Abstract
A critical step in X-chromosome inactivation (XCI), which results in the dosage compensation of X-linked gene expression in mammals, is the coating of the presumptive inactive X chromosome by the large noncoding Xist RNA, which then leads to the recruitment of other factors essential for the heterochromatinisation of the inactive X and its transcriptional silencing. In an approach aimed at identifying genes implicated in the X-inactivation process by comparative transcriptional profiling of female and male mouse gastrula, we identified the Eif1 gene involved in translation initiation and RNA degradation. We show here that female embryonic stem cell lines, silenced by RNA interference for the Eif1 gene, are unable to form Xist RNA domains upon differentiation and fail to undergo X-inactivation. To probe further an effect involving RNA degradation pathways, the inhibition by RNA interference of Rent1, a factor essential for nonsense-mediated decay and Exosc10, a specific nuclear component of the exosome, was analysed and shown to similarly impair Xist upregulation and XCI. In Eif1-, Rent1-, and Exosc10-interfered clones, Xist spliced form(s) are strongly downregulated, while the levels of unspliced form(s) of Xist and the stability of Xist RNA remain comparable to that of the control cell lines. Our data suggests a role for mRNA nuclear degradation pathways in the critical regulation of spliced Xist mRNA levels and the onset of the X-inactivation process.
Synopsis
In mammals, each cell of the female contains two X chromosomes and hence, potentially a double dose of all X-linked genes when compared to XY males, who carry a single X chromosome. X-inactivation is the mechanism that ensures the dosage-compensation of X-linked gene products between the two sexes. X-inactivation is under the control of a specific region of the X chromosome, the X inactivation center (Xic), which contains the Xist gene encoding a large noncoding RNA transcript whose upregulation is critical to the initiation of X-inactivation. Such changes in steady-state transcript level could be due to altered rates of transcription or changes in the stability and processing of the transcript. How expression of Xist RNA is regulated and the nature of the mechanisms, which lead to Xist upregulation, remain unanswered or only partially answered questions of major importance to the field. In the following article, the authors identify three genes, Eif1, Rent1, and Exosc10, involved in nuclear mRNA degradation pathway(s), which are required for Xist expression upregulation and associated X-inactivation. Inhibition of the function of one or other of these genes leads to a failure of the female cells to undergo X inactivation, suggesting that post-transcriptional nuclear mRNA degradation pathway(s) are essential for the regulation of Xist RNA metabolism and X chromosome inactivation process.
Introduction
Since upregulation of the noncoding Xist RNA is concomitant with the establishment of the onset of X-inactivation [1,2], the precise regulation of Xist transcript levels in the nucleus is likely to be critical to this process. The increase in steady-state levels of Xist with the onset of X-inactivation was thought to be solely dependent on stabilisation of Xist transcripts and therefore essentially post-transcriptional. More recently, evidence has accumulated that control at the transcriptional level, mediated by chromatin remodelling at the Xist locus induced by biallelic Tsix antisense transcription, is of critical importance [3–5] (Navarro et al., unpublished data).
In female embryonic stem (ES) cells—an extensively used ex vivo model for the study of random X-inactivation [6], the biallelic expression of Xist and its antisense Tsix present in undifferentiated cells can be visualised as pinpoints by RNA–fluorescence in situ hybridization (RNA-FISH) [7]. When initiation of X-inactivation is induced in these cells by differentiation, upregulation of the Xist transcript levels on the presumptive inactive X [1,2] is visualised as the formation of a Xist RNA domain coating the inactive X, and this occurs concomitantly with the extinction of the antisense Tsix signal [8,9]. These initial events are followed by the dynamic recruitment of multiple chromatin modifications to the inactive X, involving polycomb group proteins, extensive histone modifications, and DNA methylation, resulting in the formation of the highly stable inactive heterochromatinised structure which characterises the inactive X [10].
As an approach to identifying additional molecular species that might be implicated in X chromosome inactivation (XCI), we have applied the serial analysis of gene expression (SAGE) technique [11] to the study of male and female 6.5 d post-coitum mouse embryos in its downsized version, which makes possible the exploitation of small samples (SAGE adaptation on downsized extracts [SADE], [12]). At this developmental stage, random X-inactivation has just occurred in the epiblast of female embryos and sexual dimorphism between XX and XY embryos is limited. We reasoned that if there were differences in transcript frequencies between female and male embryos at this stage, a subset of such differences might be implicated in X-inactivation. More than 214 genes overexpressed in female compared with male were identified [13], and a subset was validated using the ES cell system. Of the candidates falling within the category of upregulated genes and which are implicated in RNA metabolism and processing, the Eif1 (other name, Sui1-rs1) gene was retained for functional analysis. Eif1 (eukaryotic initiation factor 1) plays a critical role in stringent AUG selection during eukaryotic translation [14]. Studies in yeast have demonstrated that mutations in the Eif1 gene induced a stabilization of transcripts which contain a premature stop codon, indicating that the mutation of this gene can affect both translation and nonsense-mediated mRNA decay (NMD) pathways [15]. The NMD pathway's major role is currently thought to be to proofread mRNAs and to ensure the degradation of mRNAs that contain premature stop codons [16,17]. Protein products of the Rent1 (other name, Upf1), Upf2, Upf3A, and Upf3B genes are also essential for NMD activity [18].
