Abstract
Human coronavirus NL63 (HCoV-NL63), a recently discovered member of the Coronaviridae family, has spread worldwide and is associated with acute respiratory illness in young children and elderly and immunocompromised persons. Further analysis of HCoV-NL63 pathogenicity seems warranted, in particular because the virus uses the same cellular receptor as severe acute respiratory syndrome-associated coronavirus. As there is currently no HCoV-NL63-specific and effective vaccine or drug therapy available, we evaluated several existing antiviral drugs and new synthetic compounds as inhibitors of HCoV-NL63, targeting multiple stages of the replication cycle. Of the 28 compounds that we tested, 6 potently inhibited HCoV-NL63 at early steps of the replication cycle. Intravenous immunoglobulins, heptad repeat 2 peptide, small interfering RNA1 (siRNA1), siRNA2, β-d-N4-hydroxycytidine, and 6-azauridine showed 50% inhibitory concentrations of 125 μg/ml, 2 μM, 5 nM, 3 nM, 400 nM, and 32 nM, respectively, and low 50% cytotoxicity concentrations (>10 mg/ml, >40 μM, >200 nM, >200 nM, >100 μM, and 80 μM, respectively). These agents may be investigated further for the treatment of coronavirus infections.
Coronaviruses are enveloped viruses with a positive, single-stranded RNA genome of approximately 27 to 32 kb. The 5′ two-thirds of their genome encodes a polyprotein that contains all proteins necessary for RNA replication. The 3′ one-third encodes several structural proteins, such as spike (S), envelope (E), membrane (M), and nucleocapsid (N), that, among other functions, participate in viral budding and are incorporated into the virus particle. Nonstructural accessory protein genes are also present in the 3′ part of the genome, at a position and arrangement that is characteristic for each of the different coronavirus groups.
Coronavirus infection starts with the recognition of a specific receptor on the host cell surface by an S protein, followed by virus internalization, which occurs either immediately by direct fusion with the plasma membrane or after endocytosis. Fusion of the viral membrane with the cellular membrane triggers the release of the viral RNA genome into the host cell cytoplasm. Viral RNA is copied by the viral replicase in membrane-associated replication centers (14). During the replication process, copies of the full-length genomic RNA and a nested set of subgenomic mRNAs are generated. These subgenomic mRNAs are functional templates for the translation of the structural proteins encoded in the 3′ one-third of the genome. Full-length viral RNA is encapsidated and released from the host cell as an infectious virus particle.
Human coronavirus NL63 (HCoV-NL63), a recently discovered (45, 55) member of the Coronaviridae family, has spread worldwide, is observed most frequently in the winter season, and is associated with acute respiratory illness and croup in young children, elderly people, and immunocompromised patients (2, 6, 15, 25, 29, 40, 53-56). A recent report suggested that HCoV-NL63 is the causative agent of Kawasaki disease (28), although other studies did not confirm this relationship (7, 26, 49). In the developed world, Kawasaki disease is the most common cause of acquired heart disease in children (37, 47). Further analysis of HCoV-NL63 pathogenicity seems warranted, in particular because the virus uses the same cellular receptor as severe acute respiratory syndrome-associated CoV (SARS-CoV) (34).
An effective antiviral treatment is required for HCoV-NL63-infected patients who are admitted to the intensive care unit due to acute respiratory disease. To investigate the therapeutic options, we tested several potential inhibitors that target specific steps of the coronavirus life cycle, e.g., receptor binding, membrane fusion, transcription, translation, posttranslational processing, and virus release. The compounds inhibiting the early phase of HCoV-NL63 infection appeared to be the most potent antivirals.
MATERIALS AND METHODS
Antiviral agents.
Information about all 28 tested compounds is summarized in Table 1. The 50% inhibitory concentrations (IC50s; based on the cytopathic effect [CPE] reduction assay and viral yield) and 50% cytotoxic concentrations {CC50s; based on the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] inner salt assay} were determined for each antiviral agent. Human sera obtained from healthy adults were inactivated by incubation for 30 min at 56°C and stored at −80°C until use.
TABLE 1.
Antiviral agents tested
| Agent | Source | Solventa |
|---|---|---|
| S-Nitroso-N-acetylpenicillamine | Calbiochem | DMSO |
| 2,2′-(Hydroxynitrosohydrazino) bis-ethanamine | Sigma | Water |
| Calpain inhibitor VI | Calbiochem | DMSO |
| Calpain inhibitor III | Sigma | DMSO |
| Glycyrrhizin | Sigma | DMSO |
| Chloroquine phosphate | Sigma | Water |
| Aurintricarboxylic acid | Calbiochem | 0.1 M NaOH |
| Valinomycin | Calbiochem | DMSO |
| Escin | Sigma | DMSO |
| Reserpine | Sigma | DMSO |
| Pentoxifylline | Calbiochem | Water |
| 2′,5-Dichloro- 4′-nitrosalicylanilide | Sigma | DMSO |
| 6-Azauridine | Sigma | Water |
| Hygromycin B | Sigma | Water |
| Dipyridamole | Sigma | DMSO |
| Actinomycin D | Sigma | Water |
| Ribavirin | Sigma | Water |
| HR peptide 1 | Water | |
| HR peptide 2 | Water | |
| siRNA1 | QIAGEN | Buffer (supplied) |
| siRNA2 | QIAGEN | Buffer (supplied) |
| IVIG | Sanquin | Water |
| β-d-N4-hydroxycytidine | NIH AIDS Reagent Program | DMSO |
| Ritonavir | Abbott Laboratories | DMSO |
| Nelfinavir | Agouron Pharmaceuticals | DMSO |
| Indinavir | Merck & Co. | DMSO |
| Saquinavir | Roche | DMSO |
| Amprenavir | GlaxoWellcome | DMSO |
DMSO, dimethyl sulfoxide.
Plasmid construction, bacterial protein expression, and purification.
