localization of a functional epitope-tagged version of the septin Cdc3p (Roemer et al., 1996). In the wild type and in chs5 and spa2 mutants, Cdc3p shows the typical ring structure at the neck (Kim et al., 1991). In contrast, Cdc3p is dramatically mislocalized in chs5 spa2 cells at 30 or 37°C (Figure 2). Abnormal septin structures are observed in 75% of chs5 spa2 cells at 30°C (n = 507); defects include fragmented rings, mislocalized rings not placed at the neck, and the presence of septin patches. This indicates that the lack of Chs5p and Spa2p directly or indirectly affects septin organization in yeast.
Figure 2.
Cdc3p is mislocalized in chs5 spa2 cells. Wild-type (Y1937) and chs5 spa2 (Y1940) strains containing the CDC3::3XHA allele were incubated at 30°C in YPDAS and then transferred to YPDA at 30°C for 6 h. Cells were fixed and stained with anti-HA mAb by indirect immunofluorescence (left panels). Hoechst 33258 DNA staining of the same cells is shown in the right panels.
Screen for High-Copy-Number Suppressors of the chs5 spa2 Lethality
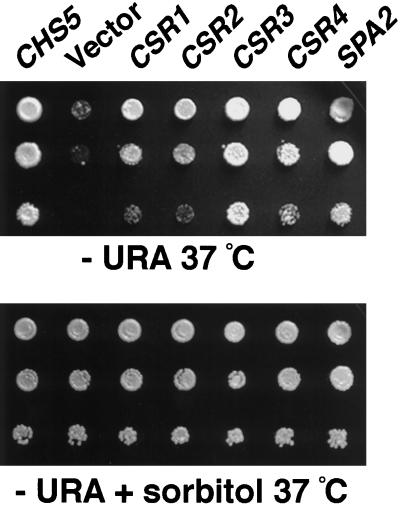
To identify additional components that may function in the same processes as CHS5 and SPA2, we searched for genes that, when present in high copy number, suppress the chs5 spa2 lethality at high temperature (see MATERIALS AND METHODS). Ten plasmids that reproducibly suppressed the lethality were identified. Six contained CHS5, whereas the other four plasmids each carried a different yeast genomic DNA fragment; therefore, each represents a different suppressor gene. These genes were designated CSR1–CSR4 (chs5 spa2 rescue); CSR3 is the strongest suppressor, whereas CSR2 is the weakest (Figure 3). None of the suppressor genes complements chs5 or spa2 single mutant defects. For instance, chs5 spa2 mutants, like chs5, are resistant to calcofluor as a result of the low levels of chitin. However, none of the plasmids suppresses this phenotype, suggesting that overexpression of these genes does not affect chitin levels. Likewise, overexpression of the CSR genes does not restore normal shmoo morphology in a spa2 mutant. Therefore, the CSR genes suppress defects specific to the chs5 spa2 double mutant.
Figure 3.
Multicopy suppressors of chs5 spa2 lethality at 37°C. chs5 spa2 strain (Y1941) transformed with vector (YEp24) or plasmids containing CHS5, SPA2, or the suppressor genes CSR1, CSR2, CSR3, or CSR4 were grown overnight at 30°C in SC-Ura plus 1 M sorbitol. Ten-fold serial dilutions starting with 10% of the saturated culture were spotted in 5-μl drops onto SC-Ura plates (top panel) or SC-Ura plus sorbitol (bottom panel) and were incubated at 37°C. The suppressor genes rescue the growth defect of chs5 spa2 cells to different degrees.
All of the suppressor plasmids contained several ORFs. The suppressing gene was identified by testing individual subcloned fragments for suppressing activity. CSR1 and CSR2 are novel genes (Saccharomyces Genome Database ORF designation YLR380w and YPR030w, respectively), CSR3 corresponds to YDR351w/SBE2, and CSR4 is identical to YIL147c/SLN1. SBE2 was isolated previously as a suppressor of bem4 (Mack et al., 1996), but no further characterization has been reported. Sln1p is a two-component signal transducer involved in the high-osmolarity glycerol (HOG) MAPK pathway (Ota and Varshavsky, 1993; Maeda et al., 1994). Characterization of CSR1, CSR2, and CSR4/SLN1 will be described elsewhere. The characterization of CSR3/SBE2 is described below.
Sbe2p and Sbe22p Are Highly Homologous Proteins
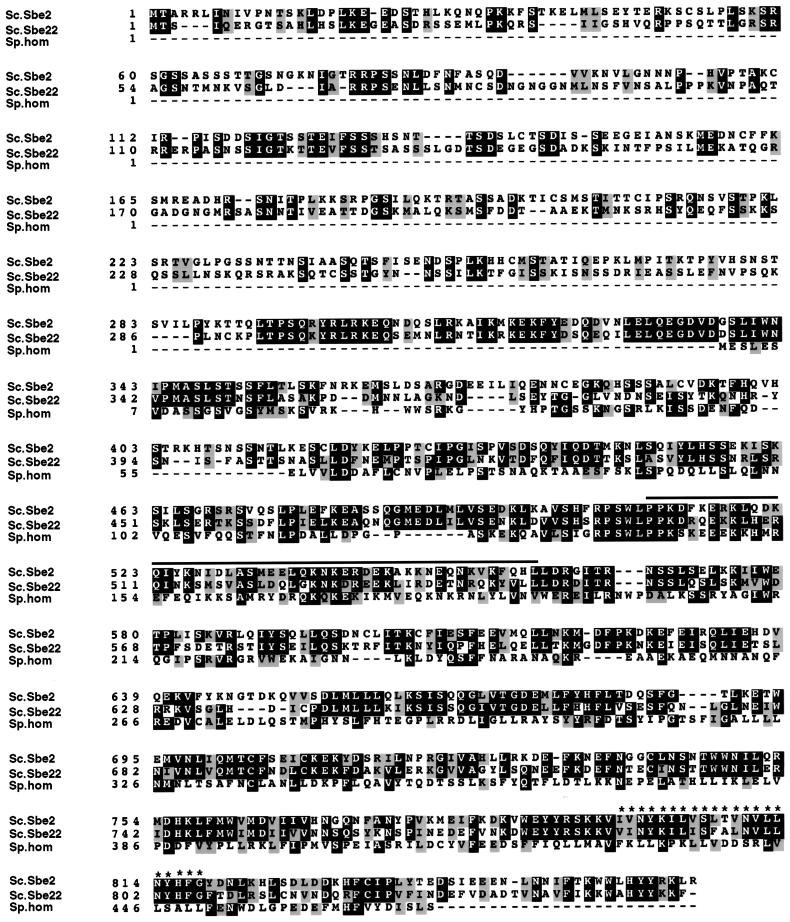
SBE2 encodes a hydrophilic protein of 864 amino acids. A search with the FASTA program revealed that S. cerevisiae possesses another gene, YHR103w/SBE22, predicted to encode a protein with high amino acid sequence identity to Sbe2p. Sbe22p is 852 amino acids in length and contains 43% identity and 63% similarity to Sbe2p over their entire length (Figure 4). They are more divergent at the N terminus (44% similarity) and more conserved at the C terminus (71% similarity). Sbe2p and Sbe22p are novel proteins and do not exhibit high homology with other known proteins in yeast or other organisms. However, they both display low-level similarity (25%) with a predicted 472-amino acid protein (C23D3.03c) of Schizosaccharomyces pombe, sharing a perfectly conserved 8-amino acid block (RPSWLPPK) that does not exist in any other protein in the databases (Figure 4).
Figure 4.
