Abstract
Bacteriophages have been used for more than a century for (unconventional) therapy of bacterial infections, for half a century as tools in genetic research, for 2 decades as tools for discovery of specific target-binding proteins, and for nearly a decade as tools for vaccination or as gene delivery vehicles. Here we present a novel application of filamentous bacteriophages (phages) as targeted drug carriers for the eradication of (pathogenic) bacteria. The phages are genetically modified to display a targeting moiety on their surface and are used to deliver a large payload of a cytotoxic drug to the target bacteria. The drug is linked to the phages by means of chemical conjugation through a labile linker subject to controlled release. In the conjugated state, the drug is in fact a prodrug devoid of cytotoxic activity and is activated following its dissociation from the phage at the target site in a temporally and spatially controlled manner. Our model target was Staphylococcus aureus, and the model drug was the antibiotic chloramphenicol. We demonstrated the potential of using filamentous phages as universal drug carriers for targetable cells involved in disease. Our approach replaces the selectivity of the drug itself with target selectivity borne by the targeting moiety, which may allow the reintroduction of nonspecific drugs that have thus far been excluded from antibacterial use (because of toxicity or low selectivity). Reintroduction of such drugs into the arsenal of useful tools may help to combat emerging bacterial antibiotic resistance.
The increasing development of bacterial resistance to traditional antibiotics has reached alarming levels, thus necessitating the development of new antimicrobial approaches. Such new antimicrobials should possess novel modes of action and/or different cellular targets compared with existing antibiotics. The unmet medical need for new antibiotics, coupled with revolutions in genomics, high-throughput screening, and medicinal chemistry, has already spurred pharmaceutical companies and emerging biotechnology companies to search for totally new agents effective in the treatment of bacterial disease caused by resistant organisms (35, 37). As a result, new classes of compounds designed to avoid defined resistance mechanisms are undergoing clinical evaluation (8, 32, 40). Classical short-term approaches include chemical modification of existing agents to improve potency or spectrum. Long-term approaches are relying on bacterial and phage genomics to discover new antibiotics that attack new protein targets that are essential to bacterial survival and to which there is no known resistance. Microbial and phage genomic sequencing is now being used to find previously unidentified genes and their corresponding proteins. Bacterial genes (and proteins) found to be essential for survival can be regarded as potential targets, while toxic phage gene products may be leads for novel antibiotics (21, 23, 36).
In both traditional and newly developed antibiotics, the target selectivity lies in the drug itself, in its ability to affect a mechanism that is unique to the target microorganism and absent in its host. As a result, many potent drugs have been excluded from use as therapeutics because of low selectivity, i.e., toxicity to the host as well as to the pathogen (13). This brings to mind the limited selectivity of anticancer drugs and recent efforts to overcome it by developing targeted therapy strategies. Immunotargeting of cancer cells is being developed since the breakthrough development of monoclonal antibodies (MAbs) (17). Since then, MAbs and derived single-chain antibodies have been used to deliver potent cytotoxic components to cancer cells (18, 41).
As the advantages of targeted therapy became more apparent, additional targeting moieties that are not antibody based, such as short peptides, nonantibody ligand-binding proteins, and even nonproteinaceous molecules such as carbohydrates are receiving increasing attention. Still, targeted therapy is mostly focused on cancer, with limited attention to immune system and autoimmune disorders. Immunoconjugates are bifunctional molecules that consist of a targeting domain that localizes in tumors coupled to a therapeutic moiety. Immunoconjugates, in the broadest sense, may utilize MAbs, MAb fragments, hormones, peptides, or growth factors to selectively localize cytotoxic drugs, plant and bacterial toxins, enzymes, radionuclides, photosensitizers, or cytokines to antigens expressed on tumor cells or on cells of the tumor neovasculature (41, 44). Key issues in designing and testing potential agents for targeted therapy include the nature of the target molecule, its abundance at the target, and its specificity for the target and the linkers used to attach the drug to the targeting moiety. So far, to the best of our knowledge, no attempt at targeted therapy against microbial diseases has been reported. This merger of technologies is the focus of our study.
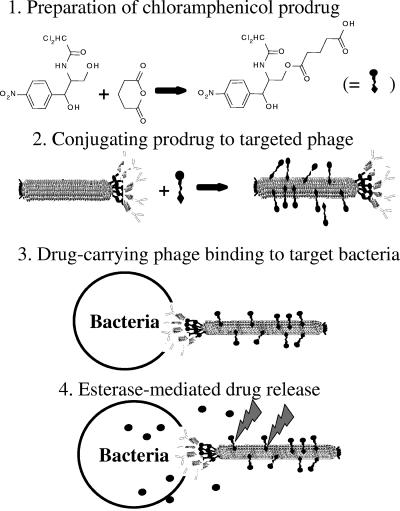
Here we describe our study that demonstrates the feasibility of using filamentous (fd or M13) phage as a targeted drug carrier. Our approach is based on three basic features. The first is amplification of the potency of nonpotent drugs by creating a microenvironment of high concentration around the targeted bacteria. This is facilitated by drug release while the phage carrier is tethered to the target cell. The second is reduction of general toxicity since the drug is inactive as long as it is linked to the phage. This leads to the third feature—reintroduction of nonselective toxic substances into active duty as drug candidates in the battle against drug-resistant bacteria. We demonstrate these principles by using the bacteriostatic nonpotent antibiotic chloramphenicol (which has a known toxicity to blood cells [42]) as a model drug. Targeting is demonstrated by using two different targeting moieties: (i) displayed target-specific peptides that are displayed on the major coat protein of the phage and (ii) immunoglobulin G (IgG) antibodies linked to the phages via an IgG-binding ZZ domain that is displayed on the minor coat protein of the phage. As a model target, we chose Staphylococcus aureus strain SH1000, a gram-positive pathogenic bacterium that causes a wide variety of human diseases ranging from superficial abscesses and wound infections to systemic infections (12, 22). The scheme of antibacterially targeted drug-carrying bacteriophages is illustrated in Fig. 1.
FIG. 1.
Schematic representation of antibacterial targeted drug-carrying bacteriophages.
MATERIALS AND METHODS
All of the chemicals used were of analytical grade and were purchased from Sigma (Israel). Unless stated otherwise, reactions were carried out at room temperature (about 25°C).
General analytic methods for preparation and evaluation of chloramphenicol prodrug.
Thin-layer chromatography (TLC) was performed with silica gel plates (Merck 60 F254); compounds were visualized by irradiation with UV light. Flash chromatography was performed with silica gel (particle size, 0.040 to 0.063 mm; Merck 60). 1H nuclear magnetic resonance (NMR) analysis was performed with a Bruker AMX 200. Chemical shifts are expressed in δ relative to tetramethylsilane (δ = 0 ppm), and the coupling constant, J, is expressed in Hertz. The spectra were recorded in methanol-d as a solvent at room temperature.
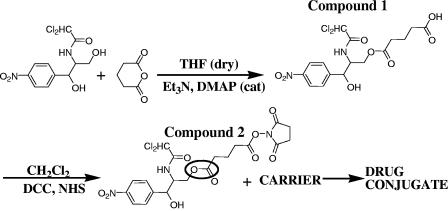
Two chemical steps were used to modify chloramphenicol, compounds 1 and 2. Compound 1 was prepared with 2 g (6.2 mmol) of chloramphenicol (molecular weight, 323.13; catalog no. C0378; Sigma) dissolved in dry tetrahydrofurane; glutaric anhydride (800 mg, 6.82 mmol), triethylamine (1.0 ml, 6.82 mmol), and a catalytic amount of dimethylaminopyridine were then added. The reaction mixture was stirred at room temperature overnight and monitored by TLC (ethyl acetate [EtOAc]-hexane ratio, 9:1). After completion, the reaction mixture was diluted with EtOAc and washed with a 1 N solution of HCl. The organic layer was dried over magnesium sulfate, and the solvent was removed under reduced pressure. The crude product was purified by column chromatography on silica gel (EtOAc-hexane ratio, 4:1) to give compound 1 (2.2 g, 81%) (Fig. 2, top) in the form of a colorless viscous oil. We named compound 1 a chloramphenicol-linker adduct. The results of the NMR analysis of compound 1 were as follows: 1H NMR (200 MHz, CD3OD) δ = 8.17 (2H, d, J = 8), 7.65 (2H, d, J = 8), 6.22 (1H, s), 5.08 (1H, d, J = 2), 4.44-4.41 (2H, m), 4.24 (1H, d, J = 2), 2.40-2.32 (4H, m), 1.92 (2H, t, J = 7).