For the production of the HR1 and HR2 peptides corresponding to amino acid residues 955 to 1064 (HR1) and 1241 to 1285 (HR2) of the HCoV-NL63 spike protein, a PCR fragment was prepared with the plasmid carrying the HCoV-NL63 spike gene (34). The primers 5′-GCGGATCCCAAGCACGACTTAACTATG-3′ and 5′-CGAATTCAAGTAATTAATCTGTCAACTTG-3′ were used for the amplification of HR1, and the primers 5′-GCGGATCCTTTAATTTAACATATCTTAATTTG-3′ and 5′-CGAATTCACAACTTCAAATCAACATATGT-3′) were used for HR2. Bacterial expression and purification were then performed essentially as described previously (12), with a few modifications. Lysozyme (100 μg/ml), dithiothreitol (DTT; 7 mM), and sarkosyl (1%) were added to phenylmethylsulfonyl fluoride (PMSF; 1 mM) prior to sonication, and Triton X-100 (2.8%) was added to the supernatant after centrifugation, prior to glutathione-Sepharose 4B purification. Production of the murine coronavirus (murine hepatitis virus [MHV] strain A59) HR2 peptide has been described previously (13).
Selection of siRNA sequences.
The small interfering RNAs (siRNAs) were designed and synthesized by QIAGEN (QIAGEN, Benelux B.V.) with the HiPerformance design algorithm (Novartis AG) and integrated with a stringent homology analysis tool (QIAGEN). The siRNAs were synthesized using a TOM amidite chemistry process, yielding >90% purity, as determined by ion-exchange high-performance liquid chromatography analysis. Oligonucleotides were provided as annealed double-strand siRNA (Table 2). The sequence and identity of each siRNA were confirmed using matrix-assisted laser desorption ionization-time of flight spectrometric analysis. The stock and working solutions were prepared according to the manufacturer's protocol (QIAGEN). As a negative control, we used siRNA targeting Rattus norvegicus mRNA collybistin 1 (GenBank accession number AJ250425). Sequences of siRNA were BLAST analyzed (using the NCBI database “Search for short, nearly exact matches” mode) against human sequences to exclude those siRNA sequences with potential targets in the human genome.
TABLE 2.
siRNA oligonucleotide sequence
| Transfected RNA | Oligonucleotide sequence |
|---|---|
| siRNA1-NL63a | |
| Sense | r(GGA AUU AUA CGU UCU UCA A)dTdT |
| Antisense | r(UUG AAG AAC GUA UAA UUC C)dAdG |
| siRNA2-NL63 | |
| Sense | r(AGG UUA AUA UAU CUC UUA A)dTdT |
| Antisense | r(UUA AGA GAU AUA UUA ACC U)dGdG |
| siRNA3 control | |
| Sense | r(GAC CAC AGU GAU UAC AGA U)dTdT |
| Antisense | r(AUC UGU AAU CAC UGU GGU C)dTdT |
Coordinates of the target sequences are nucleotides 21980 to 22000 and nucleotides 22384 to 22404 in the HCoV-NL63 genome for siRNA1 and siRNA2, respectively. The target for control siRNA3 is the collybistin 1 gene of Rattus norvegicus, nucleotides 1190 to 1208.
Cell culture.
LLC-MK2 cells were cultured in minimal essential medium (MEM; 2 parts Hanks' MEM and 1 part Earle's MEM) supplemented with 3% heat-inactivated fetal calf serum (PAA Laboratories), penicillin (100 U/ml), and streptomycin (100 μg/ml). Twenty-four hours prior to transfection, the cells were plated onto 96-well plates at a density of 2 × 104 cells/well in fresh medium (100 μl per well) and cultured at 37°C with 5% CO2. Twenty-four hours prior to the addition of the drug, cells were plated onto 96-well plates at a density of 4 × 104 cells/well in fresh medium (100 μl per well) with 100 μg of penicillin and 100 μg of streptomycin and cultured at 37°C with 5% CO2.
Cytopathic effect reduction assay.
HCoV-NL63 (isolate Amsterdam 1) was obtained from a virus culture on LLC-MK2 cells as described previously (55). The infectious titer of the virus was determined according to the Reed and Muench formula (46) on an LLC-MK2 cell monolayer. The virus stock has a titer of 2 × 105 50% tissue culture infectivity doses/ml. For the CPE reduction assay, cells were treated with serially diluted compounds and infected with HCoV-NL63 at a multiplicity of infection of 0.01. The CPE was scored visually at day 6 postinfection and confirmed with an MTS assay. Experiments were performed in quadruplicate.
Immunostaining-based HCoV-NL63 infection inhibition assay.
Virus entry inhibition by the HR2 peptide was analyzed on LLC-MK2 cells in 96-well plates (4 × 104 cells per well). Cells were inoculated with HCoV-NL63 at a multiplicity of infection of 0.5 in the presence of serial dilutions of the peptides. The MHV HR2 peptide was included as a negative control. After incubation for 24 h, cells were washed with phosphate-buffered saline (PBS), fixed with 3% formaldehyde for 20 min, and permeabilized with 1% Triton X-100 in PBS for 5 min. After they were washed twice with PBS and blocked with PBS-5% fetal calf serum, the HCoV-NL63-positive cells were detected by intracellular peroxidase staining using a human polyclonal serum (1:200) in combination with a biotinylated antihuman antibody (1:250) and the VECTASTAIN ABC kit (Vector Laboratories). The reaction mixture was developed with 3-amino-9-ethylcarbazole (AEC; Sigma) according to the manufacturer's instructions. Experiments were performed in duplicate. Infected cells were counted using a light microscope.
Drug cytotoxicity.
Cytotoxicity of the compounds was determined by measuring mitochondrial activity on day 6 posttreatment with MTS (CellTiter 96 AQueous one solution cell proliferation assay; Promega) according to the manufacturer's instructions. Cytotoxicity measurements were confirmed by determining the mRNA levels of the household gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Experiments were performed in duplicate.
Transfection of LLC-MK2 cells with siRNA.
Transfection with siRNA was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. siRNA-transfected cells were infected with HCoV-NL63 after 24 hours.
Reverse transcription-PCR and real-time quantitative PCR.
Total RNA was extracted from the medium and cells by the silica-affinity-based Boom extraction method (11) and eluted in 100 μl water. Reverse transcription was performed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) (200 U per reaction) and 10 ng of random hexamers (Amersham Biosciences) in 10 mM Tris, pH 8.3, 50 mM KCl, 0.1% Triton X-100, 6 mM of MgCl2, and 50 μM of each deoxynucleoside triphosphate at 37°C for 90 min in a total volume of 40 μl.