Here we report that the downregulation by RNA interference of several genes (Eif1, Rent1, and Exosc10 genes) involved in mRNA degradation pathways leads to downregulation of spliced Xist transcript production and blocks the onset of the X-inactivation process. Our results suggest that the NMD pathway may play a role in the post-transcriptional regulation of Xist metabolism.
Results
Generation of Stable Cell Lines Interfered for Eif1 and Rent1
In order to investigate the putative role of Eif1 in particular and the NMD pathway in general in the XCI process, we have exploited RNA interference to generate ES cell lines stably interfered for the Eif1 and Rent1 genes [19]. ES clones derived from the female cell line LF2 in which the downregulation of Eif1 and Rent1 genes was greater than 70% (E1 and E2 for Eif1 and R1 and R2 for Rent1) were used to characterise the stability of Eif1 and the Rent1 interference during ES cell differentiation (Figure 1A). The T1 clone is a control generated by integration of an empty pSuper-puro vector into the LF2 cell line. Quantitative RT-PCR (Q-RT-PCR) analysis of two independent knockdown clones for both Eif1 and Rent1 showed that the Eif1 and Rent1 expression levels are stably interfered with, both prior to and during ES cell differentiation, at d 4 (Figure 1A) and d 14 of differentiation (unpublished data).
Figure 1. The Eif1, Rent1, and Exosc10 Genes Are Essential for the Upregulation of Xist mRNA During Differentiation.
(A) Quantification of Eif1, Rent1, Exosc10, and Rasa1 RNAs using Eif1Up/Lo, Rent1Up/Lo, Exosc10Up/Lo, and Rasa1Up/Lo primers, respectively, at d 0 and d 4 of retinoic acid–induced ES cell differentiation. LF2 is the parental female ES cell line. The T1 clone is a control generated by integration of an empty pSuper-puro vector into the LF2 cell line. The E1 and E2 clones were generated by integration of a pSuper-puro vector containing a hairpin designed against the ORF of the Eif1 gene. The R1 and R2 clones were generated by integrating a pSuper-puro vector containing hairpins 1 and 2, respectively, directed against the ORF of Rent1 gene. The Ex1 and Ex2 clones were generated by integrating a pSuper-puro vector containing hairpins 1 and 2, respectively, directed against the ORF of the Exosc10 gene. The Ra1 and Ra2 clones were generated using the plasmid previously described by Rossant and colleagues [33] that contains a hairpin directed against the Rasa1 gene.
(B) Quantification, using exon1–exon3 primers, of spliced Xist RNAs at d 0 and d 4 of differentiation, in the parental female ES cell line (LF2), the T1 control clone, and clones interfered for Eif1 (E1 and E2), Rent1 (R1 and R2), Exosc10 (Ex1 and Ex2), and Rasa1 (Ra1 and Ra2).
Columns and bars show the mean ± standard deviation (SD) (n = 3), expressed in arbitrary units, of the quantity of Eif1, Rent1, Exosc10, Rasa1, and Xist transcript standardized over Rrm2 RNA.
Impact of Interference on the Xist mRNA Level
We monitored the expression of Xist using Q-RT-PCR in ES clones stably silenced for Eif1 and Rent1 expression, both before and after the induction of differentiation. As shown in Figure 1B, undifferentiated ES cells silenced for the Eif1 and Rent1 genes displayed reduced levels of Xist RNA compared to control ES cells (LF2, T1). Most strikingly and in contrast to wild-type (WT) LF2 cells, Xist expression failed to undergo upregulation during the differentiation of the Eif1 and Rent1 silenced clones (Figure 1B). To investigate the specificity of the Eif1 and Rent1 interference on Xist RNA, we generated stably silenced clones against Rasa1, a gene not known to be involved in mRNA degradation or XCI (Figure 1A). Since Xist expression was not affected in clones silenced for the Rasa1 gene (Figure 1B), we conclude that the effects of Eif1 and Rent1 knockdown on Xist RNA metabolism and inactivation are specific.
Upregulation of Eif1 Induced High Levels of Xist RNA
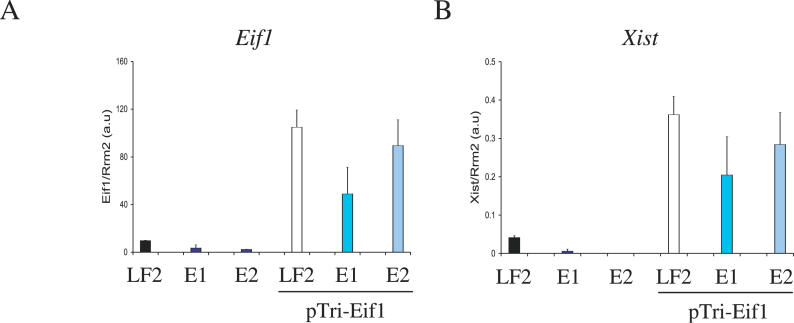
Further confirmation of the specificity of the Eif1 interference effect on Xist was provided by the transitory overexpression of Eif1 in Eif1-silenced clones (Figure 2A). Overexpression of Eif1 induced high levels of Xist RNA in transfected cells (Figure 2B). Expression of other members of the Eif gene family was unaffected by the overexpression of Eif1 (unpublished data), in agreement with these genes being unaffected in the Eif1 knockdown clones (unpublished data). We conclude that altering Eif1 expression levels leads to specific effects on Xist RNA levels.