FIG. 2.
Synthesis and evaluation of chloramphenicol prodrug for conjugation to amine groups. Two chemical steps were used to modify chloramphenicol. In the first step, the chloramphenicol primary OH group was reacted with glutaric anhydride to create an ester linkage, resulting in compound 1. In the second step, the free carboxyl group of compound 1 was activated with NHS to allow subsequent linkage to amine groups such as on lysines. At this stage, the chloramphenicol prodrug compound 2 is not toxic to bacteria and is ready for conjugation to proteins. The labile ester bond is marked by a circle. Et3N, triethylamine; DMAP, dimethylaminopyridine; DCC, N,N′-dicyclohexylcarbodiimide; cat, catalyst.
Compound 1 (2 g, 4.57 mmol) was dissolved in dichloromethane, and N,N′-dicyclohexylcarbodiimide (1.4 g, 6.86 mmol), and N-hydroxysuccinimide (NHS) (790 mg, 6.86 mmol) were added. The reaction mixture was stirred at room temperature and monitored by TLC (EtOAc-hexane ratio, 9:1). After completion of the reaction, the mixture was filtered out and the solvent was removed under reduced pressure. The crude product was purified by column chromatography on silica gel (EtOAc-hexane ratio, 4:1) to give compound 2 (Fig. 2, bottom) in the form of a white solid (1.5 g, 62%). We named compound 2 a chloramphenicol prodrug which is not toxic to bacteria and is ready for conjugation to proteins. The results of the NMR analysis of compound 2 were as follows: 1H NMR (200MHz, CD3OD) δ = 8.17 (2H, d, J = 8), 7.65 (2H, d, J = 8), 6.22 (1H, s), 5.08 (1H, d, J = 2), 4.44-4.41 (2H, m), 4.24 (1H, d, J = 2), 3.02 (4H, s), 2.91 (2H, t, J = 7), 2.68 (2H, t, J = 7), 2.20 (2H, t, J = 7), 1.43 (1H, t, J = 7).
Preparation of phages for drug conjugation.
Filamentous phages were routinely propagated in DH5-α/F′ cells by standard phage techniques as previously described (10). Phages were usually recovered from 1-liter overnight cultures of phage-carrying bacteria. The bacteria were removed by centrifugation, and the phage-containing supernatant was filtered through a 0.22-μm-pore-size filter. The phages were precipitated by addition of 20% (wt/vol) polyethylene glycol 8000 (PEG)-2.5 M NaCl, followed by centrifugation as previously described (5). The phage pellet was suspended in sterile miliQ double-distilled water at a concentration of 1013 CFU/ml and stored at 4°C.
Conjugation of chloramphenicol prodrug to phages.
A stock phage solution of 1013 CFU/ml and a stock chloramphenicol prodrug solution of 100 μM were used in all conjugations. A constant molar ratio of 3 × 105 chloramphenicol prodrug molecules/phage was used. The reaction mixture was mixed for 1 h. Next, phage precipitates were removed by centrifugation for 15 min at 14,000 rpm and 4°C in a Microfuge. Soluble phages were precipitated with PEG-NaCl as described above.
Quantifying the number of linked chloramphenicol molecules per phage.
Similar numbers of chloramphenicol-conjugated phages and unconjugated phages as a reference were incubated for 48 h in the presence of 10% rabbit serum (Sigma) at 37°C. The phage solutions were ultrafiltered with a 10-kDa cutoff ultrafiltration microcon cartridge (Millipore) to eliminate the phages, as well as large proteins and other contaminants. The filtrate optical absorbance was recorded at 280 nm, and the number of released chloramphenicol molecules was calculated by using a calibration curve of free chloramphenicol absorbance that was recorded at 280 nm.
Quantifying drug release from linker in serum.
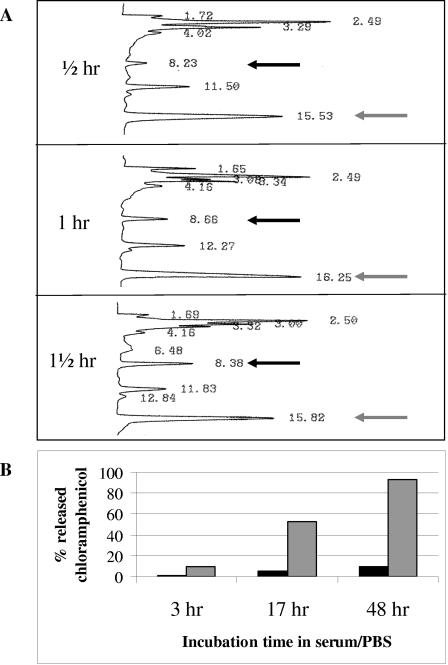
Reverse-phase high-performance liquid chromatography (HPLC) was used to measure the chloramphenicol release rate following the incubation of chloramphenicol prodrug in 10% or 99% horse serum or without serum, all in phosphate-buffered saline (PBS). A reverse-phase C18 column was used on a LabChrom L7400 Merck Hitachi machine with acetonitrile-water (30:70 ratio) in the mobile phase at a 1-ml/min flow rate. Under these conditions, free chloramphenicol is eluted 8 to 9 min after sample loading while the intact chloramphenicol prodrug is eluted 16 min after sample loading (Fig. 3). The percentage of released chloramphenicol was calculated by dividing the area of the 9-min peak by the total area of all relevant peaks and multiplying by 100.
FIG. 3.
Rate of chloramphenicol release from its linker in horse serum. (A) Reverse-phase HPLC was used to measure the rate of chloramphenicol breakdown following incubation for the indicated time periods in 99% horse serum. A reverse-phase C18 column was used on a LabChrom L7400 Merck Hitachi machine with acetonitrile-water (30:70 ratio) in the mobile phase at a 1-ml/min flow rate. Free chloramphenicol (black arrows) is eluted 8 to 9 min after sample loading, while the intact chloramphenicol prodrug (gray arrows) is eluted 16 min after sample loading. (B) Rate of chloramphenicol release from its linker in 10% horse serum (gray bars) or without serum (in PBS, black bars) was evaluated by HPLC as described for panel A. The percentage of chloramphenicol released was calculated by dividing the area of the 9-min peak by the total area of all relevant peaks.
Affinity selection of a peptide phage display library.
We used a random 12-mer peptide combinatorial phage display library based on the fth1 type 88 expression vector (11) that contains >109 random clones. In the library we used, the peptides are displayed on the N terminus of major coat protein p8 of the fd filamentous bacteriophage. Use of p8 for display of peptides typically requires the expression of two p8 genes, the wild-type p8 gene and a recombinant p8 gene (a gene fusion with the sequence coding for the displayed peptide), to generate mosaic phages. Such type 88 phage vectors contain two p8 genes in one phage genome (11, 38). Escherichia coli K91K cells (39) were used for phage propagation. The library phages were affinity selected on S. aureus cells as follows. S. aureus cells were grown in tryptic soy broth (TSB; Difco) medium at 37°C. The cells were collected by centrifugation and washed twice with 1 ml of chilled PBS containing 0.05% (vol/vol) Tween 20 (PBST). Washed cells (109) were resuspended in 1 ml of 1% (vol/vol) nonfat milk in PBS containing 1011 CFU of peptide library phages and left on ice for 1 h. The cells were then washed three times (1 ml per wash) with PBST and three times with PBS (5). The selecting S. aureus cells were then mixed with 109 E. coli K91K cells in 1 ml that were grown in 2×YT medium (5) supplemented with 50 μg/ml kanamycin with vigorous shaking at 37°C. The mixed culture was incubated for 30 min at 37°C without shaking and then incubated for 30 min at 37°C with gentle shaking that enables the selected phages to infect the E. coli K91K bacteria. Aliquots were taken for determination of phage output, and the rest of the culture was either plated on 2×YT plates (5) containing 20 μg/ml tetracycline and 50 μg/ml kanamycin to obtain single colonies for immunoscreening or amplified to obtain a phage stock to be used as input for the next affinity selection cycle.