Virus yield was determined with real-time PCR, using the Platinum quantitative PCR SuperMix-uracil-DNA glycosylase (Invitrogen). Ten microliters of cDNA was amplified in 50 μl 1× Platinum quantitative PCR SuperMix-uracil-DNA glycosylase (Invitrogen) with 125 nM of MgCl2, 10 μM of specific probe labeled with FAM (6-carboxyfluorescein) and TAMRA (6-carboxytetramethylrhodamine), and 45 μM of each primer. The following primers were used for HCoV-NL63: sense, 5′-GCGTGTTCCTACCAGAGAGGA-3′; antisense, 5′-GCTGTGGAAAACCTTTGGCA-3′; and probe, 5′-FAM-ATGTTATTCAGTGCTTTGGTCCTCGTGAT-TAMRA-3′. The reaction was carried out on a TaqMan machine (ABI). Following the denaturation step for 2 min at 50°C and 5 min at 95°C, 45 cycles of amplification were performed for 15 s at 95°C and 60 s at 60°C.
Nucleotide sequence accession numbers.
The sequence for HCoV-NL63 isolate Amsterdam 57 described in this study was deposited in GenBank under accession number DQ471450. The GenBank accession number for HCoV-NL63 isolate Amsterdam 1 is NC_005831; that for HCoV-229E is AF304460; and that for porcine epidemic diarrhea virus strain CV777 is AF353511. The sequence of the GAPDH gene (LLC-MK2 cells derived from Macaca mulatta) was deposited in GenBank under accession number DQ445913.
RESULTS
HCoV-NL63 inhibition assay.
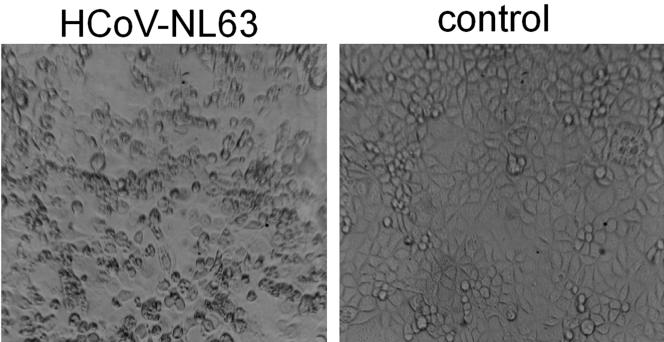
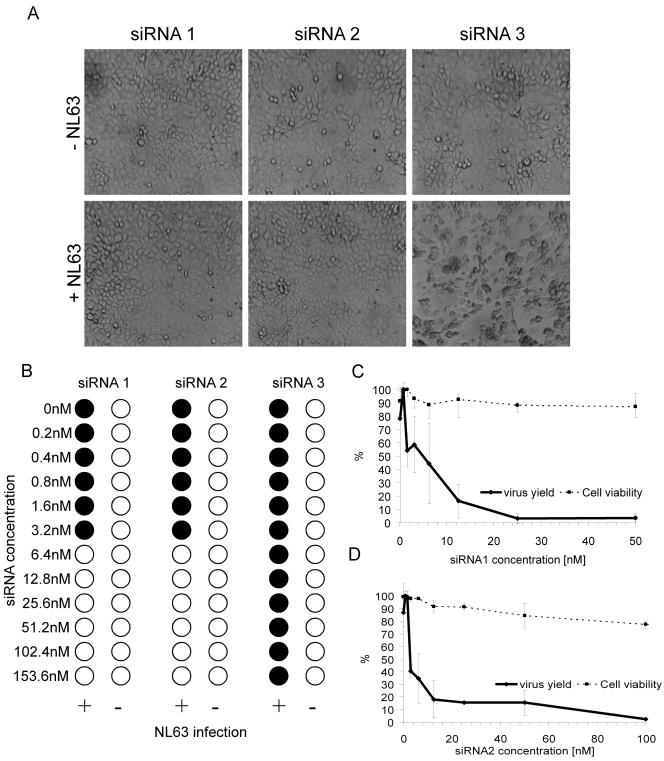
The inhibition of HCoV-NL63 infection can be determined using several assays. The most straightforward assay is based on the reduction of CPE. Six days after infection, HCoV-NL63-infected LLC-MK2 cells exhibit evident morphological changes consisting of cell enlargement, rounding, and, eventually, detachment from the surface (Fig. 1). Thus, infecting the target cells in the presence of serially diluted candidate antiviral agents provides a means of visualizing their inhibitory activity by scoring the reduction in CPE. LLC-MK2 cells seeded on the 96-well culture plate and subsequently infected with HCoV-NL63 in the presence of serially diluted compounds were scored for CPE using phase-contrast microscopy. Agents that showed antiviral activity in this initial assay were subsequently tested by measuring the virus yield at active concentrations of the compound.
FIG. 1.
CPE induced by HCoV-NL63 on LLC-MK2 cells.
Virus neutralization by purified human immunoglobulin G (IgG).
Sera from virtually all healthy adults contain HCoV-NL63 antibodies (34). To determine the neutralizing potential of these antibodies, we tested 10 randomly selected human sera from healthy adults by the CPE reduction assay. All tested sera inhibited HCoV-NL63 infection at 25- to 50-fold dilutions, illustrating that the majority of adults carry neutralizing antibodies against HCoV-NL63.
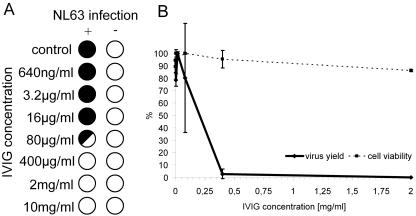
To further explore the potential of neutralizing antibodies in antiviral therapy, we tested pooled purified human IgG from healthy donors (intravenous immunoglobulin [IVIG]). In fact, IVIG is part of the effective routine treatment for Kawasaki patients (43) and several immunodeficiency syndromes (23, 31). The CPE reduction assay demonstrates that this agent is very active against HCoV-NL63 infection (Fig. 2A). To confirm that CPE reduction is an accurate measurement of virus inhibition, the virus yield was determined for each serial IVIG dilution (Fig. 2B). The virus yield also allows a more precise measurement of the IC50 value, which was 200 μg/ml for IVIG. This concentration is ∼10 times lower than the dose advised for treatment (2 g/kg of body weight) (43). The cell survival assay indicated that IVIG has a very low cytotoxicity (CC50 > 10 mg/ml), thus yielding a high selectivity index (CC50/IC50 > 50).
