Abstract
The clustering of ryanodine receptors (RyR2) into functional Ca2+ release units is central to current models for cardiac excitation-contraction (E-C) coupling. Using immunolabeling and confocal microscopy, we have analyzed the distribution of RyR2 clusters in rat and ventricular atrial myocytes. The resolution of the three-dimensional structure was improved by a novel transverse sectioning method as well as digital deconvolution. In contrast to earlier reports, the mean RyR2 cluster transverse spacing was measured 1.05 μm in ventricular myocytes and estimated 0.97 μm in atrial myocytes. Intercalated RyR2 clusters were found interspersed between the Z-disks on the cell periphery but absent in the interior, forming double rows flanking the local Z-disks on the surface. The longitudinal spacing between the adjacent rows of RyR2 clusters on the Z-disks was measured to have a mean value of 1.87 μm in ventricular and 1.69 μm in atrial myocytes. The measured RyR2 cluster distribution is compatible with models of Ca2+ wave generation. The size of the typical RyR2 cluster was close to 250 nm, and this suggests that ∼100 RyR2s might be present in a cluster. The importance of cluster size and three-dimensional spacing for current E-C coupling models is discussed.
INTRODUCTION
The ryanodine receptor subtype 2 (RyR2) is predominantly expressed in cardiac muscle (1–3) and plays a central role in the Ca2+ signaling that controls cardiac excitation-contraction (E-C) coupling. The RyR2 molecule is a protein tetramer with molecular mass of 565 kDa per subunit (4,5). Structurally, the RyR2 molecules are congregated in discrete clusters that are densely localized in the Z-disk (or Z-line as viewed in the two-dimensional image) in cardiac myocytes (6). Functionally, the RyR2 serves as an intracellular Ca2+ channel in the sarcoplasmic reticulum (SR) membrane. An increase in the cytosolic Ca2+ opens the RyR2 channel to allow Ca2+ release from the SR into the cytosol, triggering a regenerative process known as Ca2+-induced Ca2+ release (7–9). Hence the RyR2 molecules in a cluster are activated by Ca2+ in a cooperative manner, releasing quantal amounts of Ca2+ that are visualized as Ca2+ sparks (10–13). In other words, a RyR2 cluster serves as an elementary Ca2+ release unit (CRU) in cardiac myocytes. We will use these two terms interchangeably from here on.
Distribution of the discrete RyR2 clusters in three-dimensional (3-D) space provides structural arrangements for the local control of CRUs by L-type Ca2+ channels, which permits the graded control of cardiac E-C coupling (14). In further development of this idea, recent studies show that the stochastic nature of discrete CRUs also play a part in determining the spread of Ca2+ release events from CRUs to the neighboring CRUs in the form of propagating Ca2+ waves (15–17). Spontaneously arising Ca2+ waves can disrupt normal muscle contraction and electrical activity (18), leading to fibrillation and arrhythmias in ventricular and atrial myocytes. In the atrial myocytes that lack a t-tubule system, action potential-triggered Ca2+ waves also serve a physiological function by facilitating a rapid increase of global Ca2+ to cause muscle contraction (19,20).
The Ca2+ signaling system, in essence, is a controlled nonlinear dynamic system with positive feedback (due to Ca2+-induced Ca2+ release from RyR2). The control of Ca2+ wave generation is influenced by multiple factors, including the SR Ca2+ load, the endogenous Ca2+ buffers, the Ca2+-sensitivity of RyR2, and the spatial distance between the RyR2 clusters. To understand the Ca2+-signaling dynamics in cardiac myocytes, we aim to develop a quantitative model that is based on realistic parameters measured from the cells. In our earlier modeling of Ca2+ spark properties and the Ca2+ wave dynamics in atrial cells, we used a CRU transverse spacing of 2 μm as reported in a previous publication (21), along with the well-established longitudinal spacing of 2 μm. However, our modeling study suggested that Ca2+ waves could not initiate unless the transverse spacing is closer to 1 μm. This problem highlights the need for detailed knowledge on the spatial organization of CRUs, and we have therefore carried out a study to obtain accurate measurements of the 3-D distribution of RyR2 clusters in cardiac myocytes.
We used antibody labeling and confocal microscopy to visualize the RyR2 clusters in rat ventricular and atrial myocytes. To improve the accuracy in measuring the distance between adjacent clusters, we developed a new method to take confocal images of the cell's transverse plane in addition to the longitudinal plane to achieve high resolution (confocal microscope x,y resolution of 0.25 μm) for all three spatial dimensions and analyzed these data using a density-dependent filter. Thus we obtained the distribution of RyR2 clusters in three regions of interest (ROIs): 1), along the longitudinal direction, 2), within the transverse plane, and 3), as a novel finding interspersed between the Z-lines on the cell periphery (intercalated RyR2 clusters). In addition, we also performed 3-D image deconvolution and reconstruction to aid visualization of the RyR2 cluster distribution. Based on these measurements we constructed a model to reflect the 3-D geometry of CRU distribution in the ventricular and atrial myocytes. This model is used in our construction of a quantitative model to simulate the relationship between the CRU spacing and the probability of Ca2+ wave initiation and propagation, and to predict the influence of altered CRU spacing on the spontaneous Ca2+ wave generation under pathological conditions (22).
METHODS AND MATERIALS
Cell isolation
The Sprague-Dawley rats were purchased from Harlan (Indianapolis, IN). Rats were anesthetized with sodium pentobarbital (100 mg/kg injected IP, 4000 u/kg heparin). After the suppression of spinal cord reflexes, the hearts were removed via midline thoracotomy. A standard enzymatic technique (20) was used to isolate the ventricle and atrial myocytes. All animals and procedures were handled strictly in accordance with the National Institutes of Health guidelines and protocols approved by our Institutional Animal Care and Use Committee.
Chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) if not specified otherwise.
Frozen tissue section preparation
The heart was first perfused with Ca2+-free phosphate buffered saline (PBS) by injection into the left ventricle with venting from a small incision on the right atrial free wall (∼60 ml/30 s). The heart was then removed and laterally dissected to expose the ventricular walls and chambers. Tissue freezing medium was filled into the chambers to preserve the morphology during the subsequent freezing process. The tissue was flash frozen by submerging first into chilled isopentane for 10–20 s, then placed in dry ice, and finally stored in a −80°C freezer. For thin sectioning, the frozen tissue was placed in a Reichert Cryostat 2800 Frigocut-E (Bannockburn, IL) and allowed to equilibrate to the box temperature of −30°C. The tissue was then sliced into 20 μm thick sections. The frozen tissue sections were again stored in a −80°C freezer until use.
Antibody labeling of isolated cells
Freshly isolated ventricular and atrial myocytes were fixed in 1% paraformaldehyde PBS solution for 5 min, washed twice in cold PBS, then permeablized in 0.1% Triton X-100 PBS solution for 5 min. The cells were incubated in the primary antibody (1:100 dilution) solution containing 5% bovine serum albumin (BSA), 3% goat serum 0.01%, and Triton X-100 in PBS for 1.5–2 h at room temperature; washed twice in cold PBS, and then incubated in the Alexa Fluor 488 conjugated secondary antibody (1:200 dilution, Molecular Probes, Eugene, OR) solution for 1.5–2 h at room temperature. For specific labeling of RyR2, we used anti-RyR2 monoclonal antibody (mouse IgG1, clone C3-33, from Affinity BioReagents, Golden, CO) that strongly detects RyR2 and only weakly detects RyR1 (23,24). In some earlier experiments, we also used a polyclonal anti-RyR2 (aa5029) antibody (rabbit IgG, from A. R. Marks Lab, Columbia University, New York, NY) and another monoclonal antibody (mouse IgG1, clone C34 from Affinity BioReagents), which detects RyR2 as well as RyR1 and RyR3. All antibodies generated similar labeling patterns.