Sbe22p is a homologue of Sbe2p. Amino acid sequence alignment of Sbe2p and Sbe22p of S. cerevisiae and their homologue in S. pombe (ORF C23D3.03c). The alignment was performed with BCM Search Launcher (Baylor College of Medicine, Houston, TX). Shading was performed with the Boxshade 3.21 program (Swiss Institute for Experimental Cancer Research Bioinformatics Group). Some features of these proteins are annotated; both Sbe2p and Sbe22p display a predicted transmembrane domain at the C terminus (asterisks), and Sbe2p contains a potential coiled-coil region (line above the sequence).
Sbe2p and Sbe22p proteins share some common structural features. Using the TMpred program (Hofman and Stoffel, 1993), we found that Sbe2p displays a potential transmembrane domain from amino acids 798 to 818 and, similarly, Sbe22p is predicted to have a C-terminal transmembrane domain from amino acids 786 to 807. However, other programs predict that Sbe2p and Sbe22p are soluble proteins. Secondary structure analysis of Sbe2p with the use of the program COILS (Lupas, 1996) identifies a potential coiled-coil region (510–560 amino acids); this feature is not evident in Sbe22p. Despite this difference, the high level of amino acid sequence similarity between Sbe2p and Sbe22p suggests that they are likely redundant proteins with similar functions in the cell.
sbe2 sbe22 Double Mutants Display a Lysis Defect
To explore the function of SBE2 and SBE22, null mutants were generated and isogenic single and double mutants were analyzed. The sbe2 and sbe22 null mutants are viable and have growth rates in rich medium comparable to those of the isogenic wild-type strain at 25 or 30°C; however, sbe2 mutants display a slow-growth phenotype at 16 and 37°C, whereas sbe22 null strains exhibit a growth rate indistinguishable from that of wild-type cells at these temperatures (Figure 5A). Thus, SBE2 and SBE22 are not essential and are dispensable for growth.
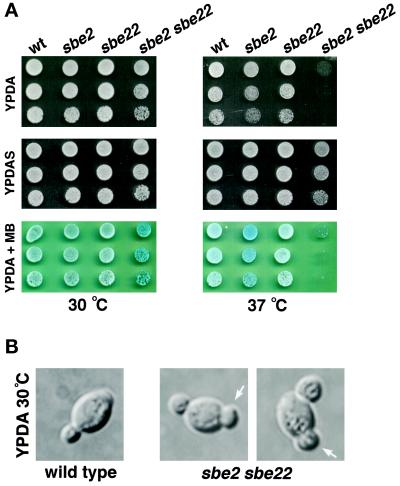
Figure 5.
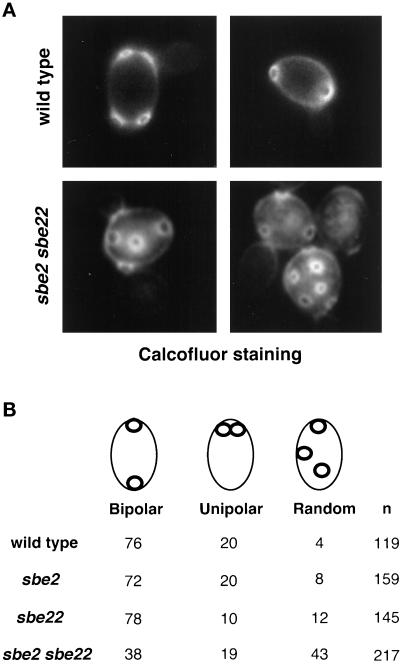
sbe2 sbe22 double mutant shows a lysis defect at 37°C. (A) Wild-type (Y270), sbe2 (Y1942), sbe22 (Y1943), and sbe2 sbe22 (Y1944) homozygous diploid strains were grown overnight at 30°C in YPDAS. Five-fold dilutions starting with 4% of the saturated culture were spotted in 5-μl drops onto YPDA (upper panel), YPDAS (middle panel), or YPDA plus 0.001% methylene blue (MB) dye (lower panel). Plates were incubated at 30°C (left panels) or 37°C (right panels). (B) Morphological defects of the sbe2 sbe22 mutant (Y1944) at 30°C, showing a characteristic bump at one of the poles (arrows).
In contrast, sbe2 sbe22 double mutants display a severe growth defect at 37°C. This defect is completely rescued by the addition of 1 M sorbitol (Figure 5A) and is stronger in diploids than in haploids. Therefore, unless mentioned otherwise, we have used homozygous diploids in all of the studies described below. The additive phenotypes of the sbe2 and sbe22 mutations suggest that the SBE2 and SBE22 genes perform redundant functions.
sbe2 sbe22 double mutants were examined for morphological defects. At 30°C, lysed cells and unbudded or small-budded cells with a large “bump” at one of the poles are frequently observed (Figure 5B). By calcofluor staining, the bump usually stains less than the rest of the cell wall, suggesting that it could represent a distended birth scar. Consistent with this interpretation, in haploids most of the cells have a bud at the same pole as the bump, as expected for the axial budding pattern of haploids cells, in which cells bud adjacent to the previous site of cytokinesis. In diploids, the bud and the bump are at opposite poles, in agreement with the bipolar budding pattern displayed by diploid cells (Figure 5B). At 37°C, the number of lysed cells, assayed by microscopic examination, increases (30% after 6 h at 37°C), but cells with elongated necks and multiple buds are also observed (5%). The vast majority of lysed cells are small budded. Microscopic examination of sbe2 sbe22 cells growing in osmotically stabilized medium (1 M sorbitol) at the restrictive temperature did not reveal any morphological defects. Thus, cells lacking SBE2 and SBE22 exhibit a strong cell lysis defect.
To confirm this phenotype, we used plates containing methylene blue, a vital dye that can be used as an indicator of cell lysis (Peppler and Rudert, 1953). Colonies turn blue when cell lysis occurs and are unstained when cells remain intact. sbe2 sbe22 double mutants turned blue at 30°C, indicative of lysis (Figure 5A, lower panels). Furthermore, sbe2 single mutants display a pale blue color at 37°C, indicative of a mild lysis defect (Figure 5A).
Cells deleted for sbe2, sbe22, or both were also analyzed for their ability to form a mating projection. When exposed to a high isotropic concentration of the α-factor mating pheromone (5 μg/ml) at 30°C, none of the mutants was found to exhibit a defect in mating projection formation. From these results, we conclude that SBE2 and SBE22 have redundant functions in the maintenance of cell integrity during vegetative growth, especially at high temperatures, although SBE2 appears to perform a more prominent role. Consistent with this interpretation, SBE22 in a multicopy plasmid does not rescue chs5 spa2 lethality at high temperature (our unpublished results).
sbe2 sbe22 Mutants Are Hypersensitive to Calcofluor and SDS
A sorbitol-remediable lysis defect phenotype has been described previously for mutants with a defective cell wall, such as those affected in β-1,3-glucan synthesis or in the Slt2p MAPK pathway (Cid et al., 1995). Therefore, we analyzed whether sbe2 sbe22 mutants exhibit cell wall defects by testing their sensitivity to calcofluor and SDS.
Calcofluor is toxic to yeast cells because of its interaction with chitin and its interference with cell wall assembly (Roncero et al., 1988). Mutants affecting cell wall integrity are susceptible to this agent (Lussier et al., 1997). The sensitivity of sbe2, sbe22, and sbe2 sbe22 mutants to 50 μg/ml calcofluor at 30°C was tested. Under these conditions, the sbe2 mutant is more sensitive and the sbe2 sbe22 double mutant is much more sensitive compared with the isogenic wild-type strain. sbe22 has no detectable defect (Figure 6A).