FIG. 2.
Inhibition of HCoV-NL63 by IVIG. (A) CPE reduction mediated by IVIG. Filled circles indicate CPE development, empty circles indicate the absence of CPE, and the half-filled circle represents the development of CPE in 50% of wells. (B) Decrease in HCoV-NL63 virus yield after IVIG treatment and cell viability assay. Numbers on the y axis represent the percentages of produced virus and percentages of viable cells.
Inhibition of cell entry.
The spike protein of HCoV-NL63 is a class I fusion glycoprotein consisting of a globular S1 domain that recognizes the receptor and a rodlike S2 domain involved in membrane fusion. After receptor binding and virus internalization, the S protein undergoes a structural switch, resulting in the exposure of the fusion peptide (13, 27). The HR1 and HR2 regions in the S2 domain rearrange and interact during the structural switch. Blocking this interaction between HR1 and HR2 provides an effective antiviral strategy (12, 13). The HCoV-NL63 spike protein contains HR1 and HR2 regions with a characteristic 7-residue periodicity. HR2 is located adjacent to the transmembrane domain, and HR1 is about 170 residues away, toward the N terminus. In all coronaviruses, HR1 is consistently larger than HR2, and all group 1 coronaviruses, including HCoV-NL63, show a remarkable insertion of two heptad repeats (14 amino acids) in both HR regions (13, 21).
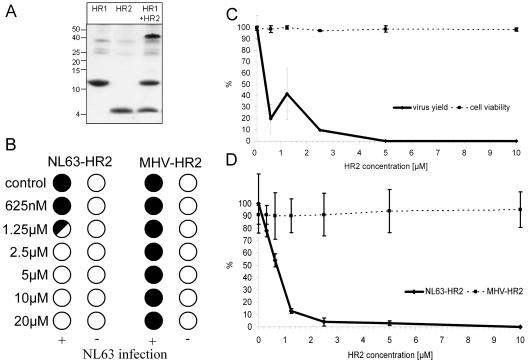
Peptides corresponding to the HCoV-NL63 HR1 and HR2 regions were prepared with the bacterial glutathione S-transferase expression system and purified by using reverse-phase high-performance liquid chromatography. It was previously shown for SARS-CoV and MHV (12, 13) that mixing HR1 and HR2 peptides leads to the assembly of an oligomeric complex that is resistant to 2% sodium dodecyl sulfate (SDS) (13). Using the same approach, we observed that the HR1 and HR2 peptides of the HCoV-NL63 spike protein behaved in a similar manner, forming an SDS-resistant oligomeric complex in an equimolar mixture (Fig. 3A).
FIG. 3.
Inhibition of HCoV-NL63 by the HR2 peptide. (A) SDS-polyacrylamide gel electrophoresis analysis of HR peptides separately and the HR1/HR2 complex formation. The molecular mass of the complex corresponds to the predicted heterohexamer. (B) CPE reduction mediated by HR2 peptide. Filled circles indicate CPE development, empty circles indicate the absence of CPE, and the half-filled circle represents the development of CPE in 50% of wells. (C) Decrease in HCoV-NL63 virus yield after HR2 treatment and cell viability assay. Numbers on the y axis represent the percentages of produced virus and percentages of viable cells. (D) Immunostaining-based HCoV-NL63 infection inhibition assay. Values on the y axis represent the percentages of infected cells.
The HR2 peptide was subsequently tested for its inhibitory potency in the CPE reduction assay. Concentration-dependent inhibition of HCoV-NL63 infection was observed with an IC50 value of ∼0.5 μM and a CC50 value of >20 μM (Fig. 3B and C). This effect is sequence specific, because no inhibition was seen with a corresponding peptide derived from the HR2 region of MHV (MHV-HR2) that is known to block MHV infection (13) (Fig. 3D).
Previous studies examined the antiviral activity of HR peptides (for coronaviruses) by immune peroxidase staining of infected cells after 24 h. Using this method, the IC50 was ∼0.5 μM (Fig. 3D), identical to the value measured in the CPE reduction assay. NL63-HR2 exhibits the same powerful antiviral potency as the MHV-HR2 peptide against MHV infection (IC50 of 0.9 μM [13]). The activity of NL63-HR2 is much higher than the inhibiting activity described for the corresponding SARS-CoV HR2 peptide (IC50 of 17 μM [12]).
Targeting the viral RNA by RNA interference.
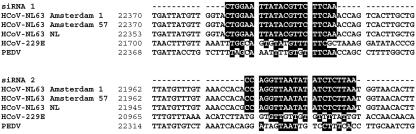
siRNA-mediated degradation of the incoming full-length HCoV-NL63 genome will prevent transcription and, thus, virus production. We selected two siRNAs that target the S gene based on an algorithm for optimal siRNA design and the lack of complementarity with host gene sequences (Table 2). The siRNAs were designed against sequences that are conserved among HCoV-NL63 isolates, thus providing a broad antiviral activity. The target sequence is absent in other coronaviruses, as illustrated in Fig. 4, for the closest group 1 relatives, HCoV-229E and porcine epidemic diarrhea virus.
FIG. 4.
Sequence alignment illustrating conservation and specificity of siRNA target sequences in different HCoV-NL63 isolates. PEDV, porcine epidemic diarrhea virus.
To examine the inhibitory capacity of HCoV-NL63-specific siRNA, we transfected cells with the synthetic siRNAs and subsequently infected them with HCoV-NL63. Transfection of siRNA1 and siRNA2 significantly inhibited the development of CPE, whereas transfection of the control siRNA3 did not (Fig. 5A). The latter result indicates that the transfection procedure does not interfere with CPE production and virus replication and that siRNA1 and siRNA2 inhibit HCoV-NL63 in a sequence-specific manner. The IC50s are ∼5 nM and ∼3 nM for siRNA1 and siRNA2, respectively (Fig. 5B). The CC50 values are higher than 200 nM. These results were confirmed by real-time reverse transcription-PCR analysis of the viral RNA load in the culture medium (Fig. 5C and D).
FIG. 5.