The quality of labeling was evaluated by the maintenance of cell morphology (i.e., clear striations, rod-like shape for ventricular cells, and spindle-like shape for atrial cells) and the uniformly bright labeling. In well-preserved and well-labeled cells, the peripheral RyR2 labeling shows a clean and smooth outline and the intercalated RyR2 units are clearly visible. In contrast, in overpermeablized cells the peripheral outline often became jagged and the intercalated RyR2 units were missing.
Antibody labeling of tissue sections
The tissue sample slides were thawed at room temperature for 5 min. The tissue sections were incubated in the blocking solution containing 5% goat serum and 3% BSA in PBS for 30 min, rinsed twice in PBS, then incubated in the primary antibody solution (1:200 dilution) for 1.5 h, rinsed twice in PBS, and then incubated in the secondary antibody solution (1:200 dilution) for another 1.5 h. The antibody-labeled tissue sections were covered with antifade and sealed under a No. 1 glass coverslip for imaging.
Confocal imaging of the longitudinal and transverse sections of cardiac myocytes
Confocal images were obtained using a BioRad (Hercules, CA) Radiance 2000 confocal microscope with a water emersion objective 63× numerical aperture 1.2, corrected for the thickness of the No. 1 glass coverslip. The pixel dimensions are 0.02 μm × 0.02 μm in the focal plane, and the z-sectioning interval was 0.2 μm. For 3-D image deconvolution, we determined the microscope's point spread function (PSF) following the method of Hollingworth et al. (25) with cubic pixels of 0.10 μm in x, y, and z. Briefly, we embedded fluorescence beads (Molecular Probes) in the agarose gel with a refractive index of 1.38 (30% sucrose, 0.5% agarose) to match the cell's refractive index and used the optical settings for Alexa Fluor 488 to obtain a series of x-y (pixel size 0.10 × 0.10 μm) images of a single fluorescent bead (φ ∼ 0.17 μm) using z-sectioning (z interval 0.1 μm), starting 20 μm below and ending 20 μm above the bead. Deconvolution and reconstruction of 3-D images was carried out using the method described previously (26).
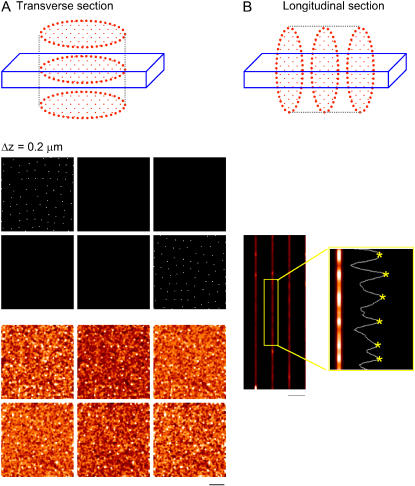
High resolution images were obtained from both the longitudinal and transverse sections of cardiac myocytes. For longitudinal sectioning, the cells were suspended in solution and allowed to settle on the glass coverslip, and hence the cell's longitudinal plane was naturally oriented in parallel to the microscope's focal plane. For transverse sectioning, we embedded the cells in agarose with a low melting temperature (<36°C, 1.5%–2% in PBS). The cells became immobilized at various orientations after cooling the agar to room temperature. This cell-in-agar “sausage” was then sliced and placed onto the glass coverslip for confocal imaging. By chance, some of the cells embedded in the agarose were oriented with the transverse plane parallel to the glass coverslip (Fig. 3 D), which allowed optical sectioning in transverse planes to take advantage of the x,y resolution of ∼0.25 μm, instead of the z-resolution of ∼1 μm.
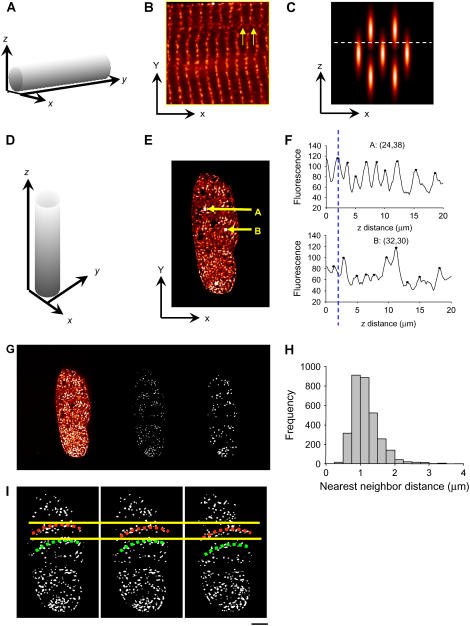
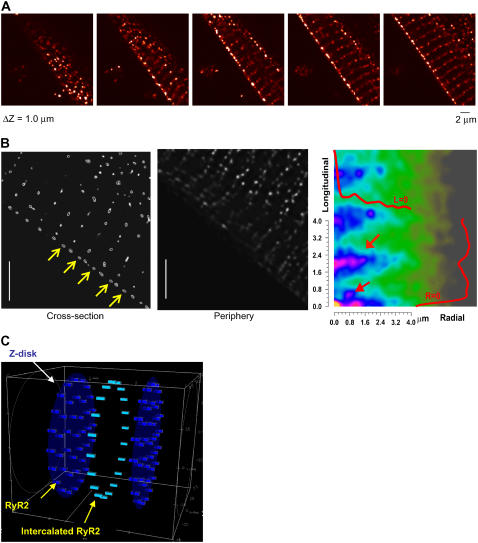
FIGURE 3.
Analysis of transverse RyR2 cluster spacing. Panel A shows the conventional orientation of confocal sectioning with respect to the cell axis. Panel B shows a typical cross section. Note the points of dislocation (arrows) and the curved distribution of RyR2 labeling. Panel C shows how 3-D distribution of RyR2 clusters, when combined with an extended section resolution, leads to an overestimate of RyR2 density. The long ellipses represent optically blurred RyR2 clusters. D shows the cell orientation used in the agarose embedding method. In this orientation, resolution of the transverse section is increased to that of the microscope's in-plane resolution. Panel E shows a typical cell cross section when the cell is oriented as shown in D. Arrows A and B show regions analyzed in panel F. Note the phase difference between the maxima of labeling intensity as the confocal section is moved along the cell. The mean distance between peaks is 2.1 ± 0.7 μm and 1.9 ± 0.6 μm for site A and B, respectively, which is in agreement with the longitudinal RyR2 spacing value obtained from the conventional longitudinal section method as shown in Fig. 1. See the movie in Supplementary Materials for an animated display of the RyR2 labeling in transverse section images along the z axis. Panel G shows image processing with a new digital filter to define the center of RyR2 cluster labeling (see Methods and Materials). The original image, the binary image after thresholding, and the density-dependent filtered image are displayed in order from left to right. In the filtered image, the in-focus RyR2 labeling is picked up and the background noise is reduced. H shows the histogram of RyR2 cluster nearest neighbor distances in the transverse plane. The mean and standard deviation are 1.05 ± 0.44 μm with a median value of 0.96 μm for the transverse spacing. Panel I shows the curvature of RyR2 cluster distribution in the transverse section. Note how the position of the wave moves across the cell with section position. Scale bar 5.0 μm.
Measurement of RyR2 separation distances in the longitudinal section images
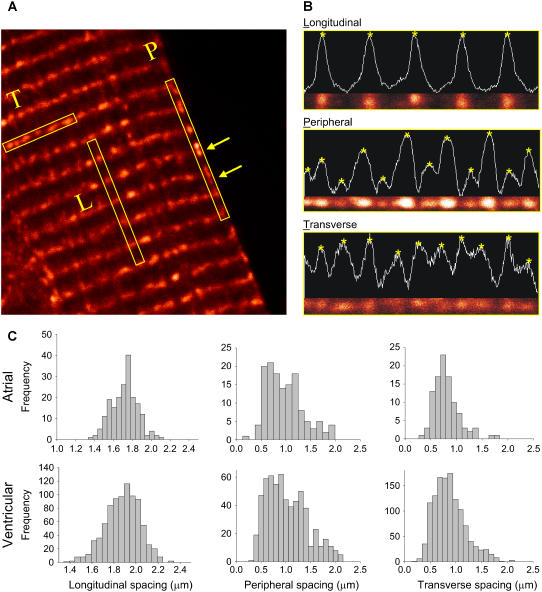
Three characteristic RyR2 cluster separation distances were measured in the isolated ventricular and atrial myocytes: 1), the longitudinal spacing, 2), the transverse spacing, and 3), the peripheral spacing. Measurements were taken only in the regions of the cell where the RyR2 were arrayed on a straight line. Since atrial cells were often curved, measurements were taken from shorter straight sections. To measure, for example, the transverse spacing, the image was rotated to make the RyR2 row horizontal. The ROI was selected (yellow box in Fig. 2 A), and the fluorescence intensity averaged along the width of the region (0.8 μm wide in 40 pixels) was plotted against the length of the region. The spacing between RyR2 clusters is defined as the distance between peaks in the average fluorescence values.
