Abstract
Assessing bacterial viability by molecular markers might help accelerate the measurement of antibiotic-induced killing. This study investigated whether rRNA could be suitable for this purpose. Cultures of penicillin-susceptible and penicillin-tolerant (Tol1 mutant) Streptococcus gordonii were exposed to mechanistically different penicillin and levofloxacin. Bacterial survival was assessed by viable counts and compared to quantitative real-time PCR amplification of either the 16S rRNA genes or the 16S rRNA, following reverse transcription. Penicillin-susceptible S. gordonii lost ≥4 log10 CFU/ml of viability over 48 h of penicillin treatment. In comparison, the Tol1 mutant lost ≤1 log10 CFU/ml. Amplification of a 427-bp fragment of 16S rRNA genes yielded amplicons that increased proportionally to viable counts during bacterial growth but did not decrease during drug-induced killing. In contrast, the same 427-bp fragment amplified from 16S rRNA paralleled both bacterial growth and drug-induced killing. It also differentiated between penicillin-induced killing of the parent and the Tol1 mutant (≥4 log10 CFU/ml and ≤1 log10 CFU/ml, respectively) and detected killing by mechanistically unrelated levofloxacin. Since large fragments of polynucleotides might be degraded faster than smaller fragments, the experiments were repeated by amplifying a 119-bp region internal to the original 427-bp fragment. The amount of 119-bp amplicons increased proportionally to viability during growth but remained stable during drug treatment. Thus, 16S rRNA was a marker of antibiotic-induced killing, but the size of the amplified fragment was critical for differentiation between live and dead bacteria.
Classical techniques to identify bacterial pathogens include phenotyping of live bacteria, specific immunostaining, and indirect measurement of the host's serologic response. However, cultures can remain negative in the case of fastidious organisms or after antibiotic administration. Immunostaining is not always specific and is highly dependent on sampling. Serology provides a delayed diagnostic answer and can also lack specificity.
Recent molecular methods may help circumvent some of these limitations. They are aimed at detecting either proteins or nucleic acids by direct molecular probing or by amplifying specific determinants, such as the rRNA genes, by PCR (1, 5, 12-14, 25, 27, 28, 35). They play an important role in the case of culture-negative infections (3, 13, 14). Moreover, they yield results within hours, compared to days or sometimes weeks with conventional phenotypic techniques. Yet, none of these methods provide information on the viability of the infecting microorganisms.
Determination of bacterial viability is critical to monitoring the bactericidal activity of antibiotics in vitro and in vivo as well as assessing the presence of viable pathogens in contaminated food or environmental samples (2, 13, 20, 21, 24). Molecular markers of viability might be useful to follow microbiological cure in a variety of clinical situations and to rapidly appraise drug efficacy. In this regard, notoriously problematic pathogens include Mycobacterium tuberculosis and Mycobacterium leprae, for which determining drug-induced killing by standard techniques takes between weeks and months (18, 31, 38).
Several physiological and molecular reporters of viability have been explored. These include measurement of cellular integrity (e.g., by vital staining), measurement of metabolic activities (e.g., active electron transport chain, transport of glucose, and esterase activity), measurement of synthetic activities (e.g., polymer synthesis and cell elongation) (37), and molecular probing or amplification of DNA, mRNA, and 16S rRNA (20). DNA amplification appeared particularly unreliable because it can remain positive for extensive periods of time, in spite of effective bacterial killing (4, 7, 15, 16, 19-21, 29, 30, 34, 36, 37). In addition, most other methods provided conflicting results that varied depending on the experimental conditions, including whether bacteria were killed by antibacterial drugs (15, 20, 36) or by physical means (e.g., heat and UV irradiation) (24, 32, 37). Thus, there are currently no reliable molecular tools for routine determination of cell viability and drug-induced killing in medical microbiology.