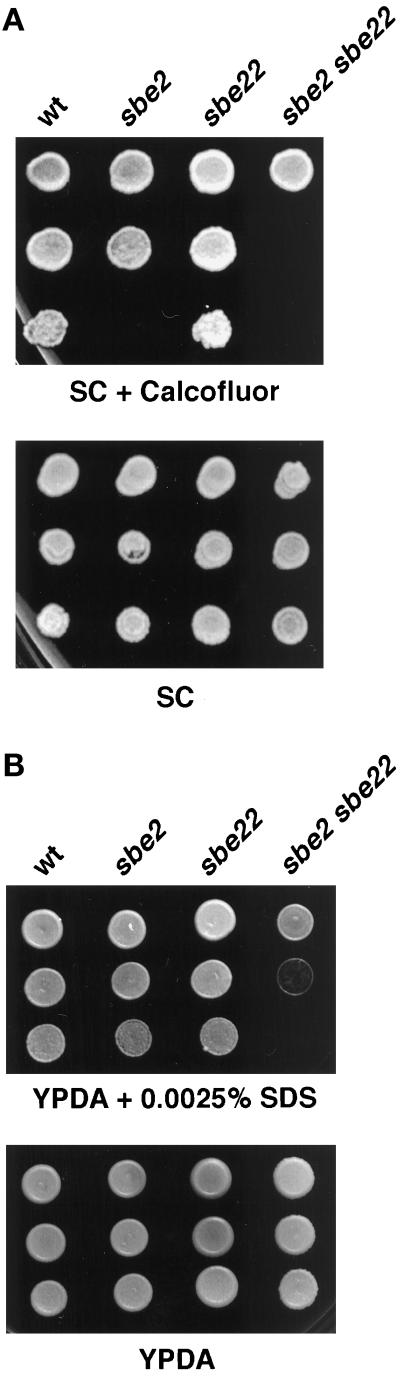
Figure 6.
sbe2 sbe22 mutants are sensitive to calcofluor and SDS. Wild-type (Y270), sbe2 (Y1942), sbe22 (Y1943), and sbe2 sbe22 (Y1944) homozygous diploid strains were grown overnight at 30°C in YPDAS. (A) Five-fold serial dilutions starting with 20% of the saturated culture were spotted in 5-μl drops onto SC and SC containing 50 μg/ml calcofluor. (B) Ten-fold dilutions starting with the saturated culture were spotted onto YPDA and YPDA containing 0.0025% SDS. All plates were incubated at 30°C.
SDS is a toxic detergent for yeast cells because it affects cell integrity. Mutants with a defective cell wall are usually more sensitive to this detergent (Shimizu et al., 1994). The sbe2 sbe22 double mutant is at least 10 times more sensitive to low levels of SDS (0.0025%) compared with wild type. The sbe2 mutant is slightly more sensitive and the sbe22 mutant behaves like the wild type (Figure 6B).
These results strongly indicate that loss of SBE2 and SBE22 function leads to a defective cell wall, suggesting the involvement of Sbe2p and Sbe22p in establishing or maintaining cell wall integrity.
sbe2 sbe22 Mutants Exhibit an Abnormal Cell Wall Ultrastructure
To further explore the requirement for Sbe2p and Sbe22p for proper cell wall architecture, the morphology of the cell wall was examined by electron microscopy. The cell wall of wild-type cells appears as a layered structure, exhibiting an electron-dense fibrillar outer layer, rich in mannoproteins, and an electron-transparent amorphous inner layer (Horisberger and Vonlanthen, 1977). The inner layer contains glucan and a small amount of chitin and may be subdivided into two different zones: one closest to the plasma membrane, which is rich in proteins, and an outer one, which contains a significant proportion of β-1,6-glucan (Kopecka et al., 1974).
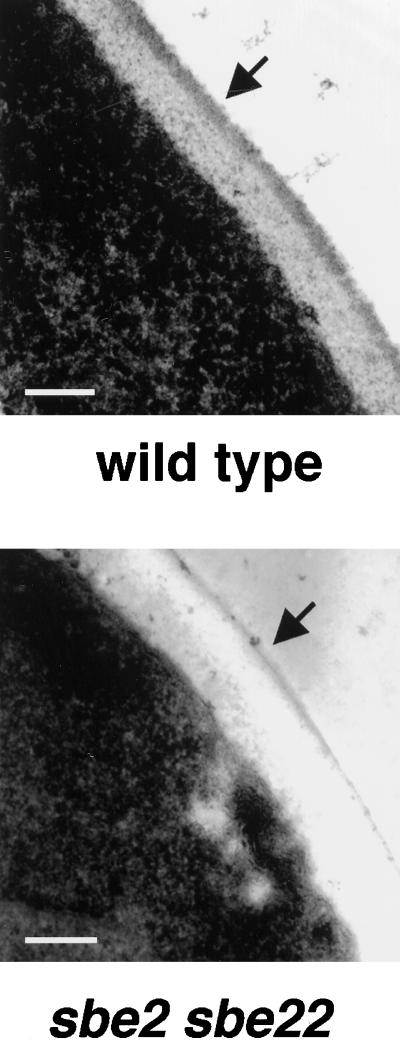
We analyzed the ultrastructure of the cell wall of sbe2, sbe22, and sbe2 sbe22 mutants growing in rich medium at 30°C. Whereas sbe2 or sbe22 cells show a normal layered structure similar to wild-type cells, the double mutant displays an aberrant cell wall architecture. The inner layer of glucan and chitin is enlarged, and the outer layer of mannoproteins is diminished or absent (Figure 7). We also observed that some small-budded cells appear to lyse at the tip of their buds because their cell walls are very thin or almost absent (data not shown). In conclusion, sbe2 sbe22 mutants exhibit an altered cell wall, with the thickness of the mannoprotein layer being very reduced.
Figure 7.
Cell wall ultrastructure of wild-type and sbe2 sbe22. Wild-type (Y270) and sbe2 sbe22 (Y1944) cells were grown overnight at 30°C in YPDAS and then diluted into YPDA and incubated at 30°C until they reached early log phase. Electron micrographs of the cell walls are presented; the outer layer of mannoproteins is indicated with arrows. More than 100 mutant cells were observed, and all display a similar phenotype. Bars, 0.1 μm.
Chs3p Is Mislocalized in the sbe2 sbe22 Mutants
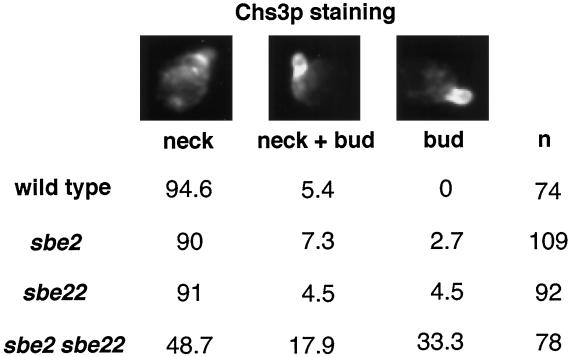
Because the sbe2 sbe22 mutant has a defective cell wall, we examined the localization of Chs3p and Chs5p, two proteins required for chitin synthesis, in the double mutant. In wild-type cells, Chs5p localizes in cytoplasmic patches (Santos and Snyder, 1997); this localization is not affected in sbe2, sbe22, or sbe2 sbe22 null mutants at 30 or 37°C. Localization of Chs3p in sbe2 and sbe22 single mutants is also similar to that in the wild type (see INTRODUCTION). In contrast, in sbe2 sbe22 null mutants, Chs3p is often found at the bud plasma membrane or diffuse throughout the bud in small-budded cells, instead of forming a ring at the bud neck like those observed in wild-type cells (Figure 8). Thus, in the absence of SBE2 and SBE22, Chs3p appears to be polarized to the correct general location but fails to assemble properly.
Figure 8.