Inhibition of HCoV-NL63 with two specific siRNAs. (A) LLC-MK2 cells transfected with 25 nM siRNA1 and siRNA2 and 50 nM siRNA3 and infected with HCoV-NL63. CPE was observed only in the culture transfected with control siRNA3. The images were taken 6 days postinfection. (B) CPE reduction mediated by siRNA1 and siRNA2. Filled circles indicate CPE development, and empty circles indicate the absence of CPE. (C) Decrease in HCoV-NL63 virus yield after siRNA1 treatment and cell viability assay. Numbers on the y axis represent the percentages of produced virus and percentages of viable cells. (D) Decrease in HCoV-NL63 virus yield after siRNA2 treatment and cell viability assay. Numbers on the y axis represent the percentages of produced virus and percentages of viable cells.
Inhibition of HCoV-NL63 with nucleoside analogues.
A rational approach to the development of drugs for the treatment of HCoV-NL63 infection in patients is to identify compounds that specifically inhibit viral RNA replication. There are several possible mechanisms of action of nucleoside analogues: chain termination resulting from incorporation into elongated RNA strands, interference of these compounds with nucleotide synthesis, or inhibition of the viral polymerase. We tested two nucleoside analogues: β-d-N4-hydroxycytidine and 6-azauridine.
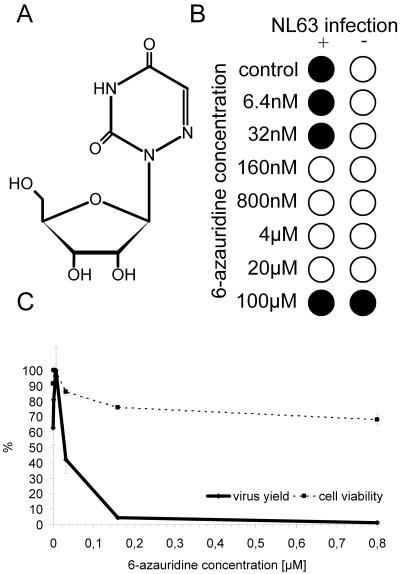
6-Azauridine is a uridine analogue with a histidine-like N-3 pKa (Fig. 6A). It is used as an antineoplastic antimetabolite as it interferes with pyrimidine biosynthesis, thereby preventing the formation of cellular nucleic acids. The antiviral effect of 6-azauridine has previously been documented for several virus types in vitro (19, 42), including coronaviruses (3, 18). This compound may thus act as a broad antiviral agent. We found that 6-azauridine is also an efficient inhibitor of HCoV-NL63 replication. The CPE reduction assay and viral yield determination indicate that the IC50 value is 35 nM (Fig. 6B and C). The CC50 value determined in the MTS assay is about 80 μM, a value consistent with previous reports (19). Thus, 6-azauridine exhibits a very high selectivity index for HCoV-NL63 inhibition.
FIG. 6.
Inhibition of HCoV-NL63 with 6-azauridine. (A) Structure of 6-azauridine. (B) CPE reduction mediated by 6-azauridine. Filled circles indicate CPE development, and empty circles indicate the absence of CPE. (C) Decrease in HCoV-NL63 virus yield after treatment with 6-azauridine and cell viability assay. Numbers on the y axis represent the percentages of produced virus and percentages of viable cells.
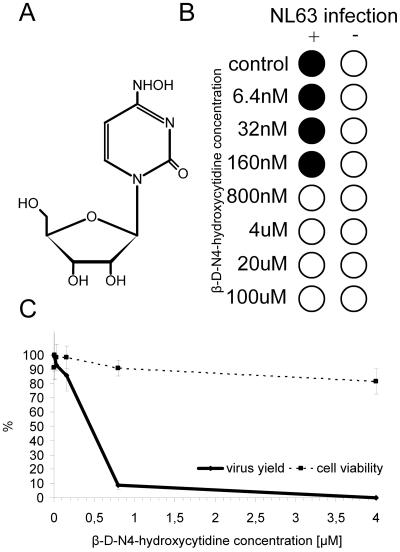
The base-modified nucleoside analogue β-d-N4-hydroxycytidine (Fig. 7A) also inhibits HCoV-NL63 replication (Fig. 7B). The virus yield measurements indicate that the IC50 value is ∼400 nM (Fig. 7C). The CC50 value determined in the MTS assay is about 80 μM.
FIG. 7.
Inhibition of HCoV-NL63 by β-d-N4-hydroxycytidine. (A) Structure of β-d-N4-hydroxycytidine. (B) CPE reduction mediated by β-d-N4-hydroxycytidine. Filled circles indicate CPE development, and empty circles indicate the absence of CPE. (C) Decrease in HCoV-NL63 virus yield after treatment with β-d-N4-hydroxycytidine and cell viability assay. Numbers on the y axis represent the percentages of produced virus and percentages of viable cells.
Agents with low inhibitory activity against HCoV-NL63.
Several other compounds were tested. We observed weak antiviral activity with ritonavir (16), the human immunodeficiency virus type 1 (HIV-1) protease inhibitor, at a concentration of >20 μM but with a very low selectivity index (CC50, ∼50 μM) (Table 3). Other HIV-1 protease inhibitors (nelfinavir, indinavir, amprenavir, or saquinavir) did not show any anti-HCoV-NL63 activity. We observed inhibition of HCoV-NL63 with aurintricarboxylic acid (32, 36), an RNase and polymerase inhibitor (22, 30), at a concentration of ∼60 μM, but we could not exclude the possibility that the effect was the result of an increased pH of the medium (Table 3). We measured no anti-HCoV-NL63 activity in the following compounds: calpain inhibitors VI and III (4), glycyrrhizin (18), valinomycin (58), escin (58), ribavirin (41), dipyridamole (3), actinomycin D (38), and pentoxifylline (8). Several of these compounds have been reported to inhibit other coronaviruses.
TABLE 3.
HCoV-NL63-inhibiting compounds
| Antiviral agent | IC50 | CC50 | Decrease in virus yielda | Selectivity index (CC50/IC50) |
|---|---|---|---|---|
| IVIG | 125 μg/ml | >10 mg/ml | 3.42 × 103 | >80 |
| Heptad repeat peptide | 2 μM | >40 μM | 5.75 × 103 | >20 |
| siRNA1 | 5 nM | >200 nM | 6.54 × 101 | >40 |
| siRNA2 | 3 nM | >200 nM | 9.12 × 101 | >66 |
| β-d-N4-hydroxycytidine | 400 nM | >100 μM | 3.73 × 103 | >250 |
| 6-Azauridine | 32 nM | 80 μM | 1.59 × 103 | 2,500 |
| Aurintricarboxylic acid | 65 μM | 300 μM | NDb | >4.6 |
| Ritonavir | 20 μM | 80 μM | ND | 4 |
With >50% of cells viable.