The present experiments explored the correlation between bacterial viability and the number of copies of 16S rRNA, as determined by reverse transcription followed by quantitative real-time PCR, during bacterial growth and antibiotic-induced killing of Streptococcus gordonii. This organism was used as a model because it is poorly lysed by penicillin and provides a well-described isogenic pair of kill-susceptible (parent) and kill-resistant (tolerant mutant Tol1) strains (6, 22). Studies were performed with two mechanistically unrelated compounds, i.e., penicillin G and the quinolone levofloxacin. A specific fragment of 16S rRNA of a specific size appeared suitable to quantify drug-induced killing.
MATERIALS AND METHODS
Microorganisms and growth conditions.
A described streptomycin-resistant S. gordonii Challis strain and its penicillin-tolerant mutant Tol1 were used as model organisms (6, 22). Streptococci were grown at 37°C without aeration either in brain heart infusion (Difco Laboratories, Detroit, MI) supplemented with 200 mg/liter of streptomycin (in order to respect the experimental conditions previously described for these isolates) (6, 22) or on Columbia agar (Becton Dickinson Microbiology Systems, Cockeysville, MD) supplemented with 3% blood. Escherichia coli XL1-Blue cells were grown at 37°C in Luria-Bertani (LB) broth (Difco) or on LB agar (Difco), supplemented with 50 mg/liter of ampicillin (Sigma Chemicals). Stocks were stored at −70°C in culture medium supplemented with 10% (vol/vol) glycerol. Bacterial growth was monitored by determining optical density at 620 nm with a spectrophotometer (Sequoia-Turner, Montainville, CA) and colony count on agar plates. When appropriate, penicillin G (Hoechst-Pharma AG, Zurich, Switzerland) and levofloxacin (Aventis Pharma Ltd., Romainville, France) were added to the medium at final concentrations of 2 mg/liter and 12.5 mg/liter, respectively, mimicking a high-dose treatment in human. The MICs of these antibacterials for the test bacteria were 0.004 and 0.5 mg/liter, respectively (6, 11).
Antibiotics and chemicals.
Streptomycin was purchased from Sigma AG (Buchs, Switzerland), penicillin G from Hoechst-Pharma AG (Zurich, Switzerland), and levofloxacin from Aventis Pharma Ltd. (Romainville, France). The restriction enzymes (Boehringer Mannheim, Germany), Taq DNA polymerase (Gibco BRL, Gaithersburg, MD), and T4 DNA ligase (Gibco) were used according to the manufacturer's recommendations. Nucleic acid sequencing and synthesis were performed by Microsynth GmbH (Balgach, Switzerland). All other chemicals were reagent-grade, commercially available products.
Antibiotic susceptibility and time-kill curves.
The MICs were determined by standard macrodilution methods (26). Time-kill curves were determined by adding appropriate concentrations of antibiotics to bacterial cultures in the exponential phase of growth at an optical density of 620 nm of 0.2 (6, 22). At various time points before and after drug addition, samples were removed and processed (i) for viable count, (ii) for DNA extraction, and (iii) for RNA extraction. For viable count, antibiotic carryover on the agar plates was avoided, as described previously (10, 11). Colonies were counted after 48 h of incubation at 37°C. DNA was extracted from frozen culture samples kept at −70°C, whereas RNA was isolated directly from fresh samples.
DNA extraction and purification.
Total DNA from 3 ml of culture samples was extracted and purified using a DNeasy tissue kit according to the manufacturer's instructions (QIAGEN GmbH, Hilden, Germany). Lysis of streptococci was performed as follows. Samples were centrifuged at 14,000 rpm for 15 min at 4°C. Pellets were washed two times with NaCl, 0.9%, and resuspended in 220 μl buffer (Tris-HCl, 20 mM [pH 8.0]; EDTA, 5 mM; 25% sucrose; and 17 μg/ml of lysozyme [Sigma]). The samples were then incubated for 30 min at 37°C before the addition of 200 μl of lysis buffer (buffer AL; QIAGEN) supplemented with final concentrations of 0.2% of sodium dodecyl sulfate (Sigma) and 1 mg/ml of proteinase K (Sigma). After an additional 30 min of incubation at 70°C, DNA was purified according to standard protocol (QIAGEN), resuspended in a 200-μl final volume, and kept frozen at −20°C. Input volumes for further amplification consisted of 1.5-μl aliquots of the stored samples.