Figure 2. Effect of Eif1 Upregulation on Xist mRNA Levels.
Quantification of Eif1 (A) and Xist (B) RNAs in undifferentiated parental female ES cell line (LF2), clones knocked down for Eif1, and clones lipofected with the pTriEx-Eif1 expression plasmid (see Methods and Materials) after 4 d of selection on G418-containing medium.
Columns and bars show the mean ± SD (n = 3), expressed in arbitrary units, of the quantity of Eif1 and Xist transcripts standardized over Rrm2 RNA.
Clones Silenced for Eif1 and Rent1 Fail to Undergo XCI
To investigate whether Xist RNA domains could be established in differentiated ES clones silenced for Eif1 and Rent1 despite the low level of Xist expressed as measured by Q-RT-PCR (Figure 1B), we performed RNA-FISH on cells differentiated for 4 d. At this stage, 50% of WT LF2 cells displayed a Xist domain that allowed the unequivocal identification of the inactive X chromosome. In striking contrast, no Xist domains could be detected in the Eif1 and Rent1 knockdown clones at either d 4 or even d 14 of differentiation (Figure 3A and unpublished data). The Eif1 and Rent1 knockdown clones maintain an X chromosome profile identical to that of undifferentiated cells with 100% of d-4 differentiated Eif1- and Rent1-silenced cells displaying two pinpoints for Xist/Tsix (probes lambda 510 and Pas34 specific for Tsix; Figure 3A and unpublished data). Phase-contrast microscopy of differentiated Eif1 and Rent1 silenced ES cell cultures (data not shown) and Q-RT-PCR analysis of the differentiation marker Pou5f1 (other name, Oct4; Figure 3C), a gene expressed in undifferentiated ES cell lines which is turned off during differentiation, confirmed that the Eif1 and Rent1 knockdown clones had been able to undergo normal differentiation.
Figure 3. Analysis by RNA-FISH and RNA-Immunofluorescence of H3K27 Methylation Enrichment on the X Chromosome and Pou5f1 mRNA Levels during Differentiation.
(A) RNA-FISH analysis of Xist and Mecp2 expression at d 4 of differentiation in a control culture (LF2) and Eif1 knockdown clone (E1), using the lambda 510 (Xist/Tsix, in green) and Mecp2 probes (in red). The presence of a single Xist RNA domain corresponding to the inactive X and a pinpoint Mecp2 signal corresponding to the active X chromosome are observed in 50% or more of cells of the control cell line (LF2). In contrast, Xist domains are not observed in the silenced cell line (E1), in which two Xist/Tsix pinpoint signals and two Mecp2 pinpoint signals can be observed, indicating that both Xs are active.
(B) Representative nuclei of differentiating ES cells hybridized with Xist RNA (green), and stained with antibodies to H3di/tri-meK27 [20], detected using a goat anti-mouse Alexa 680 secondary antibody (Molecular Probes, Eugene, Oregon, United States; red). In the control uninterfered cell line (LF2) the accumulation of Xist RNA was accompanied by enrichment of H3 di/tri-meK27 by d 4 of differentiation [20]. No enrichment of H3 tri-meK27 was observed in the interfered cell line (E1), in which 2 pinpoints of XistTsix are observed.
(C) Quantification, using Pou5f1Up/Lo primers, of spliced Pou5f1 RNAs at d 0 and d 4 of differentiation, in the parental female ES cell line (LF2), the clones interfered for Eif1 (E1), Rent1 (R1), and Exosc10 (Ex1).
Columns and bars are constructed as in Figure 1.
In order to probe further the X-inactivation status of the Eif1- and Rent1-interfered clones, we explored the expression of two X-linked genes, Mecp2 and Chic1, by RNA-FISH (Figure 3A and unpublished data). The presence of two pinpoint signals for Mecp2 and Chic1 in all knockdown clones at d 4 of differentiation indicated that both X chromosomes had remained active. Analysis of H3K27 trimethylation, a histone modification acquired by the inactive X early in the inactivation process [20], confirmed the absence of an inactive X (Figure 3B). Our results clearly implicate the Eif1 and Rent1 gene products in the establishment of XCI.