FIGURE 2.
Distance between adjacent RyR2 clusters from longitudinal section images. Panel A shows the three ROIs that were measured to obtain the longitudinal, the transverse, and the peripheral RyR2 cluster spacing, respectively. (B) Intensity profiles along each ROI. The stars show the peaks of intensity, and their distance is measured as the spacing between the RyR2 clusters. (C) Histograms of RyR2 spacing in atrial and ventricular myocytes. The mean and the standard deviation values are listed in Table 1.
Measurement of nearest neighbor distance between RyR2 clusters in the transverse section images
Among the randomly oriented cells imbedded in agar, our measurements were carried out only on cells that were vertically oriented, that is, their longitudinal axis parallel to the optical axis and their transverse plane parallel to the microscope focal plane. The distinction between vertically oriented cells from those that were tilted is based on the criteria that vertically oriented cells showed no lateral motion of the surface contour as the focal plane traveled up and down the z axis (z-sectioning), whereas the surface contour of tilted cells migrated systematically across the optical field. Examples of z-sections of a vertical and a tilted myocyte are available in the Supplementary Materials in the movies of stacked images.
To determine the nearest neighbor distance of RyR2 clusters in a transverse section, a binary image was generated by eliminating all pixels whose values were <0.8 of the maximum in that image. The binary image had “islands” of closely packed pixels with variable sizes as well as isolated “atolls” that had only a few neighboring pixels or a single pixel. We used a density-dependent filter (27) to fill in the coves in the islands and to remove atolls using a “live-or-die” algorithm. The parameters in the filter were optimized to recover the identifiable labeling in the original images and the algorithm tested using model simulations. The resulting islands are hence identified as clusters of RyR2. The distance between the center of masses of island i and every other island was calculated, and the shortest distance was the nearest neighbor distance to island i. We only used transverse sections spaced 2 μm apart to minimize the contribution of fluorescence from adjacent Z-disks.
Confocal imaging of antibody-labeled tissue sections
Confocal imaging of the antibody-labeled tissue sections was carried out using the same optical settings as for imaging isolated cells. The focal plane was placed in the middle of the 20 μm thick tissue slice to avoid the interface between the tissue and the glass. To obtain the RyR2 cluster longitudinal spacing in the tissue cross section, we selected the muscle bundles with the longitudinal axis oriented in parallel to the microscope's focal plane. The samples were chosen by randomly scanning the ventricular tissue section to find such longitudinally running muscle bundles in the optical field. We also sampled various areas in the tissue sections to obtain an average value.
Measurement of sarcomere spacing
Freshly isolated cardiac myocytes were bathed in the Tyrode solution containing (in mM): NaCl 135, KCl 4, NaHPO4 0.33, MgSO4 1, HEPES 10, Glucose 10, CaCl2 1.8, pH 7.3. The cell suspension was placed in a chamber mounted on the microscope stage (Nikon, Tokyo, Japan) with a No. 1 glass coverslip forming the bottom of the chamber. The cells were oriented with the longitudinal plane parallel to the focal plane. We used an IonOptix system (IonOptix, Milton, MA) to acquire transmitted light images of the cell, measure the optical density across the sarcomeres, and calculate the average sarcomere spacing using a fast Fourier transform algorithm.
RESULTS
Antibody labeling of RyR2 in the isolated cell and in the tissue cross section
The RyR2 labeling in rat cardiac myocytes displays punctate staining with a well-organized pattern. Fig. 1 shows pseudocolored confocal images of the RyR2 labeling in the ventricular and atrial myocytes in isolated cell preparations and in frozen tissue cross sections. The RyR2 clusters were identified by a monoclonal anti-RyR2 antibody and visualized using a secondary antibody-conjugated Alexa Fluor 488. The background fluorescence was negligible (data not shown).
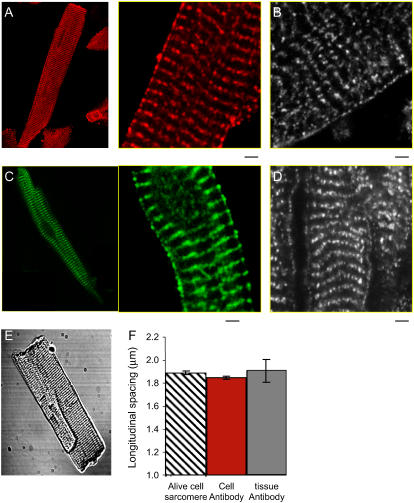
FIGURE 1.
Ryanodine receptors (RyR2) in cardiac myocytes were labeled using an anti-RyR2 monoclonal antibody and visualized using a secondary antibody conjugated Alexa Fluor 488. Images show examples of RyR2 labeling in (A) an isolated rat atrial myocyte, (B) left ventricle tissue cross section, (C) an isolated rat atrial myocyte, and (D) right atrium tissue cross section. The scale bar is 2.0 μm. The longitudinal spacing between the adjacent rows of RyR2 clusters was compared with the sarcomere length in live cells. (E) Transmission image of a typical freshly isolated rat ventricular myocyte shows clear sarcomeres. (F) The RyR2 cluster longitudinal spacing in isolated cells (shown as mean ± SE) is essentially the same as the sarcomere length in live cells. Furthermore, the RyR2 spacing in the isolated cells remains similar to that in the tissue cross section (t-test, p = 0.14).
Measurements of the RyR2 cluster spacing in isolated cells were made after taking precautions to ensure that the antibody-labeling procedure did not alter cell shape or cluster spacing. First we compared the longitudinal spacing between the adjacent rows of RyR2 clusters (located on Z-lines) in the antibody-labeled ventricular cells with the sarcomere length (which is equal to the Z-line spacing according to the known muscle ultrastructure) observed in freshly isolated live cells. The former was 1.87 ± 0.18 μm (mean ± SD, n = 151 cells) and the latter 1.89 ± 0.12 μm (n = 45 cells). The close agreement of these values suggests that antibody-labeled cells maintained the structure of live cells. We also compared the RyR2 cluster spacing in the isolated cells with those in the tissue cross sections. The longitudinal spacing in the tissue samples was 1.94 ± 0.31 μm, similar to that measured in the isolated cells (t-test, p = 0.14, Fig. 1 F).
Three distinctive features of the RyR2 cluster distribution measured from the longitudinal section image
Due to their rod-like shape, isolated cardiac myocytes settle onto the glass coverslip (mounted on the microscope stage) with their longitudinal plane oriented parallel to the microscope's focal plane; and the confocal image taken in this plane captures the cell's longitudinal section as seen in Fig. 1. In the longitudinal section, the distribution pattern of RyR2 clusters was characterized by three distinctive ROIs as illustrated in Fig. 2 A: 1), the longitudinal spacing defined as the distance between the adjacent rows of RyR2 clusters along the longitudinal direction inside the cell, 2), the peripheral spacing as the distance between the adjacent RyR2 clusters along the longitudinal direction on the cell periphery, and 3), the transverse spacing as the distance between the adjacent RyR2 clusters within the same row in the transverse direction. To measure the distance between RyR2 clusters, we took each of the ROIs from the longitudinal section of cells (Fig. 2 B), computed the optical density of the ROIs (white traces), and calculated the distance between the neighboring optical density peaks (starred). The histograms of RyR2 cluster spacing in each of the ROIs in ventricular and atrial myocytes are shown in Fig. 2 C. The mean and the standard deviation of the RyR2 cluster spacing values are listed in Table 1.