After the fourth affinity selection cycle, randomly selected single colonies of K91K cells infected with output phage were picked and used to inoculate 200 μl of 2×YT medium supplemented with 20 μg/ml tetracycline per well in U-bottom 96-well plates (Nunc). After overnight growth at 37°C with shaking at 150 rpm, the plates were centrifuged and the phage-containing supernatant from each well was mixed at a 1:1 ratio with PBST and tested for binding to S. aureus cells by phage enzyme-linked immunosorbent assay (ELISA).
Phage ELISA.
S. aureus cells were used to coat wells of 96-well polystyrene ELISA plates (Costar) as described below. Serial dilutions of the tested phages were added to the wells and allowed to bind for 1 h. When antibody-mediated binding of fUSE5-ZZ phages to S. aureus was evaluated, incubation of the phages was preceded by incubation of the cells with immune anti-S. aureus serum diluted 1,000× with PBST, followed by three washes with PBST. Following the 1-h incubation with phages, the plates were washed three times with PBST, horseradish peroxidase (HRP)-conjugated rabbit anti-M13 antibody (Amersham Biosciences) diluted 5,000 × in PBST was added (100 μl/well), and the mixture was incubated for 1 h. The plates were washed three times with PBST, and development was done with the HRP substrate 3,3′,5,5′-tetramethylbenzidine (TMB; Dako) at 100 μl/well. Following color development, the reaction was terminated with 50 μl/well 1 M H2SO4. The color signal was recorded at 450 nm.
ELISA plates were coated with bacteria as follows. Cells from a fresh overnight culture were collected by centrifugation and suspended in PBS at about 108/ml. An aliquot of 100 μl of the cell suspension was applied to each well of the plate, which was spun in a centrifuge at 4,000 rpm for 5 min at 4°C. The supernatant was carefully removed, and 100 μl of 3% glutaraldehyde in PBS was added to each well and left to fix the cells for 1 h. Next, the plate was blocked with 3% (wt/vol) skim milk powder in PBS and 5% rabbit serum (a crucial step for blocking the IgG binding of S. aureus because of cell surface protein A). E. coli strain O78 cells were used as a negative control.
Generation of ZZ domain-displaying phages.
The IgG-binding ZZ domain was initially cloned for monovalent display on the filamentous phage p3 coat protein as follows. The DNA fragment carrying the ZZ domain open reading frame was recovered by PCR with plasmid pSD-ZZ (14) as the template and primers CCGCTTCCATGGTAGACAACAAATTCAACAAAG and GGGTTTAGCGGCCGCTTTCGGCGCCTGAGCATCATTTAG. The PCR product was digested with restriction enzymes NcoI and NotI (New England Biolabs) (restriction sites are underlined in the primer sequences) and cloned into a vector fragment that was isolated from phagemid vector pCANTAB5E (6) by digestion with the same enzymes. The resulting plasmid was named pCANTAB-ZZ.
The IgG-binding ZZ domain was further cloned for polyvalent display on all copies of the filamentous phage p3 coat protein as follows. Initially, phage vector fUSE5 (http://www.biosci.missouri.edu/smithgp/PhageDisplayWebsite/vectors.doc) was modified to accept single-chain antibodies as NcoI-NotI fragments. The anti-Tac scFv coding DNA was recovered by PCR with pCANTAB5E-anti-Tac (3) as the template and primers AATTTCGGCCGACGTGGCCATGGCCCAGGTCAAACT and TATTCACAAACGAATGGATCC. The PCR product was digested with restriction enzymes SfiI and BamHI (New England Biolabs) (restriction sites are underlined in the primer sequences) and cloned into a vector fragment that was isolated from fUSE5 DNA by digestion with the same enzymes. The resulting phage vector was named fUSE5-anti-Tac. The ZZ domain was subcloned into the fUSE5 backbone by replacing the NcoI and NotI scFv fragment of fUSE5-anti-Tac with the NcoI and NotI scFv ZZ domain fragment of pCANTAB-ZZ, resulting in phage fUSE5-ZZ.
Evaluation of ZZ domain display on phages by immunoblotting.
To compare monovalent to polyvalent display of the ZZ domain, an immunoblot analysis was carried out as follows. Phage particles (1011) were separated by electrophoresis on a 12% sodium dodecyl sulfate-polyacrylamide gel. The gel was electroblotted onto a nitrocellulose membrane which was developed with an anti-p3 MAb (laboratory collection), followed by an HRP-conjugated rabbit anti-mouse antibody (Jackson ImmunoResearch Laboratories). The membrane was developed by using enhanced-chemiluminescence reagents as previously described (7).
Evaluation of the IgG-binding capacity of ZZ domain-displaying phages after drug conjugation.
IgG-binding capacity was evaluated by incubating 1012 CFU of fUSE5-ZZ phages before or following conjugation to the chloramphenicol prodrug with a 10,000 × dilution of HRP-conjugated rabbit anti-mouse IgG (Jackson ImmunoResearch Laboratories) in 1 ml of PBS for 1 h. Next, phages and phage-IgG complexes were precipitated with PEG-NaCl as described above. The pellets were suspended in 1 ml of PBS, and threefold dilutions were made in an ELISA plate containing 100 μl of PBS in each well prior to addition of phages. Development was done by addition of 50 μl of the HRP substrate TMB (Dako). Following color development, the reaction was terminated with 50 μl/well 1 M H2SO4. The color signal was recorded at 450 nm.
The possibility of protecting the ZZ domain from conjugation-induced deterioration was evaluated as follows. fUSE5-ZZ was protected by complexation with human protein A-purified IgG (Jackson Immunoresearch Laboratories) prior to chloramphenicol conjugation. A total of 1013 fUSE5-ZZ phages in 1 ml of PBS were complexed with 15 μg of human IgG for 1 h. Complexation was followed by conjugation with the chloramphenicol prodrug as described above. Precipitates were removed by centrifugation for 15 min at 21,000 × g at 4°C in a Microfuge. Next, the protecting human IgG was released from the ZZ domain by lowering the pH to 2.5 by adding 500 μl of 50 mM glycine-HCl, pH 2.2 (resulting in a pH of ∼2.5), and incubating the phages for 20 min. PEG-NaCl was used to precipitate and separate the fUSE5-ZZ phages from the free human IgG. IgG-binding capacity was evaluated by incubating the phages with HRP-conjugated rabbit anti-mouse IgG as described above.
Evaluation of the antigen-binding capacity of peptide-displaying phages after drug conjugation.
The ability of peptide-displaying phages to bind immobilized S. aureus cells before and following conjugation to the chloramphenicol prodrug was evaluated by phage ELISA as described above. Serial dilutions of phages were made starting at 1011 phages/well.
Evaluation of the effect of drug-carrying-peptide-displaying phages on the growth of S. aureus.
An overnight S. aureus culture was diluted 100 × in 1 ml of PBS. The diluted bacteria were collected by centrifugation at 21,000 × g, and the pellet was resuspended in 100 μl of PBS. A 10-μl volume of this bacterial suspension was mixed with 100 μl of PBS containing 1011 phages and then incubated for 1 h.
The incubation of the cells with the phages was followed by incubation with 50% rabbit serum for 1 h, after which they were diluted directly into 2 ml of TSB medium-20% rabbit serum in 13-ml tubes shaking at 250 rpm at 37°C. Growth was recorded after dilution into the TSB medium by recording the optical density (OD) at 600 nm.
Evaluation of the effect of antibody-targeted, drug-carrying-ZZ-displaying phages on the growth of S. aureus.