Eif1 and Rent1 Are Involved in the Post-Transcriptional Regulation of Xist RNA
In order to gain mechanistic insights into Eif1 and Rent1 function in XCI and in particular into the regulation of Xist, we first addressed the chromatin structure of the Xist/Tsix locus in relation to preinitiation complex recruitment at the Xist and Tsix promoters. We recently demonstrated that the Xist/Tsix locus is enriched for dimethylated H3K4 in undifferentiated ES cells, and that the deposition of this modification is dependent on Tsix [4]. As shown in Figure 4A, the H3K4 dimethylation profile of the locus (Figure 4C) was similar in Eif1- and Rent1-silenced cell lines and in control cells. Moreover, no significant differences in the recruitment of TFIIB, a factor that is part of the RNA polymerase II preinitiation complex, to the Tsix promoter were observed (Figure 4B). Taken together with the normal expression levels of Tsix detected by Q-RT-PCR (Figure 4D), these results indicate that Tsix transcription is unaffected by the reduction in Eif1 and Rent1 transcript levels in undifferentiated ES cells. The very low levels of TFIIB binding detected at the Xist promoter even in normal undifferentiated ES cells [4] (Figure 4B)—reflecting the transcriptional downregulation of Xist in these cells—precludes the drawing of definitive conclusions by chromatin immunoprecipitation (ChIP) analysis as to whether Eif1 and/or Rent1 are acting on Xist at the transcriptional or post-transcriptional level. To clarify this point, we have measured the levels of unspliced Xist RNA in undifferentiated and differentiated ES clones by strand-specific Q-RT-PCR using primers located in intron 1 of Xist. Strikingly, and despite the massive drop in the levels of mature Xist RNA transcripts in differentiated ES cells (Figure 1B), unspliced Xist RNA could be identified in both undifferentiated and differentiated E1 and R1 clones at levels very similar to that found in WT LF2 cells (Figure 4E). Our results strongly suggest that both Eif1 and Rent1 are affecting Xist mRNA at the post-transcriptional level.
Figure 4. ChIP Analysis of the Xist/Tsix Region in Eif1- and Rent1-Interfered Clones and Tsix mRNA Levels during Differentiation.
(A) Analysis of H3K4 dimethylation in the Xist/Tsix region by ChIP. The graph shows the percentage of immunoprecipitation obtained after normalisation using a control position corresponding to the promoter of the X-linked gene G6pd.
(B) ChIP analysis of TFIIB distribution in the Xist/Tsix region. The graph represents the percentage of immunoprecipitation obtained after normalisation using a control position located to the promoter of the Arpo gene.
(C) A schematic representation of the Xist/Tsix region. Positions of the primers used in the ChIP studies are indicated (see [4]).
(D) Quantification of spliced Tsix RNAs using exon2–exon3 primers at d 0 and d 4 of differentiation, in the parental female ES cell line (LF2) and clones knocked down for Eif1 (E1 and E2), Rent1 (R1 and R2), and Exosc10 (Ex1 and Ex2).
Columns and bars are constructed as in Figure 1.
(E) Quantification of unspliced Xist transcripts by strand-specific RT-PCR using the XistIN1DLo primer for priming the RT reaction and primers XistIN1CUp/Lo for amplification. All the primers are located in intron 1 of Xist. Control reactions without RT, or carried out in the presence of the enzyme but without the RT primer, were also performed and shown to be negative (unpublished data).
The NMD Pathway Is Implicated in Xist RNA Regulation
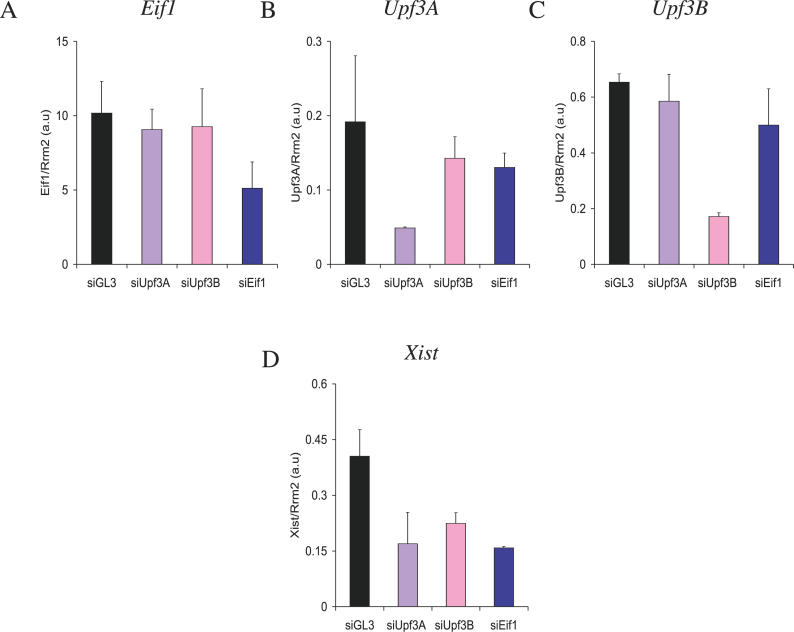
The NMD pathway, which acts post-transcriptionally, is involved in mRNA degradation (for review see [21]). To further investigate a putative role of NMD in Xist RNA regulation, we have transiently silenced two other putative members of the pathway, Upf3A and Upf3B, using short interfering RNA (siRNA) [22]. Upf3A and Upf3B are thought to be specifically involved in the NMD pathway, unlike the Eif1 and the Rent1 genes, which are known to participate in processes other than NMD. After 48 h of interference, Upf3A and Upf3B were specifically downregulated in LF2 cells, compared to control cells transfected with a siRNA directed against the exogenous luciferase gene GL3 [23] (Figure 5B and 5C). As shown in Figure 5D, undifferentiated ES cells silenced for the Upf3A and Upf3B genes (siUpf3A and siUpf3B, respectively) displayed reduced levels of Xist RNA compared to control ES cells (siGL3). Similar results were observed with an siRNA against the Eif1 gene (Figure 5A and 5D). These results confirm a role for the NMD pathway in the metabolism of Xist RNA.