Chs3p is mislocalized in small-budded sbe2 sbe22 cells. Wild-type (Y1310), sbe2 (Y1945), sbe22 (Y1946), and sbe2 sbe22 (Y1947) haploid cells containing 3Xmyc::CHS5 and CHS3::3XHA alleles were incubated at 30°C in YPDAS and then transferred to 37°C in YPDA for 4 h. Cells were fixed and stained with anti-HA antibody to visualize Chs3p by indirect immunofluorescence. The percentages of small-budded cells containing Chs3p at the mother-bud neck (neck), at the neck and around the bud (neck + bud), and primarily around the bud (bud) are presented. Representative micrographs of those three categories are shown. n, number of cells scored.
sbe2 sbe22 Diploids Display a Random Budding Pattern
To further explore the role of SBE2 and SBE22, the budding patterns of sbe2, sbe22, and sbe2 sbe22 cells were examined. Cells grown to log phase at 30°C were stained with calcofluor to visualize the chitin-rich bud scars that mark previous sites of budding (Hayashibe and Katohda, 1973). sbe2, sbe22, and sbe2 sbe22 haploid mutants show an axial budding pattern similar to the wild type (Flescher et al., 1993; Chant and Pringle, 1995). In contrast to haploids, wild-type diploid cells exhibit a bipolar budding pattern, resulting in bud scars located at both ends of the cell. sbe2 and sbe22 diploid cells display the normal bipolar budding pattern; however, sbe2 sbe22 diploids often bud randomly (Figure 9). Thus, sbe2 sbe22 diploids possess a bipolar-specific budding pattern defect.
Figure 9.
sbe2 sbe22 diploid cells display a random budding pattern. Wild-type (Y270), sbe2 (Y1942), sbe22 (Y1943), and sbe2 sbe22 (Y1944) diploid cells were stained with calcofluor to visualize bud scars. (A) In wild-type diploid cells, bud scars accumulate at both ends of the cell. In contrast, sbe2 sbe22 diploids exhibit a random budding pattern. (B) Cells with two or more bud scars were scored and classified as described in MATERIALS AND METHODS. Percentages of cells in each category are shown. n, number of cells scored.
Sbe2p Localizes to Cytoplasmic Patches and Cofractionates with Golgi Proteins
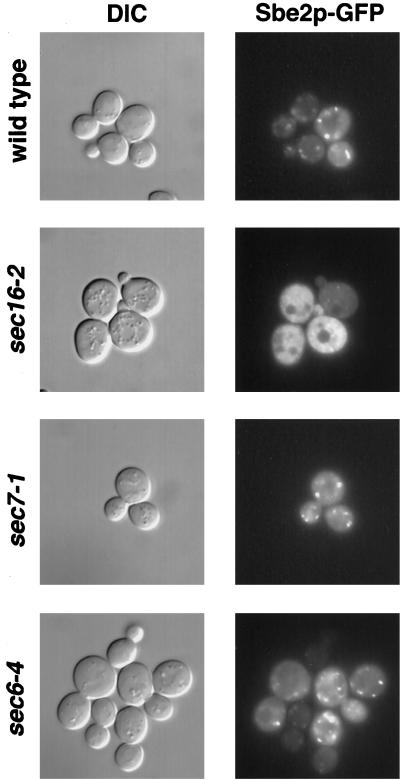
To determine the subcellular localization of Sbe2p, the GFP was fused to the N terminus of Sbe2p. This fusion protein is fully functional and rescues the thermosensitivity of sbe2 sbe22 double mutants. Localization of Sbe2p was examined in sbe2 mutant cells transformed with a centromeric plasmid containing SBE2::GFP and observed with the use of fluorescence microscopy. Sbe2p–GFP localizes to cytoplasmic patches in cells at all stages of the cell cycle. Sbe2p patches are distributed throughout the cell and are observed in both mother and daughter cells (Figure 10; top panels). Control cells without GFP do not show any signal.
Figure 10.
Localization of Sbe2p–GFP in sec mutants. Wild-type (NY13), sec6 (NY17), sec7 (NY759), and sec16 (NY415) strains transformed with pBU65 plasmid containing SBE2::GFP were grown at the permissive temperature (24°C) and then transferred to the restrictive temperature (37°C) for 2 h. Localization of Sbe2p–GFP was visualized in live cells (right panels), and corresponding differential interference contrast (DIC) images are shown (left panels). Note that in sec16 cells, which are blocked in ER-to-Golgi transport, patches are less evident and a clear increase in cytoplasmic staining is observed.
The punctate Sbe2p localization pattern is similar to that of Golgi-associated proteins (Franzusoff et al., 1991; Redding et al., 1991; Cooper and Bussey, 1992; Santos and Snyder, 1997). In spite of many attempts, colocalization experiments of Sbe2p with known Golgi proteins were not possible because we have not been able to observe Sbe2p–GFP directly with standard fixation techniques or by immunofluorescence with the use of commercial anti-GFP antibodies. We also tagged the SBE2 gene at its genomic locus with either three copies of the HA epitope coding sequence or three copies of the c-myc epitope to generate 3XHA::SBE2 or 3Xmyc::SBE2 strains. The resulting epitope-tagged proteins are functional. Immunoblot analysis of cell extracts with anti-HA or anti-myc antibodies specifically detects an ∼108-kDa protein, consistent with the predicted size of Sbe2p (Figure 11). However, indirect immunofluorescence studies of these strains with the use of anti-HA or anti-myc antibodies were unsuccessful, perhaps because of the sensitivity of Sbe2p to fixation procedures. Therefore, the cell fractionation approach was used to determine whether Sbe2p is a Golgi protein.
Figure 11.
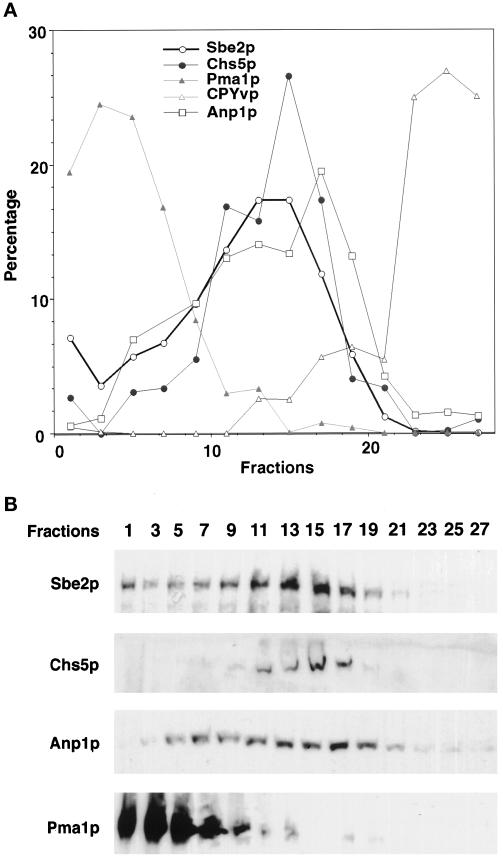
Sucrose density gradient centrifugation analysis. Cell lysates were prepared from yeast strain Y1948 containing 3Xmyc::CHS5 and SBE2::3XHA alleles and fractionated in 10–65% sucrose density gradients by centrifugation for 20 h. Fractions (abscissa in A) were collected from the bottom of the gradient and analyzed by immunoblot analysis with the use of antibodies to HA, c-myc, Pma1p, Anp1p, and CPY. Only the vacuolar isoform of the CPY protein (CPYvp) is represented. Sample blots are shown in B. The amount of protein in each lane was quantified, and the results are shown in A as percentages of the total protein.