RNA extraction and purification.
For total RNA purification, 9 ml of fresh culture samples was centrifuged at 10,000 rpm for 8 min at 4°C and processed according to the FastRNA BLUE protocol of Bio 101 (Bio 101 Inc., La Jolla, CA), modified as follows. Bacterial pellets were resuspended in 500 μl of CRSR-Blue reagent (Bio 101, Inc.) and transferred into tubes containing ceramic beads, supplemented with 500 μl of phenol acid reagent and 100 μl of a solution of 24:1 chloroform-isoamyl alcohol. The samples were further processed at 4°C by using a FastPrep FP120 apparatus (Bio 101/Savant Instruments, Inc., Holbrook, NY) for 25 s at a speed of 6.5 m/s before being centrifuged at 14,000 rpm for 10 min. The aqueous phase was collected and added to 500 μl of the chloroform-isoamyl alcohol solution. The samples were then centrifuged at 14,000 rpm for 5 min. The aqueous phase was collected and mixed with 350 μl of RLT buffer (RNeasy Mini kit; QIAGEN) supplemented with 1% β-mercaptoethanol and 250 μl RNase-free ethanol (96 to 100%). Total RNA was further purified according to the standard RNase-free DNase set protocol of QIAGEN, resuspended in a 50-μl final volume, and kept frozen at −80 C. Input volumes for further amplification consisted of 20-μl aliquots of the stored samples.
The yield of RNA was quantified in each experiment by absorbance at 260 nm (Biophotometer ThermoStat Plus; Eppendorf AG, Germany). As an additional control, the presence of rRNA in the extracts was qualitatively assessed by electrophoresis and ethidium bromide staining on a 1.2% agarose-0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) running buffer (Sigma), next to a molecular weight ladder.
Construction of molecular standards for quantitative real-time PCR.
To build standard curves for both real-time PCR and reverse transcriptase PCR (RT-PCR), it was important to first generate control molecules with known molar concentrations. First, a DNA standard was generated by cloning the PCR product corresponding to the 427-bp fragment of the 16S rRNA genes of S. gordonii into the pGemT-Easy vector system (Promega Corporation, Madison, Wis.) according to the manufacturer's instructions and the construct was transformed in E. coli by the standard technique. Effective amplicon ligation by T4 DNA ligase (Gibco BRL, Gaithersburg, Md.) was verified by standard restriction enzymes, amplification was performed by using a Wizard Midiprep kit (Promega), and the nature of the insert was controlled by DNA sequencing. Pure DNA extract was quantified and its molar concentration determined with the following formula: 1 μg of 1,000 bp DNA = 1.52 pmol = 1.52 × 10−12 mol × N molecules (where N = Avogadro's number, or 6.023 × 1023 molecules/mol). Different solutions were prepared (101 to 1012 molecules/μl) and used as standard solutions in the subsequent experiments.
Second, an RNA standard was generated by in vitro transcription of the cloned fragment described above, using a Riboprobe system T7 (Promega) according to the manufacturer's instructions. In brief, linearized plasmids containing the 427-bp sequence (and part of the T7 promoter) were separated and purified on agarose gels by using a QIAquick gel extraction kit (QIAGEN). Small fragments of RNA (539 bp) were synthesized in a 50-μl reaction using T7 RNA polymerase. DNA was removed by addition of RQ1 RNase-free DNase (1 U/1 μg template DNA; QIAGEN) and incubation for 15 min at 37°C. RNA was extracted and purified with an SV Total RNA isolation system (Promega) according to the “spin protocol” and visualized on a MOPS agarose gel. This preparation was quantified and its molar concentration determined with the following formula: 1 μg of 1,000 bp RNA = 2.94 pmol = 2.94 × 10−12 mol × N molecules (where N = Avogadro's number, or 6.023 × 1023 molecules/mol). Different solutions were prepared (101 to 1012 molecules/μl) and subjected to RT-PCR and quantitative real-time PCR.