An overnight bacterial culture was diluted 100 × into 1 ml of PBS. Diluted bacteria were incubated with human polyclonal anti-S. aureus IgG (laboratory collection) for 1 h at room temperature. Next, the cells were collected by centrifugation and resuspended in 100 μl of PBS. A 10-μl volume of this bacterial stock was mixed with 1012 CFU of fUSE5-ZZ phages in 1 ml of PBS and then incubated for 1 h. The tested phages were chloramphenicol-conjugated fUSE5-ZZ phages, chloramphenicol-conjugated fUSE5-ZZ phages that were blocked by incubation with normal (nonimmune) human IgG (Jackson ImmunoResearch Laboratories) as a negative control, and fUSE5-ZZ phages without drug as an additional negative control. Cells were incubated with the phages for 1 h, followed by incubation with 50% rabbit serum for 1 h, after which they were diluted directly into 2 ml of TSB medium-20% rabbit serum in 13-ml tubes shaking at 250 rpm at 37°C. Growth was recorded after dilution into TSB medium by recording the OD at 600 nm.
RESULTS
Preparation and evaluation of chloramphenicol prodrug.
Our initial choice for a model antibiotic was chloramphenicol, a bacteriostatic hydrophobic drug that is not routinely used clinically because of toxicity problems. It can be easily conjugated and monitored. A chloramphenicol prodrug, where chloramphenicol is linked through a labile ester bond, was prepared as described in Materials and Methods (compound 2, Fig. 2). The synthesis intermediate and final products were evaluated for purity by HPLC and NMR. The chloramphenicol prodrug did not inhibit the growth of susceptible bacteria at concentrations of up to 100 times the 50% inhibitory concentration of free chloramphenicol, showing that, indeed, the chloramphenicol was converted to a prodrug (not shown).
The rate of release of chloramphenicol from the linker was evaluated with serum as a source of esterases (16, 20) that cleave the ester bond between chloramphenicol and the linker (circled in Fig. 2). To test the chloramphenicol release rate, a reverse-phase HPLC analysis was done based on the elution time of free chloramphenicol 8 to 9 min postinjection while the chloramphenicol prodrug elutes 16 min post sample injection into the reverse-phase C18 column. The amount of chloramphenicol released was calculated from the peak areas. As shown in Fig. 3A, about 15% of the chloramphenicol was released from the linker after 1 h of incubation in 99% horse serum at 37°C with linear kinetics. A similar release rate was found after the chloramphenicol prodrug was conjugated to phages, followed by release with serum (not shown). The stability of the chloramphenicol prodrug was evaluated by a more prolonged incubation, where the release rate was monitored over 48 h in the presence of 10% horse serum or without serum. As shown in Fig. 3B, the release rate followed the same linear kinetics as with the 99% serum (after correction for the concentration difference). The spontaneous hydrolysis rate of the ester bond in PBS alone was less than 5% over the entire 48-h period.
Conjugation of chloramphenicol prodrug to filamentous phages.
The chloramphenicol prodrug was conjugated to phages, and phage-drug conjugates were recovered by two successive PEG-NaCl precipitations as described in Materials and Methods. To evaluate the number of chloramphenicol molecules that were conjugated to a single phage, we liberated the drug by prolonged incubation with serum and separated the drug from the phages by ultrafiltration. The concentration of the liberated chloramphenicol was calculated by comparing its optical absorbance at 280 nm to a calibration curve. We found (Table 1) that several consecutive conjugation steps were required to achieve maximal coverage of the phages with the drug, at >20,000 molecules/phage. However, at this conjugation level, most of the phages were lost because of precipitation (Table 1). Therefore, we routinely performed a single conjugation cycle, resulting in 2,000 to 4,000 drug molecules/phage.
TABLE 1.
Number of chloramphenicol molecules conjugated/phage in consecutive conjugation cycles and effect of conjugation on phage loss due to precipitation
| Conjugation cycle | No. of chloramphenicol molecules/phage | % of phage lost because of precipitation |
|---|---|---|
| 1 | 3,600 | <5 |
| 2 | 17,000 | ∼60 |
| 3 | 24,000 | ∼80 |
Isolation of S. aureus-binding phages.
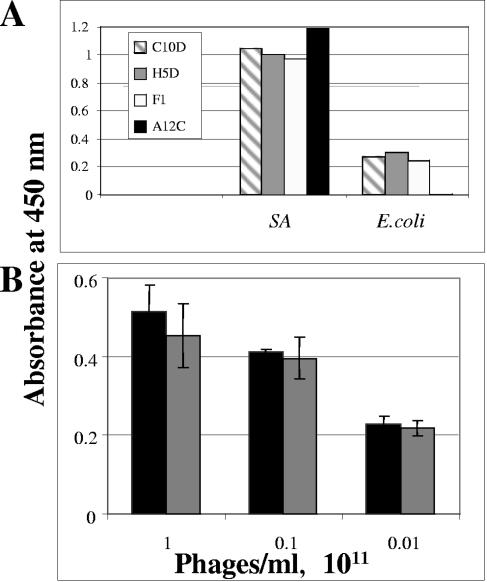
A phage peptide display library of about 109 clones of disulfide bond-constrained 12-mer random peptides was affinity selected on live S. aureus cells as described in Materials and Methods. Following four selection cycles, random clones were screened for binding to immobilized S. aureus cells by phage ELISA. The binding signals of four such clones are shown in Fig. 4A, and the deduced amino acid sequences of the peptides they display are shown in Table 2. As shown, higher ELISA signals were obtained on the S. aureus target cells in comparison with the control E. coli cells. As frequently happens following several affinity selection cycles, some of the clones were isolated more than once (Table 2). Phage A12C, which yielded the highest binding signal difference between the target and control cells and which displays a lysine-free peptide (and should not be harmed by the drug conjugation), was chosen for further evaluation as a targeted drug carrier. A12C phages were conjugated to the chloramphenicol prodrug, and their capacity to bind S. aureus was compared to that of unconjugated phages. As shown in Fig. 4B, the S. aureus-binding capacity of the phages was not affected by drug conjugation.
FIG. 4.
Binding of peptide-displaying phages to S. aureus. (A) Affinity-selected peptide-displaying phages (1010/well, names shown in inset) were applied to wells precoated with ∼107 S. aureus cells. After extensive washing, bound phages were detected with HRP-conjugated anti-M13 antibodies, followed by development with the HRP substrate TMB. OD was recorded at 450 nm. (B) Effect of conjugation of the chloramphenicol prodrug to A12C phages on their S. aureus-binding capacity evaluated in a phage ELISA as described for panel A, comparing the signal intensities due to the binding of various concentrations of untreated phages (black bars) to those of equal concentrations of drug-carrying phages (gray bars). Error bars represent the standard deviation of the data. SA, S. aureus.
TABLE 2.
S. aureus-specific phagesa
| Clone | No. of duplicates identified | Sequence | OD on S. aureus cells | OD on E. coli cells |
|---|---|---|---|---|
| C10D | 3 | SPGHYWDTKLVD | 1.05 | 0.275 |
| H5D | 4 | TYFPTMGTSFKI | 1.003 | 0.3 |
| F1 | 2 | TFLRGPSSPLVS | 0.97 | 0.24 |
| A12C | 1 | VHMVAGPGREPT | 1.2 | 0.007 |
Shown are the sequences of the peptides displayed on S. aureus-specific peptide-displaying phages and the number of duplicates that were identified in the initial screen by phage ELISA. Lysines that may be vulnerable to amine conjugation chemistry are in bold. The OD values are from Fig. 3A.
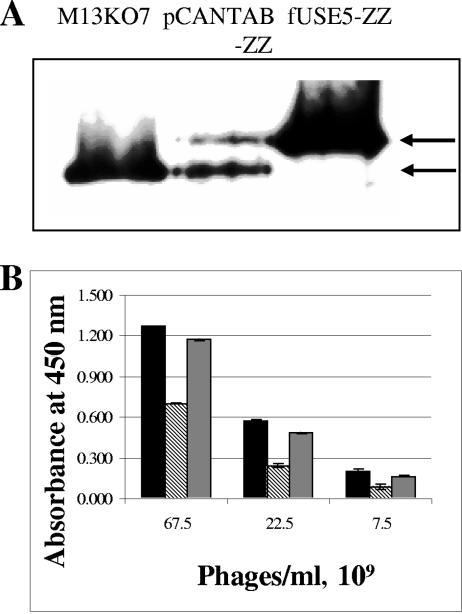
Phage fUSE5-ZZ, allowing polyvalent display of the ZZ domain on all copies of the p3 minor coat protein, was constructed as described in Materials and Methods. To compare monovalent to polyvalent display of the ZZ domain, an immunoblot analysis was carried out as described in Materials and Methods. In the immunoblot, we compared the p3 protein of the M13KO7 helper phage (Pharmacia) (where only the wild-type p3 protein is incorporated in the phage coat), that of pCANTAB-ZZ following its rescue by the M13KO7 helper phage (where both the ZZ-fused p3 protein and the wild-type p3 protein provided by the helper are incorporated in the phage coat), and fUSE5-ZZ (expected to have only ZZ-fused p3 incorporated in the phage coat). As shown in Fig. 5A, each phage had the expected pattern of p3 in its coat, verifying polyvalent display of ZZ by the fUSE5-ZZ phages.