Lysates of 3Xmyc::CHS5 3XHA::SBE2 yeast cells were prepared and subjected to 10–65% sucrose density gradient centrifugation in which the Golgi apparatus was separated from other membrane compartments such as the plasma membrane, vacuole, and ER (Santos et al., 1997). Fractions were collected and analyzed by immunoblot with the use of antibodies to HA (to detect Sbe2p–HA), c-myc (to detect Chs5p–myc), Anp1p (a Golgi marker; Chapman and Munro, 1994; Jungmann and Munro, 1998), Pma1p (a plasma membrane marker; Chang and Slayman, 1991), and CPY (a protein with three different isoforms each differentially located in the ER, Golgi, and vacuole; Stevens et al., 1982). As shown in Figure 11, Sbe2p resides predominantly in a membrane fraction that is similar in density to the Golgi compartments containing Chs5p and Anp1p. This compartment is clearly distinct from the plasma membrane (bottom of the gradient) or the vacuole (top of the gradient). A small amount of Sbe2p is also present at the bottom of the gradient, where plasma membrane fractionates. Chs5p has been shown to colocalize with Kex2p, a known TGN protein (Santos and Snyder, 1997). Together, our results strongly suggest that Sbe2p is a novel protein that resides in the Golgi apparatus.
Localization of Sbe2p in sec Mutants
To independently test the presence of Sbe2p in the Golgi apparatus, the localization of Sbe2p–GFP was examined in mutants blocked in different steps of the secretory pathway. Temperature-sensitive sec16-2, sec7-1, sec2-59, sec4-8, and sec6-4 haploid cells were transformed with a centromeric plasmid containing SBE2::GFP, incubated at the restrictive temperature, and observed with the use of fluorescence microscopy to visualize the Sbe2p–GFP protein. The sec16-2 mutant is defective in transport from the ER to the Golgi, the sec7-1 mutant is blocked from the Golgi to the secretory vesicle compartment, and sec2-59, sec4-8, and sec6-4 cells are defective in the transit from secretory vesicles to the plasma membrane (Novick et al., 1980, 1981). The punctate localization pattern of Sbe2p–GFP is not altered in sec7-1, sec2-59, sec4-8, or sec6-4 cells incubated at the restrictive temperature for 2 h or in any of the sec mutants incubated at the permissive temperature. In contrast, the punctate Sbe2p pattern is not detected in sec16-2 cells incubated at 37°C; instead, an increase in uniform cytoplasmic staining is observed (Figure 10). Ninety percent of the sec16-2 cells at the restrictive temperature show cytoplasmic staining, compared with 3–5% in wild-type, sec7-1, sec2-59, sec4-8, or sec6-4 cells. Accumulation in an ER-like pattern was not detected. These results further support the hypothesis that Sbe2p lies in a Golgi compartment.
Secretion of Invertase and Exoglucanase Is Not Severely Affected in sbe2 sbe22 Mutants
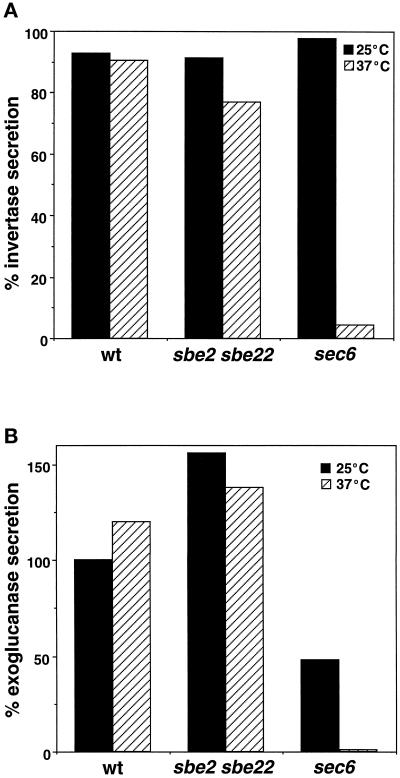
Because Sbe2p localizes to the Golgi apparatus, it is possible that the sbe2 sbe22 double mutant is defective in the general secretion and transport of proteins to the cell surface. To test this hypothesis, we examined the secretion of invertase (Novick et al., 1981) and exoglucanase (Nebreda et al., 1986) in the sbe2 sbe22 double mutant. As a control, a sec6-4 mutant that is blocked late in the secretory pathway at 37°C was included in the same experiment. Invertase is present in both the periplasmic extracellular space and the intracellular compartments. At 25°C, the fraction of invertase secreted is similar in wild-type, sbe2 sbe22, and sec6 cells. At 37°C, 95% of the invertase is extracellular in wild-type cells, compared with 77% in the sbe2 sbe22 strain and only 4% in the sec6-4 strain (Figure 12A). Thus, the delivery of invertase to the cell surface is not severely affected in the sbe2 sbe22 strain. For exoglucanase, the amount of enzyme liberated into the medium was measured. The sbe2 sbe22 double mutant does not display a defect in the secretion of exoglucanase; in fact, more enzyme is detected in the culture supernatant compared with that observed for wild-type cells (Figure 12B). The increased level in exoglucanase is most likely due to a consequence of the cell wall defect of the mutant, allowing the enzyme to be liberated more easily into the medium. In contrast, the sec6-4 mutant shows very reduced levels of secreted exoglucanase. These results demonstrate that the sbe2 sbe22 mutant does not have a large defect in general secretion and suggest a more specialized role for Sbe2p and Sbe22p in the transport of specific factors involved in cell wall formation.
Figure 12.
sbe2 sbe22 mutant is not severely affected in secretion. (A) Percentage of secreted invertase relative to intracellular enzyme in wild-type (Y270), sbe2 sbe22 (Y1944), and sec6-4 cells (NY17) after 1 h of incubation in low-glucose medium at 25 or 37°C. (B) Percentage of exoglucanase liberated into the medium by the same strains (see MATERIALS AND METHODS).
Genetic Interactions of SBE2 and SBE22
The phenotypes of sbe2 sbe22 mutants are indicative of the participation of Sbe2p and Sbe22p in two cellular processes. The thermosensitivity, lysis defect rescued by sorbitol, hypersensitivity to calcofluor and SDS, altered cell wall ultrastructure, and mislocalization of Chs3p phenotypes indicate a role for SBE2 and SBE22 in cell wall construction. In addition, the random budding pattern in sbe2 sbe22 diploids suggests a possible role in cell morphogenesis and polarity. Therefore, we tested whether the sbe2 sbe22 mutant displays genetic interactions with other mutants involved in cell wall synthesis and/or polarity.
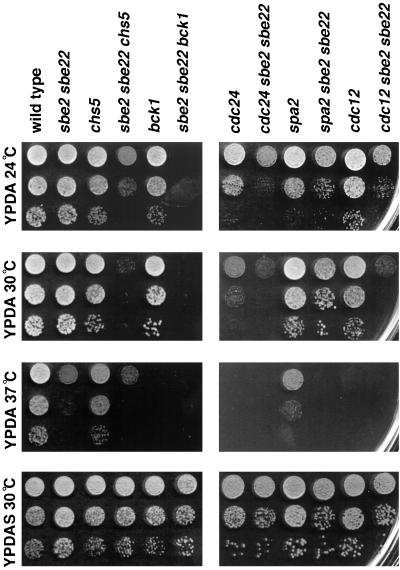
Chs5p is involved in chitin synthesis, and Bck1p is the MAPK kinase of the Slt2p pathway. We found that sbe2 sbe22 mutants exhibit severe growth defect in combination with either chs5 or bck1. sbe2 sbe22 chs5 mutants grow at 24°C, but they are unable to grow at 30°C. This defect is rescued by sorbitol (Figure 13). sbe2 sbe22 bck1 is not viable at any temperature unless sorbitol is included in the medium (Figure 13). These results corroborate a role for SBE2 and SBE22 in cell integrity.
Figure 13.