Figure 5. Transient Interference of Eif1, Upf3A, and Upf3B .
Quantification of Eif1 (A), Upf3A (B), Upf3B (C), and Xist (D) RNAs using Eif1Up/Lo, Upf3AUp/Lo, Upf3BUp/Lo, and Xist exon1–exon3 primers, respectively, after 48 h of interference using siRNA siGL3, siEif1, siUpf3A, and siUpf3B.
Columns and bars are constructed as in Figure 1.
Nuclear Exosome Pathway Is Implicated in Xist RNA Regulation
Recent studies have shown that the NMD pathway interacts with the exosome, which has been implicated in RNA degradation in both the nucleus and the cytoplasm [24,25]. The Exosc10 gene, the mouse orthologue of the yeast Rrp6 gene, has been described to be associated exclusively with nuclear exosome activity [26,27]. Based on this, we generated independent stably interfered clones for Exosc10 (Ex1, Ex2; Figure 1A). Xist RNA levels are downregulated during differentiation of these clones and X-inactivation is inhibited as observed for Eif1 and Rent1 (Figure 1B). Based on these results, it appears likely that a nuclear-specific mRNA degradation pathway is an essential player in the X-inactivation process.
Upregulation of Xist cDNA Transgene in Silenced Stable Clones
Our finding that unspliced Xist is not affected by the downregulation of Eif1 and Rent1 strongly suggested that only spliced forms of Xist are affected. To investigate whether this reflects a change in the stability of spliced Xist transcripts, we have transiently transfected Xist cDNA constructs [28] into WT ES cells and into ES cells silenced for Eif1, Rent1, and Exosc10. As shown in Figure 6A and 6B, we observed a strong upregulation of Xist RNA levels in all cells, in the absence of marked variation for Tsix RNA levels. Our results suggest that the Xist exogenous cDNA is unaffected in the interfered clones, implying rather that it is at the splicing step that Xist metabolism is perturbed in the interfered clones. To explore further a link between the transcription, splicing step, and the effects we had seen in Xist metabolism, we first treated cells (T1 and E1 clones) differentiated for 4 d with actinomycin D for 0–8 h to inhibit transcription, then isolated RNA at 2-h timepoints, and quantified by real-time RT-PCR the Xist RNA using both strand-specific and exon–exon primers. Half-lives in the control and interfered clone were similar, suggesting that the degradation pathway does not affect Xist levels in the absence of active transcription. Levels of Xist pre-mRNA also had similar half-lives in both the T1 and E1 clones (Figure S1). Taken altogether, our results suggest that the Xist processing interfered with in the Eif1 knockdown clones is closely associated with the splicing process occurring during active transcription.
Figure 6. Overexpression of Xist cDNA Transgene and Impact of Interference on ncRNAs and Imprinted RNAs.
(A–B) Quantification of Xist and Tsix RNAs using Xist exon1–exon3 and Tsix using exon2–exon3 primers in the parental female ES cell line (LF2) and knocked down clones for Eif1 (E1), Rent1 (R1), and Exosc10 (Ex1), and in the same cells 48 h after transfection with Xist cDNA transgene. Quantification of His1 (C), Bc1 (D), Ube3A (E), Peg3 (F), and H19 (G) RNAs using His1Up/Lo, Bc11Up/Lo, Ube3AUp/Lo, Peg3Up/Lo, and H19Up/Lo primers respectively, at d 0 and d 4 of retinoic acid–induced ES cell differentiation.
Columns and bars are constructed as in Figure 1.
Analysis of Other Noncoding and Imprinted RNAs
Our results pose the question as to the eventual involvement of a nuclear mRNA degradation pathway in the regulation of other noncoding RNAs (ncRNAs) or imprinted genes. To probe this, we have analysed the expression of several other ncRNAs and/or imprinted genes (Figure 6C–6G). Neither His1 and Bc1 ncRNAs nor the Ube3A and Peg3 imprinted genes were affected by the downregulation of Eif1, Rent1, and Exosc10 (Figure 6C–6F). In contrast, expression of the H19 gene, which occurs only in differentiated ES cells, was highly inhibited (Figure 6G). We conclude that Eif1, Rent1, and Exosc10 are not part of a general mechanism for regulating ncRNAs and imprinted genes, but rather affect a highly restricted subset of such genes.
Discussion
In this manuscript, we have demonstrated that mRNA degradation pathways are involved in the regulation of Xist metabolism. Using RNA interference, we generated stable female ES cell lines for the Eif1, Rent1, and Exosc10 genes. Clones silenced for each of these genes fail to upregulate Xist during cellular differentiation, and X-inactivation establishment is aborted.