FIG. 5.
Evaluation of ZZ domain display on phage and effect of drug conjugation on the ZZ domain IgG-binding capacity. (A) Evaluation of ZZ domain display by an immunoblot assay. Phage particles (each lane is identified with the corresponding phage name above it) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electroblotted onto nitrocellulose, and the p3 minor coat protein or the derived ZZ domain-p3 fused derivative was detected with an anti-p3 antibody. The upper arrow marks the position of the ZZ-p3 fusion, while the lower arrow marks the position of the wild-type p3 coat protein. (B) Comparison of the binding capacity of the phage-displayed ZZ domain to HRP-conjugated rabbit IgG that yields a color signal with the substrate TMB at 450 nm. fUSE5-ZZ phages were evaluated before conjugation (black bars), following conjugation to the chloramphenicol prodrug (striped bars), or following conjugation to the chloramphenicol prodrug while protected by complexed IgG (gray bars). Error bars represent the standard deviation of the data.
The phages were conjugated to the chloramphenicol prodrug, and their capacity to bind IgG was evaluated. As shown in Fig. 5B, at this conjugation level, the ZZ domain lost about 50% of its IgG-binding capacity. However, protection of the ZZ domain could be achieved by complexing with human IgG prior to drug conjugation.
Effect of targeted drug-carrying phages on the growth of S. aureus.
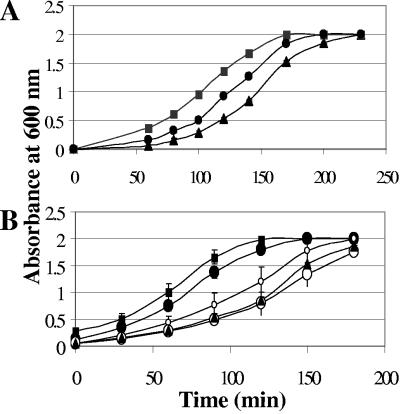
Phage A12C was chosen to assess the targeting effect of chloramphenicol-conjugated S. aureus-specific peptide-displaying phages. The phages were conjugated as described above and evaluated for their effects on bacterial growth. As shown in Fig. 6A, the growth of the S. aureus bacteria was retarded following treatment with the chloramphenicol-conjugated S. aureus-targeted phages in comparison with S. aureus treated with targeted phages that do not carry the drug. The control phages that display an irrelevant peptide also retarded bacterial growth but to a lesser extent. The equivalent concentration of free chloramphenicol did not retard bacterial growth. In Fig. 6B is shown a comparison of the growth curves that were obtained in the presence of various concentrations of free chloramphenicol to the growth of S. aureus in the presence of targeted drug-carrying A12C phages. Under these experimental conditions, the targeted A12C phages release the equivalent of 0.096 μg/ml chloramphenicol. As shown, the phages retarded S. aureus growth as efficiently as a 20× higher concentration of free chloramphenicol.
FIG. 6.
Effect of drug-carrying peptide-displaying phages on the growth of S. aureus. (A) Growth curves of S. aureus cells treated with S. aureus-specific chloramphenicol prodrug-conjugated phage A12C (triangles), chloramphenicol prodrug-conjugated irrelevant phage F2b (circles), and unconjugated S. aureus-specific phage A12C as a negative control (squares). (B) Growth curves of S. aureus cells treated with S. aureus-specific chloramphenicol prodrug-conjugated phage A12C (triangles) or with free chloramphenicol at 0.533 μg/ml (large filled circles), 1.0625 μg/ml (small open circles), or 2.125 μg/ml (large open circles) and cells grown without any growth inhibitor (filled squares). Error bars represent the standard deviation of the data.
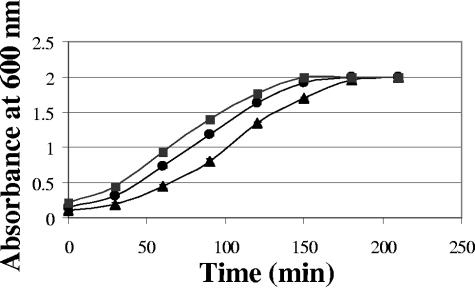
When fUSE5-ZZ was evaluated, all of the binding-targeting studies with S. aureus were done following blocking of the bacterium's own cell surface protein A with 20% rabbit serum prior to addition of the specific antibodies. The fUSE5-ZZ phages were conjugated to the chloramphenicol prodrug and evaluated for targeted drug delivery. As shown in Fig. 7, chloramphenicol-carrying fUSE5-ZZ phages retarded the growth of the target S. aureus bacteria after they were incubated with S. aureus-specific human antiserum (bound through the displayed ZZ domain) and to a lesser extent when the phages were complexed with a nonimmune human IgG that does not bind the serum-blocked S. aureus bacteria. In these experiments, targeted fUSE5-ZZ retarded S. aureus growth as effectively as a 10 × higher concentration of free chloramphenicol.
FIG. 7.
Effect of drug-carrying, antibody-targeted ZZ-displaying phages on the growth of S. aureus. Growth curves of S. aureus cells treated with chloramphenicol prodrug-conjugated phage fUSE5-ZZ targeted by anti-S. aureus antibodies (triangles) or chloramphenicol prodrug-conjugated phage fUSE5-ZZ blocked by control IgG (circles) and cells grown without any growth inhibitor (squares). Error bars represent the standard deviation of the data.
DISCUSSION
The objective of the experiments described herein was a feasibility study of applying targeted drug delivery as an antibacterial tool. The system was designed with three key components: (i) a targeting moiety, exemplified here by S. aureus-specific peptides and by immune anti-S. aureus serum complexed via the ZZ domain; (ii) a high-capacity drug carrier, exemplified here by the filamentous phage, with its ∼3,000 copies of the p8 major coat protein, each amendable to drug conjugation; and (iii) a drug linked through a labile linker that is subject to controlled release, exemplified here by chloramphenicol, which was linked through an ester bond to lysine residues on the phage coat. We chose the filamentous phage as our drug-carrying platform since it is the workhorse of display technology (2), thus readily providing the targeting arm. Other phages (such as λ, T7, and T4) have also been used for display of peptides or other binding specificity-determining proteins (albeit much less frequently than filamentous phage) (2). However, those other phages have a much smaller number of coat protein monomers than the filamentous phage (26, 34, 45); thus, their potential drug-carrying capacity is expected to be considerably smaller. Still, our study provides a model system that could be implemented with other targeted phages or other viruses.
Chloramphenicol was chosen as the initial model drug for several reasons. The first was the simplicity with which chloramphenicol can be conjugated to a linker through an ester bond. Chloramphenicol has two hydroxyl groups and only one of them is primary, and the drug is easily dissolved in most organic solvents. An additional consideration was to use a relatively nonselective drug that has serious side effect and was thus mostly excluded from systemic use. Chloramphenicol is a good example of such drug, as it is known for its hemolytic effect resulting in anemia.