ND, not determined.
DISCUSSION
The CPE reduction assay of HCoV-NL63-infected LLC-MK2 cells provides an easy and reproducible method for the evaluation of candidate antiviral compounds. We selected antiviral compounds that potentially target different stages of the HCoV-NL63 life cycle. Of the 28 compounds tested, we identified 6 compounds that effectively inhibit HCoV-NL63 replication. These compounds interfere at an early stage of virus replication: receptor binding, virus-cell membrane fusion, cytoplasmic stability of viral RNA, and transcription (Table 2).
All of the serum samples tested for the presence of neutralizing antibodies against HCoV-NL63 were positive. Thus, it is not surprising that we also measured potent inhibition with IVIG, which is consistent for 95% of pooled human IgGs isolated from the sera of healthy donors. IVIG is approved as an intravenously delivered drug by the Food and Drug Administration and is successfully used to treat several diseases, mostly primary immune deficiencies and autoimmune neuromuscular disorders but also respiratory diseases (e.g., respiratory syncytial virus) (33) and Kawasaki disease (50). The effectiveness of IVIG therapy for Kawasaki disease supports the recent claim that HCoV-NL63 is the causative agent of this disease (28), although it does not provide independent evidence for such a correlation, especially because there is accumulating evidence against such an association (7, 26, 49). IVIG treatment may be applied to severe HCoV-NL63-related diseases, as the in vitro inhibitory concentration is about 10 times lower than the therapeutic dose advised for treatment.
The spike proteins of coronaviruses are class I fusion proteins that exhibit a characteristic membrane fusion mechanism that is driven by conformational changes in the spike protein. The association of the HR1 and HR2 domains brings the fusion peptide that is located near the N terminus of HR1 in close proximity to the transmembrane domain, thereby facilitating membrane fusion. Peptides corresponding to the HR1 and HR2 domains were found to associate tightly with the prefusion complex, thus blocking the conformational switch, as has been observed previously for retrovirus and paramyxovirus fusion proteins (10, 24). An NL63-HR2 peptide was able to inhibit HCoV-NL63 infection of LLC-MK2 cells in a concentration-dependent manner. The effect is supposedly mediated by the competitive binding of NL63-HR2 to the HR1 region of the HCoV-NL63 spike protein, thus blocking the conformational switch and, consequently, the close apposition of the fusion peptide and transmembrane domain and, hence, membrane fusion. The NL63-HR2 peptide shows an antiviral potency against HCoV-NL63 similar to that of an MHV-directed HR2 peptide against MHV (13) but is more potent than the SARS-HR2 peptide (12) (IC50 values of 0.5, 0.9, and 17.0 μM, respectively). We present the first report that HR regions present in the S protein of group 1 coronaviruses, which typically contain a 14-amino-acid insert compared to group 2 coronaviruses, associate into complexes and function similarly to group 2 HR peptides (12, 13). The success of the antiviral T20 peptide against HIV-1 demonstrates the clinical potential of this class of new antivirals.
Following fusion with the host cellular membrane, viral RNA is released into the cytoplasm of the host cell. We found that targeting HCoV-NL63 RNA by employing the RNA interference machinery and transfection of cells with two siRNAs specific for HCoV-NL63 resulted in a profound inhibition of viral replication. The targeted RNA encodes the S glycoprotein, which initiates entry of the virus into susceptible cells; entry is medicated by binding to the cellular receptor, which leads to membrane fusion. The choice of the S gene as a target was also based on the theoretical sequence requirements for an effective siRNA. To avoid the possibility that sequence variation among different HCoV-NL63 strains might restrict the inhibitory effect, we chose well-conserved target sequences in the S gene. The effectiveness of siRNA against respiratory tract diseases in a therapeutic setting was demonstrated recently by the intranasal administration of siRNA targeting respiratory syncytial virus, parainfluenzavirus, and SARS-CoV, with or without transfection reagents, in mouse and monkey models (9, 39, 59). Inhaled siRNA in low doses may offer a fast, potent, and easily administered antiviral tool against HCoV-NL63 infection in humans.
The HCoV-NL63 positive-strand RNA is copied by the viral-RNA-dependent RNA polymerase via a negative-strand intermediate. We tested two pyrimidine nucleoside analogues that could potentially interfere with transcription: β-d-N4-hydroxycytidine and 6-azauridine. Nucleoside analogues may be incorporated in the new nascent strand during transcription and cause chain termination. Several pyrimidine ribonucleoside analogs, including 6-azauridine (44, 57), act as antimetabolites, exerting pharmacological effects in their monophosphate forms by inhibiting UMP synthase (17) and thereby interfering with UTP metabolism. Additionally, incorporation of the nucleoside analogues may change the processivity and fidelity of transcription. This change results in an increased mutagenicity rate that forces the replicon into “error catastrophe,” as described previously for ribavirin (1, 20). Nucleoside analogues are known for their inhibition of several types of viruses, including HIV, pestivirus, hepacivirus, flaviviruses, hepatitis C virus, West Nile virus, feline infectious peritonitis virus, and SARS-CoV (4, 5, 35, 42, 48, 51, 52). Both compounds show very potent antiviral activities, with IC50 values of 35 and 400 nM for β-d-N4-hydroxycytidine and 6-azauridine, respectively. Relatively high cytotoxicity is compensated for by low IC50 values and thus a high selectivity index.
Recent data indicate that HCoV-NL63 is the most prevalent human coronavirus that is associated with acute respiratory diseases, croup, and possibly Kawasaki disease in children. The lack of an effective vaccine or drug motivated us to design and evaluate therapeutic agents that could inhibit viral replication and thus provide a potential therapy for treating acute respiratory illness of children and immunocompromised patients. Combined with fast diagnostic tools to recognize HCoV-NL63 infection, these antivirals may provide a more appropriate therapy than the routine treatments with steroids, adrenaline, and antibiotics. The agents described in this report may be used in a mono- or multidrug therapy setting, thereby inhibiting viral infection at different stages of the replication cycle.