Measurement of the levels of spliced and unspliced Xist in Eif1- and Rent1-inhibited clones showed that unlike spliced Xist transcripts, unspliced Xist transcripts were maintained at similar levels in the silenced clones to those of the control WT female ES cell lines both before and after differentiation (Figure 4E). Since there is no evidence for alternative promoter use in Xist transcription in ES cells, we conclude that Eif1 and Rent1 cannot be affecting Xist metabolism via transcription initiation itself but are acting at the post-transcriptional level. Moreover, the dramatic decrease in spliced Xist RNA transcript levels occurs in the absence of any detectable effect on Tsix levels as analysed by Q-RT-PCR. Our results underline the specificity of this RNA degradation control for Xist, since many other ncRNAs are unaffected.
The Rent1 gene, unlike the Upf3A and Upf3B genes, participates not only in the NMD but also in nonsense-mediated altered splicing [29], Staufen1-mediated decay [30], and the efficiency of alternative splicing [31]. To provide further support for the notion that the NMD pathway is involved in the post-transcriptional control of Xist transcript levels, we therefore transiently interfered two additional genes, Upf3A and Upf3B. The downregulation of Xist RNA transcript levels in cells silenced for the Rent1, Upf3A, and Upf3B genes suggests that the effects on Xist RNA metabolism are being mediated via the NMD pathway.
To clarify whether the degradation pathway(s) is/are acting into the nucleus, we generated clones silenced for the Exosc10 gene, which is specific to the nuclear exosome pathway [26,27]. Silenced clones for the Exosc10 gene showed dramatically reduced levels of Xist RNA. Based on a parsimonious interpretation of our results, we therefore propose that a nuclear pathway, involving either all or many components of the NMD pathway, participates either directly or indirectly in the regulation of Xist RNA and XCI. We hypothesise that these effects are based around interactions occurring between the NMD machinery and elements of the splicing machinery during transcription/elongation-linked activities. The NMD pathway is known to interact directly with elements of the splicing machinery and the process of splicing itself [21]. Similarly, there is increasingly well-supported experimental evidence for the functional coupling of the transcription and splicing processes, with many active RNA polymerase II elongation complexes being found associated with splice complex and exosome components (for review see [32]). Recent experiments have shown that upregulation of Xist RNA is a critical step in the establishment of the X inactivation process, and that this is regulated transcriptionally [4,5] rather than via stabilization of the Xist-spliced form(s) as previously hypothesised [1,2]. Since in the interfered clone only spliced forms of Xist are downregulated and the stability of unspliced and spliced forms of Xist RNA and the level of unspliced Xist are comparable in control and interfered clones (T1 and E1), it is likely that it is a transcription-coupled splicing step itself that is being affected in our clones.
While further work will be necessary to consolidate our understanding of the precise mechanisms involved, and to elucidate whether the NMD components are acting directly or indirectly, our results open up another dimension to our knowledge of the pathways, that concur to ensure that levels of Xist are stringently controlled both prior to and at the onset of the X-inactivation process [3–5] (Navarro et al., unpublished data). Both the levels of Xist transcript and the spatial distribution of these transcripts are likely to be critical to this process.
Materials and Methods
Culture and in vitro differentiation of ES cells.
Female LF2 ES cell lines were cultured in Dulbecco's Modified Eagle Media (DMEM; Invitrogen, Carlsbad, California, United States), containing 15% fetal calf serum (FCS; Bio West), 1,000 U/ml LIF (Chemicon, Temecula, California, United States), 0.1 mM 2-mercaptoethanol (Invitrogen), 0.05 mg/ml streptomycin (Invitrogen), and 50 U/ml penicillin (Invitrogen) on a gelatin-coated support in the absence of feeder cells. Differentiation was induced by adding 100 nM all trans retinoic acid (Sigma, St. Louis, Missouri, United States) to LIF-free DMEM and 10% FCS medium for the first 3 d of differentiation. The culture medium was changed daily. All cells were grown at 37 °C in 8% CO2.
Construction of RNA interference vectors.
The pSUPER-puro vector (OligoEngine, http://www.oligoengine.com) was used according to the manufacturer's instrutions. Sequences coding for short hairpin RNAs were inserted as double-stranded oligos into pSUPER-puro using the BglII and HindIII sites as described previously [19]. The target sequence for the Eif1 gene was 5′-GGACGATCAGCTGAAGGTT-3′, the two target sequences for the Rent1 gene were 5′-GCGCACCGCTGAGAGAGAA-3′ (hairpin 1) and 5′-GCAGCCAATGTGGAGAAGA-3′ (hairpin 2), and the two target sequences for the Exosc10 gene were 5′-GGAGCCTCAAGGCATCATA-3′ (hairpin 1) and 5′-GCCCAGAACATAATGCAGT-3′ (hairpin 2). All target sequences have been blasted using National Center for Biotechnology Information software and shown to be specific for the gene of interest (http://www.ncbi.nlm.nih.gov/BLAST). The T1 clone is a control generated by integration of an empty pSuper-puro vector into the LF2 cell line. To test for an adverse nonspecific effect of RNAi mechanisms on Eif1, Rent1, and Exosc10 expressions, we generated stable LF2 cells interfered for the Rasa1 gene (Ra1 and Ra2) [33]. Transfection of different plasmids was carried out using Lipofectamine 2000 (Invitrogen). Stable integrants of pSuper-puro plasmids were selected on puromycin containing medium while stable integrants of the Rasa1 plasmid were selected on G418-containing medium.