TABLE 1.
RyR2 cluster nearest neighbor distance
| Nearest neighbor distance in transverse plane
|
||||
|---|---|---|---|---|
| Longitudinal distance | Peripheral distance | Transverse section | Longitudinal section | |
| Ventricular myocyte (n = 151 cells from nine hearts) | 1.87 ± 0.18 | 0.97 ± 0.42 | 1.05 ± 0.44 | 0.83 ± 0.31 |
| Atrial myocyte (n = 65 cells from eight hearts) | 1.69 ± 0.41 | 0.92 ± 0.38 | 0.97 (calculated) | 0.77 ± 0.29 |
Unit, μm; mean ± SD.
A novel finding is the presence of RyR2 labeling on the cell periphery that is interspersed between the Z-lines (box labeled P in Fig. 2 A and middle panel in Fig. 2 B). In ventricular cells, the peripheral RyR2 clusters were separated by 0.97 ± 0.42 μm and in atrial cells, 0.92 ± 0.38 μm. These “intercalated” RyR2 clusters were regularly seen only on the cell periphery between the Z-lines (yellow arrows in Fig. 2 A) and were largely absent in the cell's interior. A variation in fluorescence intensity of the intercalated RyR2 clusters was sometimes seen (middle panel of Fig. 2 B), suggesting that the alternating clusters probably locate in different focal planes (see below).
An estimate of the transverse spacing from the longitudinal section images suggested that the mean spacing between RyR2 clusters was 0.83 ± 0.31 μm and 0.77 ± 0.29 μm for ventricular and atrial cells, respectively. However, this transverse spacing is comparable to the confocal microscope's z axis resolution (∼1 μm), so the longitudinal section image contains significant fluorescence contamination from the RyR2 neighbors that are above and below the plane of focus. This problem is illustrated schematically in Fig. 3 C, which shows how optically blurred point sources (obtained by convolving the point light source with the microscope's PSF) can lead to a systematic underestimate of the transverse spacing when the RyR2 clusters are not in perfect registration in sequential Z-lines. Thus, the effect of optical blurring is to underestimate the transverse spacing when measured from the longitudinal section image. In an effort to accurately measure the transverse spacing, we developed a method to directly image the cell's transverse plane and to exploit the higher confocal x,y axis resolution of ∼0.25 μm.
Measurement of the RyR2 cluster transverse spacing using the transverse section image
By embedding cells in agarose and then selecting the cells that were oriented with the longitudinal axis perpendicular to the microscope's focal plane (Fig. 3 D), we were able to image the cell's transverse plane at a higher resolution (Fig. 3 E). The orientation of the cell was monitored by moving the focal plane along the z axis. If the edge of the cell stayed in the same area of the optical field while the focus was changed, it indicates that the cell was oriented with its longitudinal axis largely parallel to the optical axis. If the cell was tilted, the edge of the cell moved in the optical field as the focal plane was changed. (See Supplementary Materials for movies showing sample cells.) These transverse section images allowed us to measure the RyR2 transverse spacing at the x,y resolution of ∼0.25 μm, which contains less out-of-focus labeling than the longitudinal section image because the z-resolution of ∼1 μm is sufficient to reject fluorescence from the neighboring RyR2 clusters located on the adjacent Z-disks that are spaced 1.9 μm apart along the cell's longitudinal direction. Since ventricular myocytes are more rod-like and straighter than atrial myocytes, we primarily used this method for ventricular cells.
Fig. 3 E shows a typical transverse section image from a ventricular cell. To test whether the process of embedding the cells in agar altered the spatial distribution of RyR2, we measured the longitudinal RyR2 cluster spacing from a series of transverse section images along the z axis. The intensity in any selected region should vary from section to section as that region traversed sequential Z-disks and encountered RyR2 labeling (Fig. 3, E and F). For example, the mean distance between intensity peaks was 2.1 ± 0.7 μm and 1.9 ± 0.6 μm for sites A and B, respectively, which are not significantly different from the RyR2 spacing measured in longitudinal sections. We therefore conclude that the process of embedding the cells had little impact on the distribution of RyR2 labels. This view was further supported by examination of images from the cells embedded in agar that happened to lie parallel to the focal plane; in these cells there was no detectable difference in the RyR2-labeling pattern from that described above for cells in solution. We therefore conclude that the RyR2 distribution was not significantly altered by embedding the cells in agar.
To measure the nearest neighbor distances between RyR2 clusters in the transverse section images, we developed a two-step filtering method to pick out the in-focus labeling from out-of-focus labeling and background noise (see Methods and Materials). Sample image processing is illustrated in Fig. 3 G: The left-hand panel shows RyR2 labeling in a single transverse section image, and the binary representation of that labeling is shown in the middle panel. The result of applying the density-dependent filter to the binary data is shown in the right-hand panel. The resulting image contains islands of labeling that represent RyR2 clusters. From these data we calculated the nearest neighbor distances between islands of RyR2 labeling as the shortest distance between the signal mass centers. Thus we obtained the histogram of nearest neighbor distances for the transverse spacing of RyR2 clusters, including those on the cell periphery (Fig. 3 H). This histogram exhibited some kurtosis (see Discussion) with a median value of 0.96 μm and a mean value of 1.05 ± 0.44 μm.
To establish the validity of the filtering method, we tested the algorithm on a virtual cell (Fig. 4) made from a 3-D distribution of dots with longitudinal spacing of 2.0 μm and transverse spacing of 1.0 μm (randomly distributed in the transverse plane, middle panel). After blurring by convolution with the microscope PSF and adding background noise, the resulting images (lower panel) resembled the raw images obtained in experiments. (Notice that the bright spots present only on Z-disks in the model cell are now seen on subsequent image planes in the blurred cell.) After applying the same filtering algorithm with the same parameter settings as used in the above image analysis, the nearest neighbor distance was calculated to be 1.0 ± 0.2 μm, as expected from the model construction.
FIGURE 4.
Test of filter algorithm on a virtual cell. The top of panel A shows the transverse sectioning geometry of a model cell. The middle panel shows evenly spaced sections through the simulated data. The randomly placed pixels were set to appear as bright spots whose mean nearest neighbor distance was set to 1.0 μm. The lower panel shows the simulated confocal sections after blurring by convolving with the microscope PSF and adding noise. Note the bright spots appearing only on Z-lines in the unblurred myocyte (middle panel) are now seen on subsequent image planes in the blurred images (bottom panel). After processing these data with the algorithm used in Fig. 3, the mean nearest neighbor distance was 1.0 ± 0.2 μm and the median distance was 1.0 μm, as expected. Panel B shows simulated longitudinal section images generated from the same data set. The measured transverse spacing was 0.88 μm, which is shorter than the known set value of 1.0 μm, illustrating the artifactual reduction in transverse spacing seen in the longitudinal section.
To compare the difference in the transverse section and the longitudinal section measurements, we also took longitudinal section images from the blurred virtual cell (Fig. 4 B). The transverse spacing measurement was carried out in the same way as described above and shown in Fig. 2, which gave an average value of 0.88 μm. As might be expected, this value was an underestimate of the preset value of 1.0 μm in the virtual cell, confirming that limited axial resolution in the longitudinal section image leads to a systematic underestimate of true RyR2 cluster spacing in the transverse plane.
From these data we conclude that the RyR2 cluster transverse spacing has a mean value of 1.05 μm in the ventricular myocyte as obtained from the transverse section method and that the lower value obtained from the longitudinal section method (0.83 μm) is an artifact arising from the limited axial resolution of the microscope and complex sample geometry. Although we did not use the transverse section method on the atrial myocyte, one can calculate the mean transverse spacing to be 0.97 μm by scaling from the underestimated value from the longitudinal section measurement (0.77 μm).