Nonquantitative PCR amplification of 16S rRNA genes.
A 427-bp fragment (corresponding to nucleotides 88 to 514) of the S. gordonii 16S rRNA genes (GenBank accession number D38483) was amplified by PCR using the primer pair, 5′-CCA TAG ACT GTG AGT TGC GAAC-3′and 3′-CCG TCC CTT TCT GGT AAG ATAC-5′ on a 2400 GeneAmp PCR system apparatus (Perkin-Elmer) with a 1.5-μl volume from the stored DNA culture extracts, 0.5 μM of specific primers (Microsynth, Balgach, Switzerland), 0.3 mM of each deoxynucleoside triphosphate in PCR buffer (Life Technologies AG, Basel, Switzerland), 1.5 mM MgCl2, and 2 IU of Taq DNA polymerase (Life Technologies AG). Amplification was performed for 25 cycles under the following conditions: 94°C for 30 s, 50°C for 30 s, and 72°C for 20 s. Quantities of polynucleotides were assessed on an ethidium bromide-containing 1% agarose gel by using standard techniques of scanning densitometry at a wavelength of 280 with an EagleEye II still-video system and EagleSight software (Stratagene, La Jolla, CA).
Reverse transcriptase amplification.
Prior to quantification, RT-PCR was performed on all RNA samples by using an Omniscript RT kit (QIAGEN) in a total reaction volume of 20 μl according to the manufacturer's instructions. Different solutions containing between 102 and 108 RNA molecules were prepared as described above. RT-PCRs of test and standard samples were run in parallel. Test samples consisted of 20-μl aliquots of the stored materials. RT-PCR efficiency was 30% and was calculated as follows: number of molecules of cDNA/number of molecules of RNA × 100%. The number of RNA molecules of each unknown sample was calculated on the basis of both the standard curve (quantitative real-time PCR) and the RT-PCR efficiency. Each standard and test sample was run in quintuplicate and processed in parallel for further quantification in order to appraise the interexperimental relative variation. This variation was ≤20%. The results are expressed as the mean values from these quintuplicate experiments.
Quantitative evaluation by real-time PCR.
Determination of 16S rRNA gene copy numbers was performed using the probe 6-carboxyfluorescein-5′-TTG CAC CAC TAC CAG ATG GAC CTGC-3′-6-carboxytetramethylrhodamine (nucleotides 220 to 244) on a sequence detection system 5700 (Perkin-Elmer) according to the manufacturer's instructions. The fragment from nucleotides 88 to 514 (427 bp) of the 16S rRNA genes was amplified using the above-described primer pair. In certain experiments, a shorter, 119-bp internal portion of the 427-bp fragment was amplified (corresponding to nucleotides 158 to 276 of the S. gordonii 16S rRNA genes), using the primers 5′-GGA AAC GAT AGC TAA TAC CGC ATAA-3′ and 5′-AAT CGA TCA TCC ACT CCA TTG CCG AG-3′. Reactions were started on a sequence detection system 5700 (Perkin-Elmer) in a total volume of 50 μl 1× PCR buffer (Gibco) containing 0.5 μM of each primer, 120 nM of the fluorescent probe, 0.3 mM of each deoxynucleoside triphosphate, 2.5 mM MgCl2, and 1.5 IU of Taq DNA polymerase (Life Technologies AG). The following program was applied during 40 cycles: 94°C for 15 s, 52°C for 30 s, and 72°C for 30 s. Each experiment was done in triplicate.
The yield of real-time PCR was assessed by plotting the cycle threshold values versus the log10 numbers of input DNA copies. In a typical experiment (six parallel runs for each sample), the mean slopes (±standard deviations) of the plots were −3.56 ± 0.1 for the 119-bp fragment and 3.9 ± 0.04 for the 427-bp fragment. This translates into yields of 98.6% and 80.2%, respectively, for the two fragments. Although these yields were different, they did not affect the interpretation of the results because amplicons resulting from the different fragments were not compared.