Overexpression of Xist and Eif1.
The pTriEx-1.1 Neo vector (Novagen, Madison, Wisconsin, United States) was used according to manufacturer's instructions. The open reading frame (ORF) of the Eif1 gene was amplified by PCR using the following primers:
5′-AATGGTCTCGCATGTCCGCTATCCAGAACCTCCAGAACCTCCACT-3′ and
5′-CACCTGAGGTTAAAACCCATGAACCTTCAGCTGATCGTCCTTAG-3′, which contain BsaI and Bsu36I enzyme restriction sites respectively used for cloning. Plasmids pCMV-Xist [28] and pTriEx-Eif1 were transfected using Lipofectamine 2000 (Invitrogen). Eif1 and Xist overexpressing cells were selected for on G418-containing medium for 4 d and 48 h, respectively.
Real-time quantitative RT–PCR.
Total RNA was prepared using RNable (Eurobio, Courtaboeuf, France) and verified by electrophoresis. random-primed reverse transcription (RT) was carried out using the Superscript II reverse transcriptase (Invitrogen). Control reactions without enzyme were systematically performed. cDNAs were analyzed by real-time PCR using the SYBR Green Universal Mix and an ABI Prism 7700 (Perkin-Elmer Applied Biosystems, Wellesley, California, United States). Each PCR was run in triplicate to control for PCR variation. Rrm2 transcript levels were used for normalization [9]. All primer sequences are available on request. For strand-specific RT, we used the ThermoScript reverse transcriptase (Invitrogen).
Immunofluorescence and RNA-FISH analysis.
The preparation of nuclei for RNA-FISH, hybridization, and washes were performed as described [34] using DNA probes labelled by nick translation with Spectrum Red or Green dUTP (Vysis, Downers Grove, Illinois, United States). For immunofluorescence experiments performed after RNA-FISH, preparations were rinsed in phosphate-buffered saline (PBS), blocked in 0.5% bovine serum albumin (BSA) (NEB) in PBS for 15 min at room temperature, incubated with mouse monoclonal H3 di/tri-meK27 antibody at 1/100 (7B11) [20] for 45 min at room temperature, then rinsed in PBS and incubated with Texas red–conjugated goat anti-mouse secondary antibody for 45 min at room temperature. All antibodies were diluted in PBS/0.5% BSA. DNA was counterstained for 2 min with DAPI (0.2 mg/ml). Samples were mounted in 90% glycerol, 10% 1× PBS, 0.1% p-phenylenediamine (pH 9) (Sigma). A Zeiss Axioplan fluorescence microscope equiped with a Quantix CCD camera (Photometrix, Melbourne, Australia) and SmartCapture 2 software (Digital Scientific, Cambridge, United Kingdom) were used for image acquisition. All the probes used in this study have been described previously [35,36]. The results were obtained when cultures were examined after 4 and 14 d of differentiation (data not shown).
ChIP.
Immunoprecipitations were performed using a dimethylated H3 Lys-4 antibody (Upstate Biotechnology, Lake Placid, New York, United States) and TFIIB antibody (C-18; Santa-Cruz Biotechnology, Santa Cruz, California, United States), as described previously [4].
Transitory tranfection of siRNA.
siRNAs GL3 [23], Upf3A, Upf3B (Qiagen, Valencia, California, United States), and Eif1 (5′-GGTTCATGGGTTTTAAGTGdTdT-3′) were transfected using Lipofectamine 2000 (Invitrogen). Levels of interference were monitored by Q-RT-PCR as described previously.
Xist and Tsix half-life assay.
ES cells (T1, E1 clones; 105) were dispensed into each well of a six-well plate and differentiated, using retinoic acid, during 4 d. Fresh media containing 5 μg/ml actinomycin D was then added. At each time point, media was aspirated and total RNA was prepared using RNable (Eurobio). RNA levels were quantified as described in Materials and Methods by Q-RT-PCR.
Supporting Information
Xist pre-mRNA half-life(s) were measured by strand-specific, real-time RT-PCR after actinomycin D treatment. Cell culture experiments were performed in replicate, and PCR cycling reactions in triplicate. Points represent averages from duplicate experiments ± standard error of the mean.
(24 KB PDF)
Accession Numbers
The GenBank (http://www.ncbi.nlm.nih.gov/Genbank) accession numbers for the genes discussed in this paper are Eif1 (20918), Rent1 (19704), Exosc10 (50912), Xist (213742), Tsix (22097), Pou5f1 (18999), Upf3A (67031), Upf3B (68134), His1 (110153), Bc1 (12031), Ube3a (22215), Peg3 (18616), H19 (14955), Rasa1 (218397), and Rrm2 (20135).
Acknowledgments
We wish to thank A. Jacquier, C. Babinet and E. Heard for stimulating discussions, J. Rossant for the gift of the RasGAP short hairpin RNA transgene, and A. Wutz for the gift of the pCMV-Xist plasmid.