RyR2 cluster phase waves
Cursory examination of longitudinal sections suggests that the RyR2 on the Z-lines are mostly in linear register. However, on closer examination we sometimes see small deviations from linearity. For example, in Fig. 3 B we see that the Z-lines are slightly bowed out to the left and if the Z-line registration is followed carefully on the right-hand side of the image there is a discontinuity in the registration across the cell. Fig. 3 F shows that, because the fluorescence intensity rises and falls periodically along the z axis, we can assign an angular phase at each point in the transverse section, with the maximum defined (arbitrarily) to have a phase of 0°. Therefore, the phase is 0° for location A and ∼180° for location B (marked by dashed line). If the angular phases were uncorrelated at each x-y point then there would be no discernible spatial pattern in the fluorescence intensity as the focal plane travels. Instead, we see propagation of “phase waves” that arise from local regions that are in close registration (Fig. 3 I, also see Supplementary Materials). By a phase wave, we mean that the position of, say, the loci of 0° phase moves across the transverse section as the focal plane moves up. The appearance of such waves can be explained by a small angle between the (often slightly curved) planes of Z-discs and the planes of confocal sectioning.
RyR2 cluster distribution on the periphery
In addition to the quantitative measurements of RyR2 cluster spacing within the cell, we also performed 3-D image deconvolution and reconstruction to aid visualization of the 3-D geometry of RyR2 cluster distribution on the periphery of ventricular myocytes. Fig. 5 A presents a series of longitudinal section images taken at a z interval of 0.2 μm from a ventricular myocyte. After using the PSF to perform 3-D deconvolution and reconstruction of the images, Fig. 5 B shows the reconstructed images of the RyR2 labeling in the cross section and on the periphery of the cell. Note that in the cross-section image (left panel, yellow arrows) the intercalated RyR2 unit is clearly visible at the cell periphery but absent in the interior. In contrast, in the periphery image (middle panel) the intercalated RyR2 clusters line up in parallel to the Z-line to form double rows running along the surface. To analyze the periodicity of the RyR2 cluster distribution, we calculated the two-dimensional autocorrelation from these images, which had prominent peaks at ∼2 μm (longitudinal axis) and ∼1.2 μm (transverse). The repeated structure (with strong autocorrelation) in the longitudinal spacing corresponds to the resting sarcomere length, whereas the transverse spacing arises from the average spacing between clusters in a circumferential direction. Additional peaks occur at a repeated spacing of 0.8 μm and 0.6 μm in the transverse and longitudinal directions, respectively. These extra peaks are due to the loose array of intercalated RyR2s that appear to form “double rows” in some regions on the surface.
FIGURE 5.
RyR2 cluster distribution within the cell interior and the periphery. Panel A shows sequential z-section images separated by 0.2 μm. A movie showing a z-section image stack is attached in the Supplementary Materials. After processing these data by 3-D image deconvolution using the microscope PSF, B shows sections obtained across the cell (left panel) and on the cell surface (middle panel). The autocorrelation of the surface image (right panel) reveals the periodicities in the RyR2 labeling. There is a strong correlation (upper red arrow) at ∼2 μm along the longitudinal axis and at ∼1 μm along the transverse axis (labeled Radial) in correspondence with the measured longitudinal and transverse spacing. An additional peak (lower red arrow) also occurs at ∼0.6 μm and 0.8 μm at longitudinal and transverse axes, respectively. This extra peak reflects the mean position of the intercalated peripheral RyR2 clusters, which are loosely arrayed with this spacing in double rows (middle panel). The overlaid red curves show the autocorrelations at L = 0 and R = 0. Panel C shows a schematic of the 3-D geometry of RyR2 cluster distribution in cardiac myocytes as used in the mathematical model presented in the companion work (Izu et al., 2005).
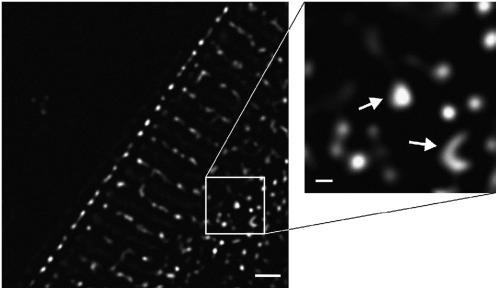
RyR2 cluster size
Most of the RyR2 labeling formed spots which were close to the resolution limit of the microscope (250 nm). However, in some places larger clusters were detected (Fig. 6). We used the higher resolution (240 nm) of the deconvolved data to investigate the apparent size of RyR2 clusters. For example, one very large cluster in this section had a maximal diameter of 410 nm, whereas the length of the other curved cluster was 960 nm (Fig. 6, inset). The extended cluster size observed in this image could not be explained by the blurring of out-of-focus clusters because the maximal stain intensity was essentially confined to this optical section. An accurate measurement of the RyR2 cluster size and spacing should provide important clues for understanding the E-C coupling mechanism, which is the topic of discussion below.
FIGURE 6.
High resolution deconvolved image of RyR2 cluster size. The panel at left shows that most clusters appear punctate with size close to the resolution limit (∼240 nm). Occasional clusters were larger, and two examples of such large clusters are shown here (arrows in right panel). One has a maximum diameter of 410 nm and the curved cluster has a length of 960 nm. The extensive size is not due to out-of-focus labeling because the maximum labeling intensity is contained within this optical section. Scale bars are 2.0 μm (left panel) and 200 nm (right panel).
DISCUSSION
Structure-function relationship of the RyR2 cluster distribution in 3-D
We set out to measure the RyR2 cluster distribution to provide accurate parameters for modeling the Ca2+ dynamics in cardiac myocytes. Now we have obtained the RyR2 cluster spacing in cardiac myocytes with 0.25-μm resolution for all three spatial dimensions. The 3-D geometry of RyR2 cluster distribution exhibits the following prominent features in three ROIs along the longitudinal direction, on the periphery, and in the transverse plane.
The measured longitudinal spacing of RyR2 clusters was 1.87 ± 0.18 μm in the isolated ventricular myocyte under control conditions. This CRU spacing is consistent with the nearest neighbor distance between Ca2+ sparks in longitudinal line scan images (10,28). To rule out possible artifacts associated with cell isolation, we also measured the RyR2 longitudinal spacing in the frozen tissue sections, which gives a value of 1.94 ± 0.31 μm in the left ventricle free wall, without significant difference from that in the isolated ventricular cells. The relationship between the CRU longitudinal spacing and the Ca2+ wave generation is studied using mathematical modeling and supercomputer simulation (22). The main result is that the longitudinal spacing critically determines the Ca2+ wave propagation. Importantly, this influence of the longitudinal spacing on wave propagation may manifest under pathological conditions (e.g., shortening of sarcomere length, t-tubule disarray, calcium overload) to drastically alter the Ca2+ dynamics in the heart.
A novel finding is the double rows of intercalated RyR2 clusters interspersed between the Z-lines on the cell periphery. Previous studies on sino-atrial node cell (29) and atrial myocytes (30,31) have also detected labeling between Z-lines at the cell surface, but without quantification. In this study, we have consistently captured these intercalated RyR2 clusters in both the isolated cardiac myocytes and the frozen tissue sections in a reproducible manner, which enabled us to quantify the spacing of intercalated clusters on the cell periphery. The mean spacing of the intercalated RyR2 cluster is 0.97 μm, which is considerably less than the longitudinal spacing between RyR2 clusters on the Z-disks within the cell (1.87 μm). It is possible that these intercalated clusters result from the trafficking of RyR2 during normal protein turnover. The RyR2 clusters may either migrate from the surface into the cell interior as t-tubules grow to invaginate the cell or be the result of RyR2 migrating from the interior toward the cell surface. In avian muscle (which lacks t-tubules) the assembling of junctions (feet structures) was clearly seen at the cell surface (32), although this does not preclude the possibility of junction assembly on the Z-disk in cells with t-tubules. If RyR2 clusters were migrating from/to the surface one might expect to see the transverse spacing retained as they spread onto/from the Z-disk.