The DNA content of each sample was determined using a standard curve obtained by processing, in parallel, samples containing fixed DNA concentrations. Determination of DNA concentrations was performed by spectrophotometry, and molar concentrations were determined using the following formula: 1 μg of 1,000 bp DNA = 1.52 pmol = 1.52 × 10−12 mol × N molecules (where N stands for Avogadro's number, or 6.023 × 1023 molecules/mol). Different solutions were then accordingly prepared (108 to 102 molecules/μl) and used as standard solutions in the subsequent experiments.
Assessment of drug-induced killing by vital staining.
The parent S. gordonii strain and its Tol1 mutant were treated with penicillin as described above. Just before and 24 h after drug addition, samples (100 μl) were removed from the cultures, washed twice with phosphate-buffered saline buffer (by centrifugation), and processed both for viable counts and for vital staining according to the manufacturer's instructions (Live/Dead BacLight; Molecular Probes, Eugene, Oregon). In brief, bacterial samples (100 μl) were mixed with 0.3 μl of a mixture of equal parts of SYTO-9 and propidium iodide and incubated in the dark for 20 min. Two microliters of the stained suspension was deposited on a glass slide and covered with a coverslip. Image acquisition was performed with a confocal scanning laser microscope (model TCS SL; Leica Lasertechnik GmbH, Heidelberg, Germany). Confocal illumination was provided by Ar and HeNe lasers (488-nm and 543-nm laser excitation, respectively). The SYTO-9 signal was collected in the range from 503 to 523 nm and the propidium iodide signal between 575 and 630 nm. The sensitivities of the photomultipliers and the laser intensity were adjusted and thereafter kept constant throughout the experiments. Randomly selected regions of each sample were imaged using a 20× oil immersion objective. The area of each section was transformed into a digital image of 512 by 512 pixels.
RESULTS
Absence of correlation between viable counts, optical density, and nonquantitative PCR of 16S rRNA genes.
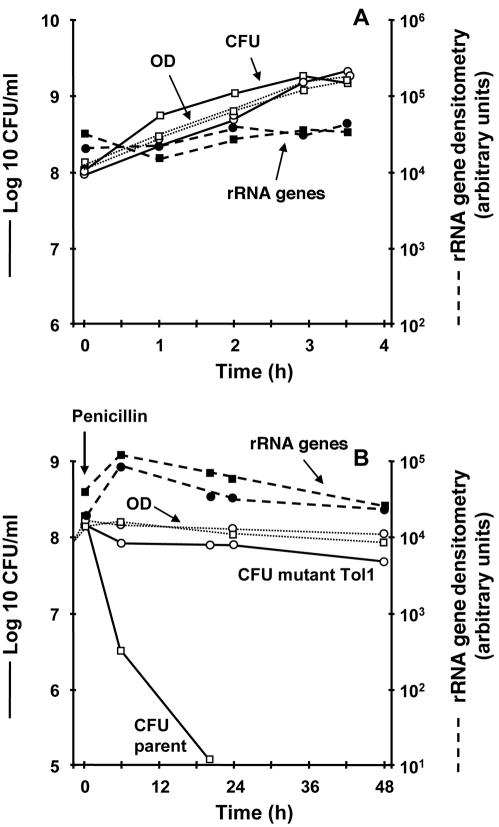
Physically intact bacteria are considered alive if they are able to give rise to progeny. Optical density is a correlate of bacterial mass, but bacteria forming this mass are not necessarily alive. Likewise, nonquantitative PCR amplification reveals the presence of specific polynucleotide fragments, but nonquantitative detection of polynucleotide fragments bears no linear correlation with bacterial mass or viability. Hence, these two methods are not likely to be good indicators of bacterial viability. This was confirmed in the present experimental setting, as a control (Fig. 1). Optical density increased proportionally to viable counts during bacterial growth (Fig. 1A). Yet, it barely decreased during penicillin treatment, in spite of a viability loss of ≥4 log10 CFU/ml within 48 h of drug exposure (Fig. 1B). Nonquantitative PCR amplification was even less representative of viability. Amplification of a 427-bp fragment of the 16S rRNA genes yielded quantities of amplicons that had no correlation with either bacterial mass or penicillin-induced killing (Fig. 1). Thus, more-precise techniques were needed.