Abbreviations
- ChIP
chromatin immunoprecipitation
- ES
embryonic stem
- ncRNA
noncoding RNA
- NMD
nonsense-mediated decay
- ORF
open reading frame
- RNA-FISH
RNA–fluorescence in situ hybridization
- siRNA
short interfering RNA
- XCI
X-chromosome inactivation
- WT
wild-type
Footnotes
Author contributions. CC, AB, MC, HCD, and PA conceived and designed the experiments. CC, AB, and CR performed the experiments. CC, MC, HCD, CR, and PA analyzed the data. CC, HC, and CR contributed reagents/materials/analysis tools. CC, MC, CR, and PA wrote the paper.
Competing interests. The authors have declared that no competing interests exist.
Funding. This work was supported by grants from the Association pour la Recherche contre le Cancer (ARC) and the French Ministry of Research under the Action Concertée Incitative contract no. 032526 to PA. Additional financial support came from the EU NoE Epigenome programme. PA, CR, and MC-T are supported by the Centre National de la Recherche Scientifique.
References
- Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- Sheardown SA, Duthie SM, Johnston CM, Newall AE, Formstone EJ et al. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Navarro P, Pichard S, Ciaudo C, Avner P, Rougeulle C. Tsix transcription across the Xist gene alters chromatin conformation without affecting Xist transcription: Implications for X-chromosome inactivation. Genes Dev. 2005;19:1474–1484. doi: 10.1101/gad.341105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- Rastan S, Robertson EJ. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Lee JT, Lu N. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell. 1999;99:47–57. doi: 10.1016/s0092-8674(00)80061-6. [DOI] [PubMed] [Google Scholar]
- Morey C, Arnaud D, Avner P, Clerc P. Tsix-mediated repression of Xist accumulation is not sufficient for normal random X inactivation. Hum Mol Genet. 2001;10:1403–1411. doi: 10.1093/hmg/10.13.1403. [DOI] [PubMed] [Google Scholar]
- Heard E. Recent advances in X-chromosome inactivation. Curr Opin Cell Biol. 2004;16:247–255. doi: 10.1016/j.ceb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Virlon B, Cheval L, Buhler JM, Billon E, Doucet A, et al. Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci U S A. 1999;96:15286–15291. doi: 10.1073/pnas.96.26.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdet A, Ciaudo C, Zakin L, Elalouf JM, Rusniol C, et al. A SAGE approach to identifying novel trans-acting factors involved in the X inactivation process. Cytogenet Genome Res. 2006;113:325–335. doi: 10.1159/000090849. [DOI] [PubMed] [Google Scholar]
- Yoon HJ, Donahue TF. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA (iMet) recognition of the start codon. Mol Cell Biol. 1992;12:248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Gonzalez CI, Kinzy TG, Dinman JD, Peltz SW. Mutations in the MOF2/SUI1 gene affect both translation and nonsense-mediated mRNA decay. RNA. 1999;5:794–804. doi: 10.1017/s1355838299982055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- Wagner E, Lykke-Andersen J. mRNA surveillance: The perfect persist. J Cell Sci. 2002;115:3033–3038. doi: 10.1242/jcs.115.15.3033. [DOI] [PubMed] [Google Scholar]
- Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae . Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Rougeulle C, Chaumeil J, Sarma K, Allis CD, Reinberg D, et al. Differential histone H3 Lys-9 and Lys-27 methylation profiles on the X chromosome. Mol Cell Biol. 2004;24:5475–5484. doi: 10.1128/MCB.24.12.5475-5484.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay in mammals. J Cell Sci. 2005;118:1773–1776. doi: 10.1242/jcs.01701. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Raijmakers R, Schilders G, Pruijn GJ. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur J Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- Brouwer R, Allmang C, Raijmakers R, van Aarssen Y, Egberts WV, et al. Three novel components of the human exosome. J Biol Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′ UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, et al. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Reed R. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol. 2003;15:326–331. doi: 10.1016/s0955-0674(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Kunath T, Gish G, Lickert H, Jones N, Pawson T, et al. > 2003. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype Nat Biotechnol 21 559 61 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Okamoto I, Heard E. X-chromosome inactivation in mouse embryonic stem cells: analysis of histone modifications and transcriptional activity using immunofluorescence and FISH. Methods Enzymol. 2004;376:405–419. doi: 10.1016/S0076-6879(03)76027-3. [DOI] [PubMed] [Google Scholar]
- Clerc P, Avner P. Role of the region 3′ to Xist exon 6 in the counting process of X-chromosome inactivation. Nat Genet. 1998;19:249–253. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- Debrand E, Chureau C, Arnaud D, Avner P, Heard E. Functional analysis of the DXPas34 locus, a 3′ regulator of Xist expression. Mol Cell Biol. 1999;19:8513–8525. doi: 10.1128/mcb.19.12.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Xist pre-mRNA half-life(s) were measured by strand-specific, real-time RT-PCR after actinomycin D treatment. Cell culture experiments were performed in replicate, and PCR cycling reactions in triplicate. Points represent averages from duplicate experiments ± standard error of the mean.
(24 KB PDF)