A possible physiological function of intercalated RyR2 is revealed by model simulations as acting like relay stations to facilitate the Ca2+ wave spread along the cell surface (22). This model prediction is consistent with the experimental data that the Ca2+ waves were frequently seen at the cell periphery (18).
Using the newly developed transverse section method, we measured the mean RyR2 cluster transverse spacing to be 1.05 μm in the ventricular myocytes and calculated it (by scaling from the value measured in the longitudinal section using the ratio obtained in the ventricular cell) to be 0.97 μm in the atrial myocytes. These values concur with the model prediction that a transverse spacing of ∼1 μm is required for initiating Ca2+ waves given 2.0-μm longitudinal spacing. Our estimate of 0.97-μm transverse spacing in rat atrial myocytes is about half of the 2 μm value reported in the cat atrial myocytes (21); the latter probably resulted from missing some middle units. Our measured value of 1.05 μm in rat ventricular myocytes is larger than the report of 0.76 μm for the transverse distance of nearest Ca2+ sparks (33). However this discrepancy can be explained by the method of measurement; Ca2+ sparks were recorded by orienting the confocal line scan along the transverse direction in the cell's longitudinal plane, so the distance between nearest Ca2+ sparks should reflect the CRU transverse spacing measured from the longitudinal section images whose resolution is limited by the axial response of the confocal microscope. As shown above, this leads to a systematic underestimate of CRU spacing and it should be noted that the Ca2+ spark distance is in good agreement with our estimate derived from images obtained in the longitudinal section (0.83 μm). Similarly, we had estimated 1.15 spark per μm or 0.87 μm between spark sites on average when measured in the longitudinal plane (34). Using a fast slit-scanning confocal microscope, Cleemann et al. (35) measured a spark density of 0.78 per μm2, which for a resting sarcomere spacing of 1.9 μm (from this study) suggests a mean distance of 0.66 μm between Ca2+ spark sites. This density is higher than that reported by Parker et al. (33), but may be explained by the poorer axial response of the slit-scanning system as compared to a point-scanning confocal. Nevertheless, all values obtained directly from measurements performed on longitudinal plane scans suffer from the poor axial resolution of confocal systems so that they overestimate (to varying degrees) the spark site density. Our transverse sectioning method reduces this problem so our estimate of 1.05-μm transverse spacing is the most reliable value to use for modeling.
The center-to-center RyR2 cluster spacing of 1.05 μm is also in reasonable agreement with the distance separating junctional areas measured from electron micrographs. Franzini-Armstrong et al. (6) measured the minimum edge-to-edge distance to be ∼414 nm, which when combined with a typical junction extent of 400 nm suggests a center-to-center spacing of ∼814 nm. Although this value is also consistent with our estimate of ∼0.83 μm derived from longitudinal section images, it should be noted that gluteraldehyde fixation used in an electron micrograph study could lead to specimen shrinkage of up to 20% (36). After allowing for cell shrinkage and the fact that the CRU spacing in the electron micrograph study was estimated from the combination of serial sections which would tend to reduce the minimum distance between CRUs, we suggest that there is reasonable agreement between our value of 1.05 μm derived from the transverse sectioning method and that reported by analysis of electron micrographs (6).
Using the CRU spacing values measured in this study, one can estimate the total number of CRUs in a typical ventricular myocyte of 100 × 20 × 10 μm3 to be in the order of 10,000 or, if intercalated RyR2 clusters act as CRUs, 13,000. This range is in accord with a previous estimate based on Ca2+ spark measurements (28).
Cluster size and number of RyR2 channels involved in a Ca2+ spark
Discrete RyR2 clusters underlie the quantal Ca2+ release events recorded as Ca2+ sparks. To understand the control of RyR2 gating, it is important to know the total number of channels in a cluster and how many of them are involved in generating a spark. The RyR2 cluster size measured in this study ranges from 250 nm to 960 nm across, which is in reasonable agreement with the electron micrographic analysis of cluster distribution by Franzini-Armstrong et al. (6). If most RyR2 clusters are ∼250 × 250 nm2 and the average area occupied by a single RyR2 molecule is 29 × 29 nm2, such a cluster would contain ∼74 RyR2s with the largest clusters (250 × 960 nm2) containing up to 285 RyR2s. Franzini-Armstrong et al. (6) estimated that the typical rat RyR2 cluster contained ∼250 RyR2s, a value larger than our estimated typical value. Since our resolution limit prevents quantification of areas <250 × 250 nm2, some RyR2 clusters might be even smaller, which would widen the discrepancy with the data of Franzini-Armstrong et al. (6).
The cause of this discrepancy may lie in the method used to calculate the number of RyR2s. In our study, we assume that the extent of label area delimits the cluster. If the junctional region was highly curved within the resolution limit, we would underestimate the total number. In the worst case, if the RyR2s were within a circle of diameter equal to the resolution limit, we would underestimate the number by a factor of π/2, suggesting an upper bound of ∼116 RyR2s in a typical cluster. However most electron micrographs suggest that junctional area is an incomplete circle (6,37). On the other hand, in electron micrographs, the number of RyR2s was calculated on the assumption that the junctional area was circular and the entire junctional area was occupied by RyR2s (6,32), which would tend to overestimate the total number. For the discussion below we will assume the typical cluster contains 100 RyR2s.
The flux (current) of Ca2+ underlying a Ca2+ spark is Ispark = N × Po × iRyR, where N is the total number of RyR2s in a cluster, Po the single RyR2 channel open probability, and iRyR the single channel Ca2+ current. If iRyR is ∼0.5 pA (38) and Ispark ∼ 10 pA as back calculated from the amount of Ca2+ in a spark (13,39,40), then the number of RyR2 channels that open during a spark is 20. This value is also in accord with the estimation of 17–20 RyR2s as derived from noise analysis of spark amplitude variability (41) and with the estimation of more than 10 RyR2s based on RyR2 inhibition studies (39). However, by analyzing the rate of rise of Ca2+ sparks, Cheng's group suggested a different estimate of 1–3 RyR2s underlying a triggered Ca2+ spark (42,43). Although more work is needed to resolve these discrepancies and to refine the estimates, an important insight can be gained by analyzing the relationship between the number of RyR2s involved in a spark and the CRU firing probability. In an extreme case, when only 1 out of 100 RyR2s channels in a cluster opens, the single channel open probability is Po = 0.01, from which the probability of a CRU failing to have any RyR2 opening, the null probability, is Po(0) = (1 − Po)N = 0.37. In this scenario, since all RyRs may close with high probability, the CRU either deactivates or does not respond to the trigger at all. Indeed, this was shown to be the case in the analysis of failures by Cheng's group. However, if 20 RyR2s open during the spark, then Po = 0.2 and Po(0) = 2 × 10−10. In this scenario, the CRU not only has high probability to be activated but also to maintain that activation by having regenerative Ca2+-induced Ca2+ release. On the other hand, robust CRU shutdown must reside in some other mechanism such as terminal depletion in this case. It is important to note that the critical factor here is NPo. For example, if only 1 RyR2 from a cluster of 50 or 250 RyR2s opens during the spark, the null probability Po(0) remains very large (0.36 and 0.37 respectively); if 20 RyR2s open, the Po(0) remains very small (8 × 10−12 and 9 × 10−10, respectively). This sensitivity of Po(0) to NPo can be understood by noticing that Po(0) = (1 − Po)N ≅ e−NPo, when Po is small and N is large.