Acknowledgments
We thank Joost Haasnoot for helpful comments and Wim van Est for photographic support.
We declare no competing interests.
REFERENCES
- 1.Anderson, J. P., R. Daifuku, and L. A. Loeb. 2004. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 58:183-205. [DOI] [PubMed] [Google Scholar]
- 2.Arden, K. E., M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2005. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 75:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlough, J. E., and B. L. Shacklett. 1994. Antiviral studies of feline infectious peritonitis virus in vitro. Vet. Rec. 135:177-179. [DOI] [PubMed] [Google Scholar]
- 4.Barnard, D. L., V. D. Hubbard, J. Burton, D. F. Smee, J. D. Morrey, M. J. Otto, and R. W. Sidwell. 2004. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARSCoV) by calpain inhibitors and beta-D-N4-hydroxycytidine. Antivir. Chem. Chemother. 15:15-22. [DOI] [PubMed] [Google Scholar]
- 5.Barreiro, P., T. Garcia-Benayas, A. Rendon, S. Rodriguez-Novoa, and V. Soriano. 2004. Combinations of nucleoside/nucleotide analogues for HIV therapy. AIDS Rev. 6:234-243. [PubMed] [Google Scholar]
- 6.Bastien, N., K. Anderson, L. Hart, P. van Caeseele, K. Brandt, D. Milley, T. Hatchette, E. C. Weiss, and Y. Li. 2005. Human coronavirus NL63 infection in Canada. J. Infect. Dis. 191:503-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belay, E. D., D. D. Erdman, L. J. Anderson, T. C. Peret, S. J. Schrag, B. S. Fields, J. C. Burns, and L. B. Schonberger. 2005. Kawasaki disease and human coronavirus. J. Infect. Dis. 192:352-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bermejo Martin, J. F., J. L. Jimenez, and A. Munoz-Fernandez. 2003. Pentoxifylline and severe acute respiratory syndrome (SARS): a drug to be considered. Med. Sci. Monit. 9:SR29-SR34. [PubMed] [Google Scholar]
- 9.Bitko, V., A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 11:50-55. [DOI] [PubMed] [Google Scholar]
- 10.Blacklow, S. C., M. Lu, and P. S. Kim. 1995. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry 34:14955-14962. [DOI] [PubMed] [Google Scholar]
- 11.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosch, B. J., B. E. Martina, R. van der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 101:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch, B. J., R. van der Zee, C. A. M. de Haan, and P. J. M. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu, S. S., K. H. Chan, K. W. Chu, S. W. Kwan, Y. Guan, L. L. Poon, and J. S. Peiris. 2005. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 40:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu, C. M., V. C. Cheng, I. F. Hung, M. M. Wong, K. H. Chan, K. S. Chan, R. Y. Kao, L. L. Poon, C. L. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2004. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax 59:252-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cihak, A., and B. Rada. 1976. Uridine kinase: properties, biological significance and chemotherapeutic aspects (a review). Neoplasma 23:233-257. [PubMed] [Google Scholar]
- 18.Cinatl, J., B. Morgenstern, G. Bauer, P. Chandra, H. Rabenau, and H. W. Doerr. 2003. Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus. Lancet 361:2045-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crance, J. M., N. Scaramozzino, A. Jouan, and D. Garin. 2003. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antivir. Res. 58:73-79. [DOI] [PubMed] [Google Scholar]
- 20.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Groot, R. J., W. Luytjes, M. C. Horzinek, B. A. van der Zeijst, W. J. Spaan, and J. A. Lenstra. 1987. Evidence for a coiled-coil structure in the spike proteins of coronaviruses. J. Mol. Biol. 196:963-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dreyer, C., and P. Hausen. 1978. Inhibition of mammalian RNA polymerase by 5,6-dichlororibofuranosylbenzimidazole (DRB) and DRB triphosphate. Nucleic Acids Res. 5:3325-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durandy, A., V. Wahn, S. Petteway, and E. W. Gelfand. 2005. Immunoglobulin replacement therapy in primary antibody deficiency diseases—maximizing success. Int. Arch. Allergy Immunol. 136:217-229. [DOI] [PubMed] [Google Scholar]
- 24.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebihara, T., R. Endo, X. Ma, N. Ishiguro, and H. Kikuta. 2005. Detection of human coronavirus NL63 in young children with bronchiolitis. J. Med. Virol. 75:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebihara, T., R. Endo, X. Ma, N. Ishiguro, and H. Kikuta. 2005. Lack of association between New Haven coronavirus and Kawasaki disease. J. Infect. Dis. 192:351-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 28.Esper, F., E. D. Shapiro, C. Weibel, D. Ferguson, M. L. Landry, and J. S. Kahn. 2005. Association between a novel human coronavirus and Kawasaki disease. J. Infect. Dis. 191:499-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallick, R. B., B. K. Chelm, P. W. Gray, and E. M. Orozco, Jr. 1977. Use of aurintricarboxylic acid as an inhibitor of nucleases during nucleic acid isolation. Nucleic Acids Res. 4:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanson, L. A., J. Bjorkander, C. Wadsworth, and B. Bake. 1982. Intravenous immunoglobulin in antibody deficiency syndromes. Lancet i:396. [DOI] [PubMed] [Google Scholar]