FIG. 1.
Growth (A) and penicillin-induced killing (B) of the parent S. gordonii strain (squares) and its tolerant mutant Tol1 (circles) monitored by optical density (OD), viable counts, and nonquantitative PCR of 16S rRNA genes. (A) During growth, both optical densities and viable counts increased in parallel, while nonquantitative rRNA gene amplification remained stable. After addition of 500× MIC of penicillin G (B), the viable counts decreased sharply in the parent strain while remaining stable in the Tol1 mutant. In contrast, optical densities and rRNA gene densitometries remained steady for both organisms. The experiments were performed in triplicate on three independent occasions. Interexperimental variation was ≤20%.
Limited correlation between viable counts and quantitative real-time PCR of 16S rRNA genes.
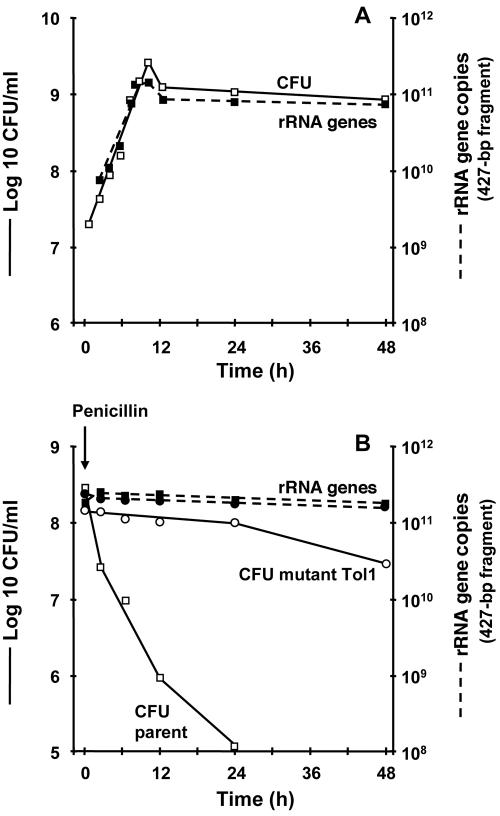
To improve quantitative assessment, we repeated the experiments using real-time PCR. Amplification of the same 427-bp fragment of the 16 rRNA genes yielded quantities of amplicons that increased proportionally to bacterial mass during logarithmic growth and stabilized after entry into stationary phase (Fig. 2). This correlated well with bacterial viability in the nontreated cultures (Fig. 2A) but not with penicillin-induced killing (Fig. 2B), as the quantities of real-time PCR products remained stable in spite of an extensive loss of viability of ≥4 log10 CFU/ml/48 h. Thus, the real-time PCR products were a good correlate of bacterial mass but not of bacterial viability sensu stricto. This confirms the good stability of DNA during nonlytic killing of bacteria by antibiotics (4, 7, 15, 16, 19-21, 29, 30, 34, 36, 37).
FIG. 2.
Bacterial growth (A) and penicillin-induced killing (B) monitored by viable counts and quantitative PCR of 16S rRNA genes. (A) Both viable counts and numbers of rRNA gene copies increased in parallel and stabilized in the stationary phase (data shown are for the parent S. gordonii strain). (B) After penicillin addition, viable counts decreased in the parent strain while remaining stable in the Tol1 mutant. Yet, quantitative PCR of rRNA genes remained steady for both organisms. Details and experimental reproducibility are described in the legend for Fig. 1.
Good correlation between viable counts and quantitative real-time PCR of 16S rRNA.