Junctional and nonjunctional RyR2 clusters
In adult mammalian ventricular myocytes, the majority of RyR2 clusters are thought to form junctional coupling with L-type Ca2+ channels on the sarcolemma and in the t-tubules. The junctional coupling serves as a “couplon” (44) to initiate intracellular Ca2+ signaling in response to membrane excitation. Such junctional RyR2 has been visualized in the electron micrograph in the “feet” structure that is closely associated with the sarcolemma or t-tubules in the form of “peripheral couplings” and “dyads” (6). The junctional coupling was also inferred from the optical colocalization of antibody-labeled RyR2 and L-type Ca2+ channel. The arrangement for RyR2 coupling is markedly different in cells lacking a t-tubule system such as adult atrial myocytes (20), sino-atrial node cells (29), Purkinje cells (45), neonatal cardiac myocytes (46), and avian ventricular myocytes (32,47), where the junctional coupling exists at the cell periphery immediately under the sarcolemma. Since the interior RyR2 clusters are not directly coupled to surface membrane L-type Ca2+ channels, they are “nonjunctional”. In cells lacking internal junctions, the peripheral junctions provide the means to achieve E-C coupling as Ca2+ released from junctional RyR2 (20,45,48) diffuses to form a centripetally propagating Ca2+ wave. In some cases, this wave can initiate release from the nonjunctional RyR2 clusters but this is not always the case as it depends on the excitability of the internal RyR2 clusters (20,45).
In the ventricular myocyte, the action potential triggers a synchronized global Ca2+ elevation via junctional RyR2 couplings distributed throughout the cell. Although the resulting synchronous release would preclude initiation of a spontaneous Ca2+ wave, under certain pathological conditions, spontaneous Ca2+ waves arise during diastole to interfere with Ca2+ signaling and trigger irregular electrical activity via Na+/Ca2+ exchange (18,48). As an essential step in developing realistic models for E-C coupling which may explain the transition to such nonuniform behavior, we have measured the CRU spacing at a resolution sufficient to allow model construction. The 3-D geometry of CRU distribution presented in this model (Fig. 5 C) is applicable to both ventricular and atrial myocytes with the spacing values adjusted to each cell type, regardless of the different arrangements for the junctional coupling between RyR2 and L-type Ca2+ channels in these cells.
Dislocation in RyR2 cluster distribution
In some regions within the cell interior we found dislocations in the transverse structure which result in a phase shift of periodicity in the longitudinal direction. It seems likely that such dislocations are the consequence of local nonuniformities in growth during maturation of the myocyte as sarcomeres are added (49). Such dislocation may cause the effective distance between RyR2 clusters to be reduced in the longitudinal direction, which may bring the cell closer to the threshold for Ca2+ wave initiation. For example, our modeling studies suggest that if the CRU longitudinal spacing were reduced to 1.6 μm, the probability of spontaneous Ca2+ wave initiation would increase drastically (22). Hence, the Ca2+ wave would tend to spread along the dislocation region and from there across the cell. On the other hand, the normal longitudinal spacing of 1.9 μm would serve to limit the spread of a Ca2+ wave. It is possible that cardiac hypertrophy may make such dislocations more common, in which case the probability of Ca2+ wave generation and the likelihood of triggered arrhythmias might increase. This intriguing possibility should be examinable with the methods we have presented here.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
Our transverse sectioning technique was inspired by Dr. Hugo Gonzalez-Serratos's experiments viewing Ca2+ release down the long axis of skeletal muscle fibers. We are grateful to Drs. Jeanine A. Ursitti, Robert J. Bloch, and Shawn W. Robinson for kindly providing the instruments and technical advice for immunohistochemistry experiments; and to Drs. Andrew R. Marks, Steven R. Reiken, and Xander H. T. Wehrens for providing the polyclonal anti-RyR antibody. Our gratitude also goes to June Clopein for administrative support and to William T. Sinclair for machine shop support.
Financial support was provided by an American Heart Association National Scientist Development Grant to Y.C. (AHA 0335250N), a National Institutes of Health K25 grant to L.T.I. (NIH 1K25HL68704), an NIH R01 grant to C.W.B. and L.T.I. (RO1HL071865), NIH grants RO1HL68733 and HL50435-05 to C.W.B., a Veterans Affairs Merit Review Award to C.W.B. and L.T.I. (VA MCB00006N), and a National Center for Supercomputing Applications Grant to L.T.I.
References
- 1.Nakai, J., T. Imagawa, Y. Hakamat, M. Shigekawa, H. Takeshima, and S. Numa. 1990. Primary structure and functional expression from cDNA of the cardiac ryanodine receptor/calcium release channel. FEBS Lett. 271:169–177. [DOI] [PubMed] [Google Scholar]
- 2.Kuwajima, G., A. Futatsugi, M. Niinobe, S. Nakanishi, and K. Mikoshiba. 1992. Two types of ryanodine receptors in mouse brain: skeletal muscle type exclusively in Purkinje cells and cardiac muscle type in various neurons. Neuron. 9:1133–1142. [DOI] [PubMed] [Google Scholar]
- 3.Giannini, G., A. Conti, S. Mammarella, M. Scrobogna, and V. Sorrentino. 1995. The ryanodine receptor/calcium channel genes are widely and differentially expressed in murine brain and peripheral tissues. J. Cell Biol. 128:893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otsu, K., H. F. Willard, V. K. Khanna, F. Zorzato, N. M. Green, and D. H. MacLennan. 1990. Molecular cloning of cDNA encoding the Ca2+ release channel (ryanodine receptor) of rabbit cardiac muscle sarcoplasmic reticulum. J. Biol. Chem. 265:13472–13483. [PubMed] [Google Scholar]
- 5.Tunwell, R. E. A., C. Wickenden, B. M. A. Bertrand, V. I. Shevchenko, M. B. Walsh, P. D. Allen, and F. A. Lai. 1996. The human cardiac muscle ryanodine receptor-calcium release channel: identification, primary structure and topological analysis. Biochem. J. 318:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franzini-Armstrong, C., F. Protasi, and V. Ramesh. 1999. Shape, size, and distribution of Ca2+ release units and couplons in skeletal and cardiac muscles. Biophys. J. 77:1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabiato, A., and F. Fabiato. 1972. Excitation-contraction coupling of isolated cardiac fibers with disrupted or closed sarcolemmas. Calcium-dependent cyclic and tonic contractions. Circ. Res. 31:293–307. [DOI] [PubMed] [Google Scholar]
- 8.Fabiato, A., and F. Fabiato. 1978. Calcium-induced release of calcium from the sarcoplasmic reticulum of skinned cells from adult human, dog, cat, rabbit, rat, and frog hearts and from fetal and new-born rat ventricles. Ann. N. Y. Acad. Sci. 307:491–522. [DOI] [PubMed] [Google Scholar]
- 9.Fabiato, A. 1992. Two kinds of calcium-induced release of calcium from the sarcoplasmic reticulum of skinned cardiac cells. Adv. Exp. Med. Biol. 311:245–262. [DOI] [PubMed] [Google Scholar]
- 10.Lopez-Lopez, J. R., P. S. Shacklock, C. W. Balke, and W. G. Wier. 1994. Local, stochastic release of Ca2+ in voltage-clamped rat heart cells: visualization with confocal microscopy. J. Physiol. 480:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Lopez, J. R., P. S. Shacklock, C. W. Balke, and W. G. Wier. 1995. Local calcium transients triggered by single L-type calcium channel currents in cardiac cells. Science. 268:1042–1045. [DOI] [PubMed] [Google Scholar]
- 12.Cheng, H., W. J. Lederer, and M. B. Cannell. 1993. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 262:740–744. [DOI] [PubMed] [Google Scholar]