Genetic interactions of SBE2 and SBE22. Wild-type (Y604), sbe2 sbe22 (Y1949), chs5 (Y1950), sbe2 sbe22 chs5 (Y1951), bck1 (Y760), sbe2 sbe22 bck1 (Y1952), cdc24 (Y1954), sbe2 sbe22 cdc24 (Y1955), spa2 (Y602), sbe2 sbe22 spa2 (Y1953), cdc12 (Y1956), and sbe2 sbe22 cdc12 (Y1957) strains were grown overnight at 24°C in YPDAS. Ten-fold dilutions starting with 1% of the saturated culture were spotted in 5-μl drops onto YPDA (three upper panels) or YPDAS (lower panel). Plates were incubated at different temperatures, as indicated in the figure.
Cdc24p is an essential protein required for polarity establishment. cdc24 conditional mutants are unable to bud and form large multinucleate cells at the restrictive temperature (Sloat et al., 1981). The cdc24 sbe2 sbe22 mutant has a lower restrictive temperature (30°C) for growth than either cdc24 or sbe2 sbe22 mutants (37°C). The presence of sorbitol in the medium rescues this defect. Spa2p is involved in cell polarity processes (Snyder, 1989). The spa2 sbe2 sbe22 mutant grows similar to sbe2 sbe22, indicating a lack of genetic interaction between them (Figure 13). Cdc12p is one of the septins involved in cytokinesis (Longtine et al., 1996). The cdc12-1 sbe2 sbe22 mutant shows a stronger growth defect than either cdc12-1 or sbe2 sbe22 mutants; they do not grow at 30°C. In addition, whereas cdc12-1 cells grown at the restrictive temperature show elongated buds, in the cdc12-1 sbe2 sbe22 strain these buds are shorter, even in the presence of sorbitol (our unpublished results). Thus, sbe2 sbe22 mutants also display synthetic lethal interactions with mutants affected in polarity components, such as cdc24 and cdc12-1, consistent with the involvement of SBE2 and SBE22 in morphogenesis as well as cell integrity.
DISCUSSION
Genetic Interaction between CHS5 and SPA2
We have shown that chs5 spa2 double mutants are not viable at high temperature. Several nonexclusive possibilities may account for this phenotype. Like Spa2p, Chs5p is required for cell fusion during mating, and this function is independent of its role in chitin synthesis (Santos et al., 1997). However, chs3 spa2 mutants have a growth defect similar to chs5 spa2 mutants (our unpublished results), suggesting that the lethality is primarily due to the lack of Spa2p and chitin, not another function of Chs5p.
Another possibility to account for chs5 spa2 lethality is the formation of aberrant septin structures. Lack of Chs5p and Spa2p causes septin mislocalization, affecting their localization at the neck and resulting in abnormal rings. One proposed function for the septin ring is to provide a structure on which assembly of other proteins can take place at appropriate times to promote bud-site selection and to direct cytokinesis (Flescher et al., 1993; Longtine et al., 1996). Several lines of evidence suggest that there are synergistic defects caused by mutations in septins, chitin, and Spa2p. spa2 deletion is lethal in conjunction with cdc10-10 (Flescher et al., 1993), and mutants defective in chitin synthesis, such as chs4, display synthetic lethal interactions with cdc12-5 (DeMarini et al., 1997). In addition, the chs5 cdc12-1 double mutant has a stronger growth defect than chs5Δ or cdc12-1 single mutants (our unpublished results).
Another possible explanation for the chs5 spa2 growth defect is that, in the absence of Spa2p, a perfectly constructed cell wall is required. It has been shown that spa2 mutants are colethal with mutants defective in cell wall structure, such as those implicated in the Slt2p pathway (Costigan and Snyder, 1994). In addition, overexpression of SBE2, a gene involved in cell wall integrity, allows chs5 spa2 cells to grow. This possibility is also in agreement with the fact that sorbitol, an osmotic stabilizer, rescues the growth defect, although how this occurs is not clear. It is possible that sorbitol triggers signaling pathways in the cell that lead to the formation of a stronger cell wall. It has been suggested that a basal level of signaling through the HOG MAPK pathway is required for the proper localization of a glycosyltransferase, Mnn1p, to the Golgi complex (Reynolds et al., 1998); this suggests that the activity of the HOG pathway may regulate the compartmental distribution of glycosyltransferases, which are involved in mannan synthesis, within the Golgi to provide a novel mechanism to regulate the composition of the cell wall. Additionally, we have found SLN1, encoding one of the receptors of the HOG pathway (Maeda et al., 1994), as a multicopy suppressor of chs5 spa2 lethality, suggesting that signaling throughout this cascade can help chs5 spa2 cells to survive at high temperature.
Sbe2p Localizes to the Golgi Apparatus
SBE2 and SBE22 genes encode highly homologous proteins. Several lines of evidence suggest that Sbe2p localizes to the Golgi apparatus. First, Sbe2p localizes to cytoplasmic patches with a punctate pattern similar to that of Golgi-associated proteins (Franzusoff et al., 1991). Second, in sucrose gradient experiments, Sbe2p cofractionates with proteins demonstrated to be in the Golgi apparatus, such as Chs5p and Anp1p (Santos and Snyder, 1997; Jungmann and Munro, 1998). Preliminary results indicate that Sbe22p displays a profile similar to that of Sbe2p in sucrose gradients, suggesting that Sbe22p might also be a Golgi protein (our unpublished results). Third, Sbe2p is mislocalized in a sec16 mutant that blocks protein transport from the ER but not in other mutants defective in the secretory pathway but containing a normal Golgi apparatus, such as sec2, sec4, sec6, and sec7. This protein is not retained in the ER in the sec16 mutant; the same behavior has been described for Chs5p, another Golgi protein also involved in cell wall construction. One possible explanation is that these proteins assemble directly in the Golgi apparatus independently of the ER.
It has been reported that most proteins in the Golgi are either integral membrane proteins (mainly glycosyltransferases or proteases) or peripheral membrane proteins (Munro, 1998). Sbe2p and Sbe22p are predicted to be membrane proteins and localize to the Golgi apparatus but do not show similarity with glycosyltransferases or proteases, suggesting that they perform a different function.
Sbe2p and Sbe22p Are Involved in Cell Wall Integrity
Several lines of evidence suggest that Sbe2p and Sbe22p are involved in cell wall integrity. First, the sbe2 sbe22 double mutant has a lysis defect at high temperature that is rescued by the presence of sorbitol in the medium. This phenotype has been described for several mutants defective in cell wall structure, including those affecting the Slt2p pathway (Cid et al., 1995). Second, the sbe2 sbe22 double mutant displays sensitivity to compounds such as SDS and calcofluor. SDS induces lysis of cells with fragile cell walls, and calcofluor is a dye that blocks chitin polymerization, resulting in a weakened cell wall. Sensitivity to these products has proved to be a powerful tool in revealing cell wall defects (Ram et al., 1994; Lussier et al., 1997). Hypersensitivity to calcofluor may also be an indication of an increased amount of chitin in the cell wall. Third, electron microscopic analysis of the sbe2 sbe22 mutants demonstrates a strongly reduced electron-dense mannoprotein layer in the cell wall. This layer is believed to both contribute to the structural integrity of the cell wall and serve to exclude hydrolytic enzymes.