- 32.He, R., A. Adonov, M. Traykova-Adonova, J. Cao, T. Cutts, E. Grudesky, Y. Deschambaul, J. Berry, M. Drebot, and X. Li. 2004. Potent and selective inhibition of SARS coronavirus replication by aurintricarboxylic acid. Biochem. Biophys. Res. Commun. 320:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hemming, V. G., W. Rodriguez, H. W. Kim, C. D. Brandt, R. H. Parrott, B. Burch, G. A. Prince, P. A. Baron, R. J. Fink, and G. Reaman. 1987. Intravenous immunoglobulin treatment of respiratory syncytial virus infections in infants and young children. Antimicrob. Agents Chemother. 31:1882-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 102:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollecker, L., H. Choo, Y. Chong, C. K. Chu, S. Lostia, T. R. McBrayer, L. J. Stuyver, J. C. Mason, J. Du, S. Rachakonda, J. Shi, R. F. Schinazi, and K. A. Watanabe. 2004. Synthesis of beta-enantiomers of N4-hydroxy-3′-deoxypyrimidine nucleosides and their evaluation against bovine viral diarrhoea virus and hepatitis C virus in cell culture. Antivir. Chem. Chemother. 15:43-55. [DOI] [PubMed] [Google Scholar]
- 36.Hunt, D. M., and R. R. Wagner. 1975. Inhibition by aurintricarboxylic acid and polyethylene sulfonate of RNA transcription of vesicular stomatitis virus. J. Virol. 16:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawasaki, T., F. Kosaki, S. Okawa, I. Shigematsu, and H. Yanagawa. 1974. A new infantile acute febrile mucocutaneous lymph node syndrome (MLNS) prevailing in Japan. Pediatrics 54:271-276. [PubMed] [Google Scholar]
- 38.Lewis, E. L., D. A. Harbour, J. E. Beringer, and J. Grinsted. 1992. Differential in vitro inhibition of feline enteric coronavirus and feline infectious peritonitis virus by actinomycin D. J. Gen. Virol. 73:3285-3288. [DOI] [PubMed] [Google Scholar]
- 39.Li, B. J., Q. Tang, D. Cheng, C. Qin, F. Y. Xie, Q. Wei, J. Xu, Y. Liu, B. J. Zheng, M. C. Woodle, N. Zhong, and P. Y. Lu. 2005. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 11:944-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moes, E., L. Vijgen, E. Keyaerts, K. Zlateva, S. Li, P. Maes, K. Pyrc, B. Berkhout, L. van der Hoek, and M. Van Ranst. 2005. A novel pancoronavirus RT-PCR assay: frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 5:6. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgenstern, B., M. Michaelis, P. C. Baer, H. W. Doerr, and J. Cinatl, Jr. 2005. Ribavirin and interferon-beta synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 326:905-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrey, J. D., D. F. Smee, R. W. Sidwell, and C. Tseng. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antivir. Res. 55:107-116. [DOI] [PubMed] [Google Scholar]
- 43.Oates-Whitehead, R. M., J. H. Baumer, L. Haines, S. Love, I. K. Maconochie, A. Gupta, K. Roman, J. S. Dua, and I. Flynn. 2003. Intravenous immunoglobulin for the treatment of Kawasaki disease in children. Cochrane Database Syst. Rev. 2003(4):CD004000. [Online.] doi: 10.1002/14651858.CD004000. [DOI] [PMC free article] [PubMed]
- 44.Pasternak, C. A., G. A. Fischer, and R. E. Handschumacher. 1961. Alterations in pyrimidine metabolism in L5178Y leukemia cells resistant to 6-azauridine. Cancer Res. 21:110-117. [PubMed] [Google Scholar]
- 45.Pyrc, K., M. F. Jebbink, B. Berkhout, and L. van der Hoek. 2004. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 47.Rowley, A. H., and S. T. Shulman. 1999. Kawasaki syndrome. Pediatr. Clin. N. Am. 46:313-329. [DOI] [PubMed] [Google Scholar]
- 48.Schinazi, R. F., J. Mellors, H. Bazmi, S. Diamond, S. Garber, K. Gallagher, R. Geleziunas, R. Klabe, M. Pierce, M. Rayner, J.-T. Wu, H. Zhang, J. Hammond, L. Bacheler, D. J. Manion, M. J. Otto, L. Stuyver, G. Trainor, D. C. Liotta, and S. Erickson-Viitanen. 2002. DPC 817: a cytidine nucleoside analog with activity against zidovudine- and lamivudine-resistant viral variants. Antimicrob. Agents Chemother. 46:1394-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu, C., H. Shike, S. C. Baker, F. Garcia, L. van der Hoek, T. W. Kuijpers, S. L. Reed, A. H. Rowley, S. T. Shulman, H. K. Talbot, J. V. Williams, and J. C. Burns. 2005. Human coronavirus NL63 is not detected in the respiratory tracts of children with acute Kawasaki disease. J. Infect. Dis. 192:1767-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stiehm, E. R., E. Ashida, K. S. Kim, D. J. Winston, A. Haas, and R. P. Gale. 1987. Intravenous immunoglobulins as therapeutic agents. Ann. Intern. Med. 107:367-382. [DOI] [PubMed] [Google Scholar]
- 51.Stuyver, L. J., S. Lostia, M. Adams, J. S. Mathew, B. S. Pai, J. Grier, P. M. Tharnish, Y. Choi, Y. Chong, H. Choo, C. K. Chu, M. J. Otto, and R. F. Schinazi. 2002. Antiviral activities and cellular toxicities of modified 2′,3′-dideoxy-2′,3′-didehydrocytidine analogues. Antimicrob. Agents Chemother. 46:3854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuyver, L. J., T. Whitaker, T. R. McBrayer, B. I. Hernandez-Santiago, S. Lostia, P. M. Tharnish, M. Ramesh, C. K. Chu, R. Jordan, J. Shi, S. Rachakonda, K. A. Watanabe, M. J. Otto, and R. F. Schinazi. 2003. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 47:244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki, A., M. Okamoto, A. Ohmi, O. Watanabe, S. Miyabayashi, and H. Nishimura. 2005. Detection of human coronavirus-NL63 in children in Japan. Pediatr. Infect. Dis. J. 24:645-646. [DOI] [PubMed] [Google Scholar]
- 54.Vabret, A., T. Mourez, J. Dina, L. van der Hoek, S. Gouarin, J. Petitjean, J. Brouard, and F. Freymuth. 2005. Human coronavirus NL63, France. Emerg. Infect. Dis. 11:1225-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vesely, J., and A. Cihak. 1973. Resistance of mammalian tumour cells toward pyrimidine analogues. A review. Oncology 28:204-226. [DOI] [PubMed] [Google Scholar]
- 58.Wu, C. Y., J. T. Jan, S. H. Ma, C. J. Kuo, H. F. Juan, Y. S. Cheng, H. H. Hsu, H. C. Huang, D. Wu, A. Brik, F. S. Liang, R. S. Liu, J. M. Fang, S. T. Chen, P. H. Liang, and C. H. Wong. 2004. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 101:10012-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, W., H. Yang, X. Kong, S. Mohapatra, H. Juan-Vergara, G. Hellermann, S. Behera, R. Singam, R. F. Lockey, and S. S. Mohapatra. 2005. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11:56-62. [DOI] [PubMed] [Google Scholar]