- 13.Izu, L. T., J. R. Mauban, C. W. Balke, and W. G. Wier. 2001. Large currents generate cardiac Ca2+ sparks. Biophys. J. 80:88–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern, M. D. 1992. Theory of excitation-contraction coupling in cardiac muscle. Biophys. J. 63:497–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keizer, J., and G. D. Smith. 1998. Spark-to-wave transition: saltatory transmission of calcium waves in cardiac myocytes. Biophys. Chem. 72:87–100. [DOI] [PubMed] [Google Scholar]
- 16.Keizer, J., G. D. Smith, S. Ponce-Dawson, and J. E. Pearson. 1998. Saltatory propagation of Ca2+ waves by Ca2+ sparks. Biophys. J. 75:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izu, L. T., W. G. Wier, and C. W. Balke. 2001. Evolution of cardiac calcium waves from stochastic calcium sparks. Biophys. J. 80:103–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berlin, J. R., M. B. Cannell, and W. J. Lederer. 1989. Cellular origins of the transient inward current in cardiac myocytes. Role of fluctuations and waves of elevated intracellular calcium. Circ. Res. 65:115–126. [DOI] [PubMed] [Google Scholar]
- 19.Berlin, J. R. 1995. Spatiotemporal changes of Ca2+ during electrically evoked contractions in atrial and ventricular cells. Am. J. Physiol. 269:H1165–H1170. [DOI] [PubMed] [Google Scholar]
- 20.Kirk, M. M., L. T. Izu, Y. Chen-Izu, S. L. McCulle, W. G. Wier, C. W. Balke, and S. R. Shorofsky. 2003. Role of the transverse-axial tubule system in generating calcium sparks and calcium transients in rat atrial myocytes. J. Physiol. 547:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kockskamper, J., K. A. Sheehan, D. J. Bare, S. L. Lipsius, G. A. Mignery, and L. A. Blatter. 2001. Activation and propagation of Ca2+ release during excitation-contraction coupling in atrial myocytes. Biophys. J. 81:2590–2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izu, L. T., S. Means, J. Shadid, Y. Chen-Izu, and C.W. Balke. 2006. Interplay of ryanodine receptor distribution and calcium dynamics. Biophys. J. 91:▪▪▪–▪▪▪. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, F. A., Q. Y. Liu, L. Xu, A. Hashem, N. R. Kramarcy, R. Sealock, and G. Meissner. 1992. Amphibian ryanodine receptor isoforms are related to those of mammalian skeletal or cardiac muscle. Am. J. Physiol. 263:C365–C372. [DOI] [PubMed] [Google Scholar]
- 24.McPherson, P. S., and K. P. Campbell. 1993. Characterization of the major brain form of the ryanodine receptor/Ca2+ release channel. J. Biol. Chem. 268:19785–19790. [PubMed] [Google Scholar]
- 25.Hollingworth, S., C. Soeller, S. M. Baylor, and M. B. Cannell. 2000. Sarcomeric Ca2+ gradients during activation of frog skeletal muscle fibres imaged with confocal and two-photon microscopy. J. Physiol. 526:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soeller, C., and M. B. Cannell. 1999. Examination of the transverse tubular system in living cardiac rat myocytes by 2-photon microscopy and digital image processing techniques. Circ. Res. 84:266–275. [DOI] [PubMed] [Google Scholar]
- 27.Izu, L. T., W. G. Wier, and C. W. Balke. 1998. Theoretical analysis of the Ca2+ spark amplitude distribution. Biophys. J. 75:1144–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannell, M. B., H. Cheng, and W. J. Lederer. 1995. The control of calcium release in heart muscle. Science. 268:1045–1049. [DOI] [PubMed] [Google Scholar]
- 29.Rigg, L., B. M. Heath, Y. Cui, and D. A. Terrar. 2000. Localisation and functional significance of ryanodine receptors during [beta]-adrenoceptor stimulation in the guinea-pig sino-atrial node. Cardiovasc. Res. 48:254–264. [DOI] [PubMed] [Google Scholar]
- 30.Tijskens, P., G. Meissner, and C. Franzini-Armstrong. 2003. Location of ryanodine and dihydropyridine receptors in frog myocardium. Biophys. J. 84:1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carl, S. L., K. Felix, A. H. Caswell, N. R. Brandt, W. J. Ball Jr., P. L. Vaghy, G. Meissner, and D. G. Ferguson. 1995. Immunolocalization of sarcolemmal dihydropyridine receptor and sarcoplasmic reticular triadin and ryanodine receptor in rabbit ventricle and atrium. J. Cell Biol. 129:673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Protasi, F., X. H. Sun, and C. Franzini-Armstrong. 1996. Formation and maturation of the calcium release apparatus in developing and adult avian myocardium. Dev. Biol. 173:265–278. [DOI] [PubMed] [Google Scholar]
- 33.Parker, I., W. J. Zang, and W. G. Wier. 1996. Ca2+ sparks involving multiple Ca2+ release sites along Z-lines in rat heart cells. J. Physiol. 497:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cannell, M. B., H. Cheng, and W. J. Lederer. 1994. Spatial non-uniformities in [Ca2+]i during excitation-contraction coupling in cardiac myocytes. Biophys. J. 67:1942–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cleemann, L., W. Wang, and M. Morad. 1998. Two-dimensional confocal images of organization, density, and gating of focal Ca2+ release sites in rat cardiac myocytes. Ann. N. Y. Acad. Sci. 95:10984–10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jearanaikoon, S., and J. V. Abraham-Peskir. 2005. An x-ray microscopy perspective on the effect of glutaraldehyde fixation on cells. J. Microsc. 218:185–192. [DOI] [PubMed] [Google Scholar]
- 37.Forbes, M. S., and E. E. van Neil. 1988. Membrane systems of guinea pig myocardium: ultrastructure and morphometric studies. Anat. Rec. 222:362–379. [DOI] [PubMed] [Google Scholar]
- 38.Mejia-Alvarez, R., C. Kettlun, E. Rios, M. Stern, and M. Fill. 1999. Unitary Ca2+ current through cardiac ryanodine receptor channels under quasi-physiological ionic conditions. J. Gen. Physiol. 113:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukyanenko, V., I. Gyorke, S. Subramanian, A. Smirnov, T. F. Wiesner, and S. Gyorke. 2000. Inhibition of Ca2+ sparks by ruthenium red in permeabilized rat ventricular myocytes. Biophys. J. 79:1273–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soeller, C., and M. B. Cannell. 2002. Estimation of the sarcoplasmic reticulum Ca2+ release flux underlying Ca2+ sparks. Biophys. J. 82:2396–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bridge, J. H., P. R. Ershler, and M. B. Cannell. 1999. Properties of Ca2+ sparks evoked by action potentials in mouse ventricular myocytes. J. Physiol. 518:469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, S.-Q., L.-S. Song, E. G. Lakatta, and H. Cheng. 2001. Ca2+ signalling between single L-type Ca2+ channels and ryanodine receptors in heart cells. Nature. 410:592–596. [DOI] [PubMed] [Google Scholar]
- 43.Wang, S. Q., M. D. Stern, E. Rios, and H. Cheng. 2004. The quantal nature of Ca2+ sparks and in situ operation of the ryanodine receptor array in cardiac cells. Ann. N. Y. Acad. Sci. 101:3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rios, E., and M. D. Stern. 1997. Calcium in close quarters: microdomain feedback in excitation-contraction coupling and other cell biological phenomena. Annu. Rev. Biophys. Biomol. Struct. 26:47–82. [DOI] [PubMed] [Google Scholar]
- 45.Cordeiro, J. M., K. W. Spitzer, W. R. Giles, P. E. Ershler, M. B. Cannell, and J. H. B. Bridge. 2001. Location of the initiation site of calcium transients and sparks in rabbit heart Purkinje cells. J. Physiol. 531:301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sedarat, F., L. Xu, E. D. W. Moore, and G. F. Tibbits. 2000. Colocalization of dihydropyridine and ryanodine receptors in neonate rabbit heart using confocal microscopy. Am. J. Physiol. 279:H202–H209. [DOI] [PubMed] [Google Scholar]
- 47.Junker, J., J. R. Sommer, M. Sar, and G. Meissner. 1994. Extended junctional sarcoplasmic reticulum of avian cardiac muscle contains functional ryanodine receptors. J. Biol. Chem. 269:1627–1634. [PubMed] [Google Scholar]
- 48.Loughrey, C. M., K. E. MacEachern, P. Neary, and G. L. Smith. 2002. The relationship between intracellular [Ca2+] and Ca2+ wave characteristics in permeabilised cardiomyocytes from the rabbit. J. Physiol. 543:859–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, J. G., and B. Russell. 2005. Cardiomyocyte remodeling and sarcomere addition after uniaxial static strain in vitro. J. Histochem. Cytochem. 53:839–844. [DOI] [PubMed] [Google Scholar]