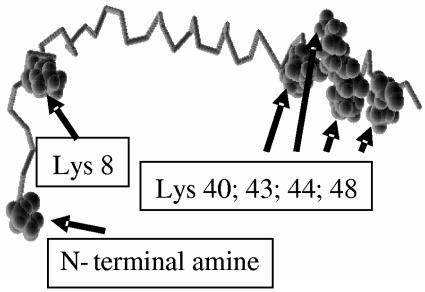
Several features of chloramphenicol were not anticipated and made its use particularly challenging. One was that, because of its high hydrophobicity, its conjugation at high density to a biological molecule would lead to a serious reduction in solubility or complete precipitation in an aqueous solution. Indeed, in the case of our phages, conjugation of more then ∼3,000 chloramphenicol molecules (out of a potential drug-carrying capacity of ∼20,000 molecules/phage, based on the total number of lysines on all copies of the p8 coat protein) led to irreversible precipitation of the phages. As shown in Fig. 8, a monomer of the p8 coat protein has two exposed amine groups (the N-terminal lysine and lysine 8) and four C-terminal lysines that are buried within the phage coat and involved in interaction with the negatively charged phage single-stranded DNA genome (15). We found that high-density conjugation of p-nitrophenyl-activated alkyne compounds to phage amines led to rapid and massive deterioration of the phage structure (data not shown). We believe that high-density conjugation of hydrophobic moieties via the coat amines attacks the buried lysines, leading to phage destabilization, followed by precipitation.
FIG. 8.
Model of the phage p8 coat protein monomer. A structural model of a monomer of filamentous phage p8 major coat protein is shown with all of the positions of amine-containing side chains marked by arrows. The coat of each phage particle is composed of some 3,000 copies of p8. Based on Protein Data Bank file 2CPS.
Second, the bacteriostatic properties and low potency of chloramphenicol, combined with a limited conjugation capacity, resulted in only partial retardation of bacterial growth rather than annihilation.
We chose a simple drug conjugation chemistry (Fig. 2) that was made possible by the fact that chloramphenicol has a single reactive primary hydroxyl group. An ester linkage was made by reacting chloramphenicol with glutaric anhydride, leading to the generation of an ester bond between the chloramphenicol primary hydroxyl and the newly exposed carboxylic end of glutaric anhydride. At the second synthesis step, the second carboxyl end of the linker was activated with a good leaving group, NHS ester, which undergoes spontaneous hydrolysis in aqueous solutions. The final product, chloramphenicol-linker-NHS (chloramphenicol prodrug, compound 2, Fig. 2), and the median product, chloramphenicol-linker adduct (compound 1, Fig. 2), were evaluated by HPLC and NMR analysis. This chloramphenicol prodrug provided for a rapid, simple conjugation process between the chloramphenicol prodrug and the phages that is completed within 1 h.
The rapid rate of spontaneous NHS hydrolysis made it necessary to apply three successive conjugations to the phages to saturate its binding capacity, at >20,000 chloramphenicol prodrug molecules/phage (Table 1). With regard to conjugation to phage lysines, a carrying capacity in excess of 104 biotin molecules/phage was previously reported in phage biotinylation studies by NHS chemistry (29, 30). The theoretical carrying capacity of the phage is based on the number of free exposed amine residues of the phage p8 major coat protein (Fig. 8). In our case, however, we had to proceed with a lower level of conjugation (about 3,000 molecules/phage) to prevent phage loss because of precipitation that is probably caused by the high density of the hydrophobic molecule on the phage coat. From these experiments and others where we conjugated other hydrophobic molecules to the phage by NHS chemistry, we can conclude that amine conjugation of hydrophobic molecules does not allow full arming of phages with drug. This limitation may be overcome by modifying the conjugation chemistry to include solubility-enhancing modules in the linker attached to the drug (our unpublished data).
By HPLC analysis, we found that the rate of release of chloramphenicol from the linker with serum as a source of esterases was about 15% of the conjugated chloramphenicol released after 1 h of incubation in 99% horse serum at 37°C with linear kinetics (Fig. 3A). A similar release rate was found after the chloramphenicol prodrug was conjugated to phages, followed by release with serum (not shown). No significant spontaneous drug release occurred during short incubation periods (Fig. 3B). Such slow kinetics of drug release should allow the targeted drug-carrying phages to localize to the target site prior to efficient release of the drug, a form of delayed release. We chose to evaluate two targeting modalities that should allow drug-carrying phages to bind the S. aureus target bacteria. Peptide-displaying phages provide for polyvalent display of target-specific peptides, which individually bind with low affinity but when densely displayed on the phage coat enjoy the benefit of avidity. The 88 display format we used displays several hundred copies of the peptide on the phage coat (38). By affinity selecting a phage library on live S. aureus cells, we isolated several specific S. aureus binders (Fig. 4A). We chose to proceed with phage A12C, which yielded the highest binding signal difference between the target and control cells and which displays a lysine-free peptide and was not supposed to be harmed by the drug conjugation. Indeed, following conjugation of the chloramphenicol prodrug to A12C phages, their S. aureus-binding capacity was fully preserved (Fig. 4B).
The effect of targeted drug-carrying phages on the growth of S. aureus bacteria was evaluated by mixing drug-carrying phages with bacteria, applying serum for drug release, and diluting the bacteria in growth medium for monitoring of their growth rate at 37°C. We chose a concentration of drug-carrying phages such that, upon drug release, the concentration of free chloramphenicol will be below that required for untargeted growth retardation. As shown in Fig. 6A, the growth of the S. aureus bacteria was retarded following treatment with the chloramphenicol prodrug-conjugated S. aureus-targeted phages in comparison with S. aureus treated with targeted phages that do not carry the drug. The control phages that display an irrelevant peptide also retarded S. aureus growth, but to a lesser extent. This may have resulted from nonspecific binding of phages to bacteria. Another possible explanation is that even a minimal interaction of the presented control (seen as negligible background in the phage ELISA) could perhaps deliver sufficient drug to the bacteria.
The major finding of our study is the comparison we made of phage-mediated growth retardation with that mediated by free chloramphenicol. As shown in Fig. 6B, under our experimental conditions, the targeted A12C phages (which release the equivalent of 0.096 μg/ml chloramphenicol) retarded S. aureus growth as efficiently as a 20 × higher concentration of free chloramphenicol. These results demonstrate a targeting effect that results from the specific binding of the phages via the displayed peptide to the target bacteria and the release of the antibiotic in their vicinity.
The second targeting modality we evaluated was antibody-mediated targeting provided by the fUSE5-ZZ phages, which avidly bind immunoglobulins. The ZZ domain, a tandem repeated, mutated domain B derived from S. aureus (31), had been used in a multitude of biotechnological applications, including its display on filamentous phage (9, 19). In these studies, binding of an HRP-conjugated rabbit IgG complex to the phage particles confirmed that the fusion proteins possessed functional IgG-binding domains. ZZ-displaying phages were shown to interact with immobilized human IgG and its various subclasses, which interaction (as proof of specificity) could be abolished in the presence of an excess of soluble protein A. In previously published studies, monovalent display of ZZ was done with a phagemid-helper phage combination. This format results in a phage population where ∼90% of the particles do not display the intended molecule on their coat (6). To get display on all particles, and possibly improved binding because of avidity, we opted for polyvalent display with the fUSE5 phage vector which was designed for display on all five copies of the p3 minor coat protein (http://www.biosci.missouri.edu/smithgp/PhageDisplayWebsite/PhageDisplayWebsiteIndex.html). As shown in Fig. 5A, polyvalent display of ZZ on all copies of p3 was achieved, while the surplus of wild-type p3 over the ZZ-p3 fusion protein seen with the pCANTAB-ZZ phage is, indeed, typical of monovalent display.
fUSE5-ZZ phages were conjugated to the chloramphenicol prodrug, and their capacity to bind IgG was compared to that of untreated phages. As shown in Fig. 5B, at a conjugation level of about 3,000 drug molecules/phage, the ZZ domain lost about 50% of its IgG-binding capacity. This loss could be anticipated, since ZZ contains 12 lysine residues, most of which are surface exposed and vulnerable to NHS chemistry. That binding loss could be prevented by complexation of the phages with human IgG prior to drug conjugation (Fig. 5B). However, because the protection protocol is cumbersome, involving multiple PEG-NaCl precipitations, and because we figured that at 50% binding capacity, with five ZZ molecules displayed on each phage, most of our particles would bind antibodies, we carried out the growth inhibition experiments with phages that were conjugated without protection. We are considering other antibody display formats for targeting drug-carrying phages (such as scFvs) that may also be vulnerable to conjugation chemistry-induced deterioration (4). It is clear that any targeting moiety will have to be matched with the particular drug conjugation chemistry that will be used.