Several genetic screens have identified yeast mutations that cause defects in the synthesis of mannan, including the mnn (mannan; Ballou, 1990), och (outer chain; Nagasu et al., 1992), ngd (N-glycosylation defective; Lehle et al., 1995), and ldb (low dye binding; Manas et al., 1997) set of mutants. Like sbe2 sbe22 double mutants, mnn6 mutants are hypersensitive to calcofluor (Wang et al., 1997). Lack of Cwp2, another mannoprotein, confers an increased sensitivity to calcofluor and strongly reduces the electron-dense layer of the outside of the cell wall (Van Der Vaart et al., 1995). Mannan is not essential for viability; however, survival without mannan is dependent on cells being able to sense a cell wall defect (Jungmann et al., 1999). Yeast cells are endowed with the capacity to compensate for alterations in the structure and/or composition of the cell wall matrix. It has been proposed that the cell increases the amount of chitin and chitin-bound β-1,6-glucosylated proteins as a rescue mechanism in response to cell wall weakening (Kapteyn et al., 1997; Popolo et al., 1997). The putative compensatory mechanism might be mediated by the PKC-dependent signal transduction pathway (Kamada et al., 1995). Thus, mutations that result in defective mannans show synthetic lethality with components of the PKC pathway, which regulates the expression of cell wall components (Rayner and Munro, 1998). This is in agreement with the fact that sbe2 sbe22 mutants, which display a reduced mannoprotein layer, are synthetic lethal with mutants affected in chitin synthesis, such as chs5, and mutants in the Slt2p pathway, such as bck1. The lethality between sbe2 sbe22 and bck1 also suggests that Sbe2p and Sbe22p are not part of the Slt2p pathway and may act in a parallel pathway involved in cell wall construction and/or cell integrity. In addition, Chs3p, the catalytic component of CSIII, is mislocalized in sbe2 sbe22 mutants. This defect in Chs3p polarization is similar to that of chs4 or bni4 mutants that prevent the interaction between Chs3p and the septins (DeMarini et al., 1997).
SBE2 and SBE22 Are Involved in Polarity Processes
Several lines of evidence suggest that SBE2 and SBE22 genes may play a role in yeast cell polarity. sbe2 sbe22 mutants posses a bipolar-specific budding pattern defect. Mutations in SPA2, RVS161, RVS167, ACT1, BNI1, and BUD6 affect bud-site selection in diploid cells but not in haploids cells (Madden and Snyder, 1998). Most of these proteins are implicated in morphogenesis and polarized growth in yeast. Thus, Sbe2p and Sbe22p may have a role in morphogenesis. mnn10/bed1 mutants, defective in mannan deposition like sbe2 sbe22 mutants, are also defective in bud emergence; mutant cells are larger and rounder, indicative of a role in polarized growth (Mondesert and Reed, 1996).
It is possible that the cell wall defects of sbe2 sbe22 mutants affect bud-site selection. Several mutants with a defective cell wall also display defects in the budding pattern. HKR1 is an essential gene that regulates β-glucan synthesis; hkr1 mutants display an altered axial budding pattern in haploids (Yabe et al., 1996). In addition, rot1-1, rot2-1, and big1 strains also have defective cell walls and show a random budding pattern in haploid cells (Bickle et al., 1998).
Consistent with the function of Sbe2p and Sbe22p in polarized cell growth, sbe2 sbe22 mutants display synthetic lethality with polarity mutants such as cdc24 and cdc12. The sbe2 sbe22 cdc12 triple mutant does not form as elongated buds at the restrictive temperature, as does the cdc12 mutant. Mutants defective in apical bud growth, like spa2 mutants, display a random budding pattern and also fail to form elongated buds in the cdc12 background under the same conditions (Sheu, Barral, and Snyder, unpublished data). sbe2 sbe22 mutants also show these phenotypes, suggesting that Sbe2p and Sbe22p may also participate in apical bud growth.
Possible Role of Sbe2p and Sbe22p Proteins
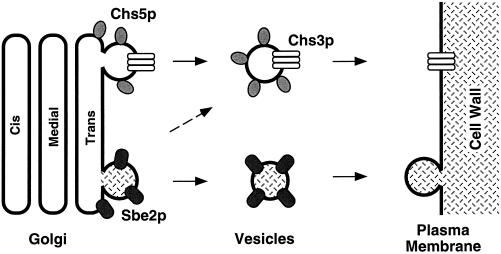
One possible role for Sbe2p and Sbe22p proteins is that they are part of the machinery involved in the transport of cell wall components to sites of growth. Several lines of evidence suggest the existence of several classes of secretory vesicles carrying different cargo. First, sec6-4 strains accumulate two types of 100-nm vesicles containing different proteins (Harsay and Bretscher, 1996). Second, myo2 and act1 mutants accumulate vesicles without affecting the secretion of well-known extracellular proteins (Johnston et al., 1991; Govindan et al., 1995; Mulholland et al., 1997). Third, by analogy with the well-studied secretory pathway, we have proposed previously that Chs5p is involved in the formation of a distinct set of vesicles required to transport Chs3p, and possibly other polarity components, to the bud neck region of the cell (Santos and Snyder, 1997; Madden and Snyder, 1998). sbe2 sbe22 mutants are not severely defective in the secretion of invertase or exoglucanase, suggesting a specialized pathway for polarized secretion of cell wall components. Because sbe2 sbe22 mutants have chitin, SBE2 overexpression is not able to rescue chs5 resistance to calcofluor, and chs5 and sbe2 sbe22 mutants are synthetic lethal, we propose that Sbe2p and Sbe22p could help to form secretory vesicles involved in the transport of mannoproteins to the cell wall. We suggest the existence of at least two pathways transporting cell wall components to the cell surface: one involving Chs5p that is important for chitin synthesis, and the other involving Sbe2p and Sbe22p that is involved in the synthesis of the mannoprotein layer (Figure 14). Because Chs3p is mislocalized in sbe2 sbe22 mutants, there also may be some connection between these two pathways. Preliminary evidence indicates that sbe2 sbe22 mutants are not affected in the incorporation of cell wall proteins such as Cwp1 or Pir2p (Van Der Vaart et al., 1995; Kapteyn et al., 1999) into the cell wall. We suggest that Sbe2p and Sbe22p may be required for either the proper localization of specific mannoproteins within the cell wall or the transport of a specific subset of proteins required for cell wall organization to the cell surface. Additional experiments will be required to elucidate more detailed roles of the Sbe2p and Sbe22p proteins.
Figure 14.
A model for the function of Sbe2p and Sbe22p. We propose that Sbe2p and Sbe22p are part of the machinery that transports cell wall components to growth sites. By analogy with the well-known secretory pathway, Sbe2p and Sbe22p could be involved in the formation of secretory vesicles containing cell wall components such as mannoproteins or help to direct the fusion of these vesicles with the plasma membrane. This pathway is independent of the pathway involving Chs5p, which functions in the transport of Chs3p to the cell surface. Nevertheless, these two pathways may still be interconnected (see DISCUSSION).
ACKNOWLEGMENTS
We thank Gertien Smits and Frans Klis at the University of Amsterdam, who did the experiments concerning incorporation of cell wall proteins. We thank B. Manning, P. San-Segundo, Y.-J. Sheu, and two reviewers for critical comments on the manuscript. C. Slayman and S. Munro provided antibodies. P. Novick provided strains, reagents for invertase assay, and anti-CPY antibodies. C. Walch-Solimena provided help with the invertase assay. C.R. Vázquez de Aldana provided advice with the exoglucanase experiment. B. Piekos provided help on the electron microscopy experiments. This research was supported by National Institutes of Health grant GM36494 to M.S. B.S. was supported in part by a postdoctoral fellowship from the Ministerio de Educación y Ciencia, Spain.