When fUSE5-ZZ was evaluated, all of the binding-targeting studies with S. aureus were done following blocking of the bacterium's own cell surface protein A with 20% rabbit serum prior to addition of the specific antibodies. This was done to avoid depletion of the targeting antibodies by the bacterium's own protein A. This step is not really necessary to demonstrate specificity, since antibodies that are bound to S. aureus bacteria through their own protein A cannot be bound by the ZZ domain that binds to the same site on the antibodies' Fc. As shown in Fig. 7, chloramphenicol prodrug-carrying fUSE5-ZZ phages retarded the growth of the target S. aureus bacteria after they were incubated with S. aureus-specific human antiserum (bound through the displayed ZZ domain) and to a lesser extent when the phages were complexed with a nonimmune human IgG that does not bind the serum-blocked S. aureus bacteria. In these experiments, targeted fUSE5-ZZ retarded S. aureus growth as effectively as a 10 × higher concentration of free chloramphenicol. We saw further evidence of specificity by treating nontarget Streptococcus pyogenes bacteria (which have a sensitivity to chloramphenicol similar to that of S. aureus bacteria) with the same drug-carrying fUSE5-ZZ-antibody combination, which did not yield any specific growth retardation (not shown).
To conclude, our study demonstrated a proof of principle of targeted, drug-carrying filamentous bacteriophage as antibacterial agents. To further pursue the potential of this approach, several points will need to be considered.
Increasing drug-carrying capacity.
The hydrophobic nature of chloramphenicol limited us to fewer than 3,000 drug molecules/phage out of a theoretical capacity of ∼20,000 by amine chemistry. We are making progress in that direction, as we saw with irreversible conjugation of ∼10,000 molecules of an aminoglycoside/phage. This was done by 1-ethyl-3(3-dimethylaminopropyl)-carbodiimide chemistry, which generated an amide bond between the amine group of the aminoglycoside (which is water soluble) and carboxyl groups on the phage coat (data not shown).
Modifying the mechanism or the rate of drug release.
The ester linkage reported here allowed the release of 15% of the conjugated drug after 1 h in 99% serum. We may need to modify that rate to fully exploit the large drug-carrying capacity of the phages. Such modification is possible by modifying the linker (i.e., to include a diester linkage which is degraded more rapidly). Other forms of modification and release may be considered, even with chloramphenicol itself that was previously converted to a monoester prodrug that could be activated by a catalytic antibody (27). It is also conceivable that the concentration of serum esterases may be rate limiting for drug release. A possible remedy could be the addition of purified esterases that were previously reported to be used for prodrug activation (1), which will result in the drug release being more controlled than delayed as it currently is. Modification of the drug release rate could even be provided by the disease state itself, as it has reported that certain bacterial infections cause elevation of serum esterases (24, 33). That could be a two-edged sword if enhanced drug release causes premature release and elevated toxicity. Experimentation with in vivo models may provide data concerning such issues.
Nonspecific binding.
We believe that targeting efficiency can be improved by using higher-affinity targeting moieties than the ones we used here. Examples are displayed peptides selected under more stringent conditions, higher-titer immune sera, or affinity-purified antibodies (polyclonal, monoclonal, or recombinant antibody fragments) rather than the sera we used here (which contained many antibodies that are not relevant to our target bacteria). This will be evaluated in future studies.
Pharmacokinetics, biodistribution, and immunogenicity.
Pharmacokinetics, biodistribution, and immunogenicity are key issues in current phage therapy studies (25, 46). Basically, phages are immunogenic on the one hand and upon intravenous injection are removed quickly by the reticuloendothelial system on the other. Attempts to modulate phage pharmacokinetics were based on isolating long-circulating mutants of phage lambda (25), but no such studies were done with filamentous phages. We believe that chemical modification of the phage coat (as we do during drug conjugation) should modulate pharmacokinetics, biodistribution, and immunogenicity in comparison with bare phages. An example could be drawn from the PEGylation of therapeutic proteins (43). In one study, the pharmacokinetics and processing of native and receptor-targeted phage in mice were investigated. It was shown that the in vivo behavior of the phages was dramatically altered by chemically modifying the phages by conjugation of either galactose or succinic acid groups (28). An interesting observation we made in the course of our study was that, following NHS conjugation, the phage was no longer recognized in ELISA by a commercial anti-fd phage antibody (Pharmacia) that binds to the p8 major coat protein. This is an initial indication of modulation of antigenicity by drug conjugation. We plan to compare the immunogenicity, pharmacokinetics, and biodistribution of unconjugated and drug-carrying phages in future animal studies.
Acknowledgments
We thank Jonathan M. Gershoni (Tel-Aviv University) for providing peptide-phage display libraries. We thank George Georgiou (University of Texas at Austin) for plasmid pSD-ZZ. We thank Yair Aharonowitz (Tel-Aviv University) for S. aureus SH1000 cells. We thank Eliora Z. Ron (Tel-Aviv University) for E. coli strain O78 cells and for critical reading of the manuscript.
REFERENCES
- 1.Azema, J., B. Guidetti, M. Malet-Martino, R. Martino, and C. Roques. 2006. Efficient approach to acyloxymethyl esters of nalidixic acid and in vitro evaluation as intra-ocular prodrugs. Bioorg. Med. Chem. 14:2569-2580. [DOI] [PubMed] [Google Scholar]
- 2.Benhar, I. 2001. Biotechnological applications of phage and cell display. Biotechnol. Adv. 19:1-33. [DOI] [PubMed] [Google Scholar]
- 3.Benhar, I., R. Azriel, L. Nahary, S. Shaky, Y. Berdichevsky, A. Tamarkin, and W. Wels. 2000. Highly efficient selection of phage antibodies mediated by display of antigen as Lpp-OmpA′ fusions on live bacteria. J. Mol. Biol. 301:893-904. [DOI] [PubMed] [Google Scholar]
- 4.Benhar, I., U. Brinkmann, K. O. Webber, and I. Pastan. 1994. Mutations of two lysine residues in the CDR loops of a recombinant immunotoxin that reduce its sensitivity to chemical derivatization. Bioconjug. Chem. 5:321-326. [DOI] [PubMed] [Google Scholar]
- 5.Benhar, I., and Y. Reiter. 2002. Phage display of single-chain antibodies, p. 10.19B.1-10.19B.30. In J. E. Coligan, B. E. Bierer, D. H. Margulis, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 6.Berdichevsky, Y., E. Ben-Zeev, R. Lamed, and I. Benhar. 1999. Phage display of a cellulose binding domain from Clostridium thermocellum and its application as a tool for antibody engineering. J. Immunol. Methods 228:151-162. [DOI] [PubMed] [Google Scholar]
- 7.Berdichevsky, Y., R. Zemel, L. Bachmatov, A. Abramovich, R. Koren, P. Sathiyamoorthy, A. Golan-Goldhirsh, R. Tur-Kaspa, and I. Benhar. 2003. A novel high throughput screening assay for HCV NS3 serine protease inhibitors. J. Virol. Methods 107:245-255. [DOI] [PubMed] [Google Scholar]
- 8.Coates, A., Y. Hu, R. Bax, and C. Page. 2002. The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discov. 1:895-910. [DOI] [PubMed] [Google Scholar]
- 9.Djojonegoro, B. M., M. J. Benedik, and R. C. Willson. 1994. Bacteriophage surface display of an immunoglobulin-binding domain of Staphylococcus aureus protein A. Biotechnology 12:169-172. [DOI] [PubMed] [Google Scholar]
- 10.Enshell-Seijffers, D., and J. M. Gershoni. 2002. Phage display selection and analysis of Ab-binding epitopes, p. 9.8.1-9.8.27. In J. E. Coligan, B. E. Bierer, D. H. Margulis, E. M. Shevach, and W. Strober (ed.), Current protocols in immunology. John Wiley & Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 11.Enshell-Seijffers, D., L. Smelyanski, and J. M. Gershoni. 2001. The rational design of a ′type 88′ genetically stable peptide display vector in the filamentous bacteriophage fd. Nucleic Acids Res. 29:E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farr, B. M. 2004. Prevention and control of methicillin-resistant Staphylococcus aureus infections. Curr. Opin. Infect. Dis. 17:317-322. [DOI] [PubMed] [Google Scholar]
- 13.Forget, E. J., and D. Menzies. 2006. Adverse reactions to first-line antituberculosis drugs. Expert Opin. Drug Saf. 5:231-249. [DOI] [PubMed] [Google Scholar]
- 14.Freeman, A., N. Cohen-Hadar, S. Abramov, R. Modai-Hod, Y. Dror, and G. Georgiou. 2004. Screening of large protein libraries by the cell immobilized on adsorbed bead approach. Biotechnol. Bioeng. 86:196-200. [DOI] [PubMed] [Google Scholar]
- 15.Hunter, G. J., D. H. Rowitch, and R. N. Perham. 1987. Interactions between DNA and coat protein in the structure and assembly of filamentous bacteriophage fd. Nature 327:252-254. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski, M. 1969. Common and species-specific serum esterases of Equidae. I. Horse and donkey. Biochim. Biophys. Acta 191:611-620. [DOI] [PubMed] [Google Scholar]
- 17.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 18.Kreitman, R. J. 1999. Immunotoxins in cancer therapy. Curr. Opin. Immunol. 11:570-578. [DOI] [PubMed] [Google Scholar]
- 19.Kushwaha, A., P. S. Chowdhury, K. Arora, S. Abrol, and V. K. Chaudhary. 1994. Construction and characterization of M13 bacteriophages displaying functional IgG-binding domains of staphylococcal protein A. Gene 151:45-51. [DOI] [PubMed] [Google Scholar]
- 20.Liederer, B. M., and R. T. Borchardt. 2005. Stability of oxymethyl-modified coumarinic acid cyclic prodrugs of diastereomeric opioid peptides in biological media from various animal species including human. J. Pharm. Sci. 94:2198-2206. [DOI] [PubMed] [Google Scholar]
- 21.Liu, J., M. Dehbi, G. Moeck, F. Arhin, P. Bauda, D. Bergeron, M. Callejo, V. Ferretti, N. Ha, T. Kwan, J. McCarty, R. Srikumar, D. Williams, J. J. Wu, P. Gros, J. Pelletier, and M. DuBow. 2004. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22:185-191. [DOI] [PubMed] [Google Scholar]
- 22.Lowy, F. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 23.McDevitt, D., and M. Rosenberg. 2001. Exploiting genomics to discover new antibiotics. Trends Microbiol. 9:611-617. [DOI] [PubMed] [Google Scholar]
- 24.Menschikowski, M., A. Rosner-Schiering, R. Eckey, E. Mueller, R. Koch, and W. Jaross. 2000. Expression of secretory group IIA phospholipase A2 in relation to the presence of microbial agents, macrophage infiltrates, and transcripts of proinflammatory cytokines in human aortic tissues. Arterioscler. Thromb. Vasc. Biol. 20:751-762. [DOI] [PubMed] [Google Scholar]
- 25.Merril, C. R., B. Biswas, R. Carlton, N. C. Jensen, G. J. Creed, S. Zullo, and S. Adhya. 1996. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA 93:3188-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikawa, Y. G., I. N. Maruyama, and S. Brenner. 1996. Surface display of proteins on bacteriophage lambda heads. J. Mol. Biol. 262:21-30. [DOI] [PubMed] [Google Scholar]
- 27.Miyashita, H., Y. Karaki, M. Kikuchi, and I. Fujii. 1993. Prodrug activation via catalytic antibodies. Proc. Natl. Acad. Sci. USA 90:5337-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molenaar, T. J., I. Michon, S. A. de Haas, T. J. van Berkel, J. Kuiper, and E. A. Biessen. 2002. Uptake and processing of modified bacteriophage M13 in mice: implications for phage display. Virology 293:182-191. [DOI] [PubMed] [Google Scholar]
- 29.Nakamura, M., K. Tsumoto, K. Ishimura, and I. Kumagai. 2001. A visualization method of filamentous phage infection and phage-derived proteins in Escherichia coli using biotinylated phages. Biochem. Biophys. Res. Commun. 289:252-256. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura, M., K. Tsumotob, K. Ishimuraa, and I. Kumagaib. 2002. The effect of an agglutogen on virus infection: biotinylated filamentous phages and avidin as a model. FEBS Lett. 520:77-80. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson, B., T. Moks, B. Jansson, L. Abrahmsen, A. Elmblad, E. Holmgren, C. Henrichson, T. A. Jones, and M. Uhlen. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1:107-113. [DOI] [PubMed] [Google Scholar]
- 32.Norrby, S. R., C. E. Nord, and R. Finch. 2005. Lack of development of new antimicrobial drugs: a potential serious threat to public health. Lancet Infect. Dis. 5:115-119. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa, M., N. Sato, S. Endo, M. Kojika, Y. Yaegashi, Y. Kimura, K. Ikeda, and T. Iwaya. 2005. Group IIA-soluble phospholipase A2 levels in patients with infections after esophageal cancer surgery. Surg. Today 35:912-918. [DOI] [PubMed] [Google Scholar]
- 34.Olson, N. H., M. Gingery, F. A. Eiserling, and T. S. Baker. 2001. The structure of isometric capsids of bacteriophage T4. Virology 279:385-391. [DOI] [PubMed] [Google Scholar]
- 35.Payne, D., and A. Tomasz. 2004. The challenge of antibiotic resistant bacterial pathogens: the medical need, the market and prospects for new antimicrobial agents. Curr. Opin. Microbiol. 7:435-438. [DOI] [PubMed] [Google Scholar]
- 36.Projan, S. J., and D. M. Shlaes. 2004. Antibacterial drug discovery: is it all downhill from here? Clin. Microbiol. Infect. 10(Suppl. 4):18-22. [DOI] [PubMed] [Google Scholar]
- 37.Sefton, A. M. 2002. Mechanisms of antimicrobial resistance: their clinical relevance in the new millennium. Drugs 62:557-566. [DOI] [PubMed] [Google Scholar]
- 38.Siman-Tov, D. D., L. Navon-Perry, N. L. Haigwood, and J. M. Gershoni. 2005. Differentiation of a passive vaccine and the humoral immune response toward infection: analysis of phage displayed peptides. Vaccine 24:607-612. [DOI] [PubMed] [Google Scholar]
- 39.Smith, G. P., and J. K. Scott. 1993. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217:228-257. [DOI] [PubMed] [Google Scholar]
- 40.Spellberg, B., J. H. Powers, E. P. Brass, L. G. Miller, and J. E. Edwards, Jr. 2004. Trends in antimicrobial drug development: implications for the future. Clin. Infect. Dis. 38:1279-1286. [DOI] [PubMed] [Google Scholar]
- 41.Trail, P. A., H. D. King, and G. M. Dubowchik. 2003. Monoclonal antibody drug immunoconjugates for targeted treatment of cancer. Cancer Immunol. Immunother. 52:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turton, J. A., C. M. Andrews, A. C. Havard, and T. C. Williams. 2002. Studies on the haemotoxicity of chloramphenicol succinate in the Dunkin Hartley guinea pig. Int. J. Exp. Pathol. 83:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veronese, F. M., and G. Pasut. 2005. PEGylation, successful approach to drug delivery. Drug Discov. Today 10:1451-1458. [DOI] [PubMed] [Google Scholar]
- 44.White, C. A., R. L. Weaver, and A. J. Grillo-Lopez. 2001. Antibody-targeted immunotherapy for treatment of malignancy. Annu. Rev. Med. 52:125-145. [DOI] [PubMed] [Google Scholar]
- 45.Yanagida, M. 1977. Molecular organization of the shell of T-even bacteriophage head. II. Arrangement of subunits in the head shell of giant phages. J. Mol. Biol. 109:515-537. [DOI] [PubMed] [Google Scholar]
- 46.Zou, J., M. T. Dickerson, N. K. Owen, L. A. Landon, and S. L. Deutscher. 2004. Biodistribution of filamentous phage peptide libraries in mice. Mol. Biol. Rep. 31:121-129. [DOI] [PubMed] [Google Scholar]