REFERENCES
- Ballou CE. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990;185:440–470. doi: 10.1016/0076-6879(90)85038-p. [DOI] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle M, Delley P-A, Schmidt A, Hall MN. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 1998;17:2235–2245. doi: 10.1093/emboj/17.8.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E, Drgonova J, Drgon T. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu Rev Biochem. 1998;67:307–333. doi: 10.1146/annurev.biochem.67.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chang A, Slayman C. Maturation of the yeast plasma membrane [H+] ATPase involves phosphorylation during intracellular transport. J Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. The functioning of the Golgi apparatus requires an ER protein encoded by ANP1, a member of a new family of genes affecting the secretory pathway. EMBO J. 1994;13:4896–4907. doi: 10.1002/j.1460-2075.1994.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Durán A, del Rey F, Snyder MP, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Bussey H. Yeast Kex1p is a Golgi-associated membrane protein: deletions in a cytoplasmic targeting domain result in mislocalization to the vacuolar membrane. J Cell Biol. 1992;119:1459–1468. doi: 10.1083/jcb.119.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigan C, Snyder M. SLK1, a homolog of MAP kinase activators, mediates nutrient sensing independently of the yeast cAMP-dependent protein kinase pathway. Mol Gen Genet. 1994;243:286–296. doi: 10.1007/BF00301064. [DOI] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AEM, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flescher EG, Madden K, Snyder M. Components required for cytokinesis are important for bud site selection in yeast. J Cell Biol. 1993;122:373–386. doi: 10.1083/jcb.122.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung S, Snyder M. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J Cell Biol. 1990;111:1451–1464. doi: 10.1083/jcb.111.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A, Lampen JO. β-d-Fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. In: Abelson JN, Simon MI, editors. Methods in Enzymology. Vol. 194. San Diego, CA: Academic Press; 1991. pp. 1–933. [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1996;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashibe M, Katohda S. Initiation of budding and chitin-ring. J Gen Appl Microbiol. 1973;19:23–39. [Google Scholar]
- Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescent resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- Hofman K, Stoffel W. Tmbase: a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Holthuis JCM, Nichols BJ, Pelham HRB. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol Biol Cell. 1998;9:3383–3397. doi: 10.1091/mbc.9.12.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horisberger M, Vonlanthen M. Localization of mannan and chitin on thin sections of budding yeasts with gold markers. Arch Microbiol. 1977;115:1–7. doi: 10.1007/BF00427837. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Rayner JC, Munro S. The Saccharomyces cerevisiae protein Mnn10/Bed1p is a subunit of a Golgi mannosyltransferase complex. J Biol Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Jung US, Piotrowski J, Levin DE. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- Kapteyn JC, Ram AFJ, Groos EM, Kollar R, Montijn RC, Van Den Ende H, Llobell A, Cabib E, Klis FM. Altered extent of cross-linking of β1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β-1,3-glucan content. J Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn JC, VanEgmond P, Sievi E, VanDenEnde H, Makarow M, Klis FM. The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and β-1,6-glucan-deficient mutants. Mol Microbiol. 1999;31:1853–1844. doi: 10.1046/j.1365-2958.1999.01320.x. [DOI] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecka M, Phaff HJ, Fleet GH. Demonstration of a fibrillar component in the cell wall of the yeast Saccharomyces cerevisiae. J Cell Biol. 1974;62:66–76. doi: 10.1083/jcb.62.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehle L, Eiden A, Lehnert K, Haselbeck A, Kopetzki E. Glycoprotein biosynthesis in Saccharomyces cerevisiae: ngd29, an N-glycosylation mutant allelic to och1 having a defect in the initiation of outer chain formation. FEBS Lett. 1995;370:41–45. doi: 10.1016/0014-5793(95)00789-c. [DOI] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Lussier M, et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Nishimura K, Dennehey BK, Arbogast T, Parkinson J, Toh-e A, Pringle JR, Bender A, Matsui Y. Identification of the bud emergence gene BEM4 and its interactions with Rho-type GTPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4387–4395. doi: 10.1128/mcb.16.8.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden K, Sheu Y, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- Manas P, Olivero I, Avalos M, Hernandez LM. Isolation of new nonconditional Saccharomyces cerevisiae mutants defective in asparagine-linked glycosylation. Glycobiology. 1997;7:487–497. doi: 10.1093/glycob/7.4.487. [DOI] [PubMed] [Google Scholar]
- Mondesert G, Reed SI. BED1, a gene encoding a galactosyltransferase homologue, is required for polarized growth and efficient bud emergence in Saccharomyces cerevisiae. J Cell Biol. 1996;132:137–151. doi: 10.1083/jcb.132.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretory vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasu T, Shimma YI, Nakanishi Y, Kuromitsu J, Iwana K, Nakayama KI, Suzuki K, Jigami Y. Isolation of new temperature-sensitive mutants of Saccharomyces cerevisiae deficient in mannose outer chain elongation. Yeast. 1992;8:535–547. doi: 10.1002/yea.320080705. [DOI] [PubMed] [Google Scholar]
- Nebreda AR, Villa TG, Villanueva JR, del Rey F. Cloning of genes related to exoglucanase production in Saccharomyces cerevisiae: characterization of an exo-β-glucanase structural gene. Gene. 1986;47:245–259. doi: 10.1016/0378-1119(86)90068-5. [DOI] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for posttranslational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Novick P, Schekman R. Secretion and cell surface growth are blocked in a temperature sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P. Biogenesis of yeast cell wall and surface components. In: Pringle J, Broach J, Jones E, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1997. pp. 229–362. [Google Scholar]
- Ota IM, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- Peppler HJ, Rudert FJ. Comparative evaluation of some methods for estimation of the quality of active dry yeast. Cereal Chem. 1953;30:146–152. [Google Scholar]
- Popolo L, Gilardelli D, Bonfante P, Vai M. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J Bacteriol. 1997;179:463–469. doi: 10.1128/jb.179.2.463-469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–601. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Ram AFJ, Wolters A, Ten Hoopen R, Klis FM. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Munro S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J Biol Chem. 1998;273:26836–26843. doi: 10.1074/jbc.273.41.26836. [DOI] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Hopkins BD, Lyons MR, Graham TR. The high osmolarity glycerol response (HOG) MAP kinase pathway controls localization of a yeast Golgi glycosyltransferase. J Cell Biol. 1998;143:935–946. doi: 10.1083/jcb.143.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Vallier L, Sheu Y-J, Snyder M. The Spa2-related protein, Sph1p, is important for polarized growth in yeast. J Cell Sci. 1998;111:479–494. doi: 10.1242/jcs.111.4.479. [DOI] [PubMed] [Google Scholar]
- Roemer T, Vallier LG, Snyder M. Selection of polarized growth sites in yeast. Trends Cell Biol. 1996;6:434–441. doi: 10.1016/s0962-8924(96)10039-8. [DOI] [PubMed] [Google Scholar]
- Roncero C, Valdivieso MH, Ribas JC, Duran A. Effect of calcofluor white on chitin synthases from Saccharomyces cerevisiae. J Bacteriol. 1988;170:1945–1949. doi: 10.1128/jb.170.4.1945-1949.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Durán A, Valdivieso MH. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J Cell Biol. 1997;136:95–110. doi: 10.1083/jcb.136.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of PCR epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Durán A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu Y-J, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu J, Yoda K, Yamasaki M. The hypo-osmolarity-sensitive phenotype of the Saccharomyces cerevisiae hpo2 mutant is due to a mutation in PKC1, which regulates expression of β-glucanase. Mol Gen Genet. 1994;242:641–648. doi: 10.1007/BF00283417. [DOI] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloat BF, Adams A, Pringle JR. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981;89:395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Trilla JA, Durán A, Roncero C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1153–1163. doi: 10.1083/jcb.145.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso MH, Mol PC, Shaw JA, Cabib E, Durán A. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J Cell Biol. 1991;114:101–109. doi: 10.1083/jcb.114.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Vaart JM, Caro LHP, Chapman JW, Klis FM, Verrips CT. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Nakayama K, Shimma Y, Tanaka A, Jigami Y. MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J Biol Chem. 1997;272:18117–18124. doi: 10.1074/jbc.272.29.18117. [DOI] [PubMed] [Google Scholar]
- Yabe T, Yamada-Okabe T, Kasahara S, Furuichi Y, Nakajima T, Ichishima E, Arisawa M, Yamada-Okabe H. HKR1 encodes a cell surface protein that regulates both cell wall β-glucan synthesis and budding pattern in the yeast Saccharomyces cerevisiae. J Bacteriol. 1996;178:477–483. doi: 10.1128/jb.178.2.477-483.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]