We next tested whether RNA would be more sensitive than DNA to monitor drug-induced killing. Both mRNA and rRNA were potential targets. However, mRNA has a very short half-life in bacteria. Hence, drugs inhibiting DNA transcription (e.g., rifampin) may result in a rapid drop in mRNA, irrespective of their bacteriostatic or bactericidal effects (15).
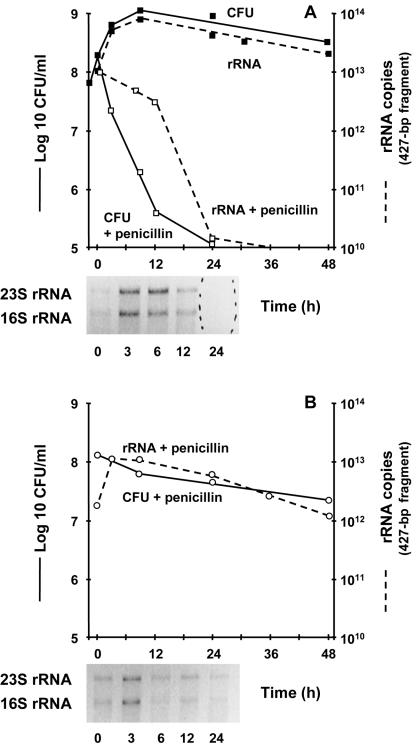
rRNA might be more appropriate. Therefore, we tested the stability of 16S rRNA as a potential marker of drug-induced killing. Total RNA was reverse transcribed, and the quantities of 16S rRNA were assessed by real-time PCR amplification of the 427-bp fragment from cDNA. This yielded quantities of amplicons that correlated very closely with viable counts during logarithmic growth and stationary phase and also correlated with drug-induced killing during penicillin treatment (Fig. 3). Moreover, the system could readily differentiate between the extensive drug-induced killing of the S. gordonii parent (Fig. 3A) and the kill-resistant phenotype of mutant Tol1 (Fig. 3B).
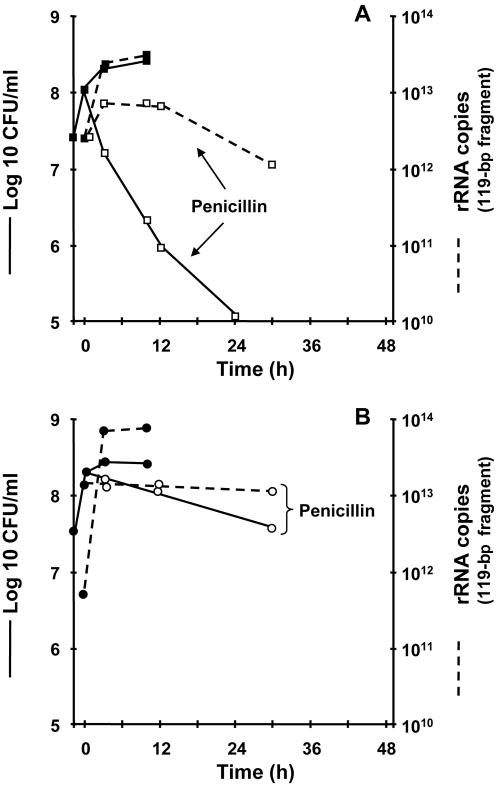
FIG. 3.
Growth and penicillin-induced killing of the S. gordonii parent (A) and penicillin-induced killing of the Tol1 mutant (B) monitored by viable counts and quantification of a 427-bp fragment of 16S rRNA. In both cases, the number of copies of rRNA was proportional to the viable counts. The insets at the bottom of the graphs depict crude 23S and 16S rRNA prepared from culture aliquots removed at various times during penicillin treatment and separated by agarose gel electrophoresis. In the kill-susceptible parent (A), the bands disappeared after 24 h of drug treatment. In the kill-resistant Tol1 mutant (B), the bands remained visible. Details and experimental reproducibility are described in the legend for Fig. 1.
Good correlation between real-time PCR of 16S rRNA and quinolone-induced killing.
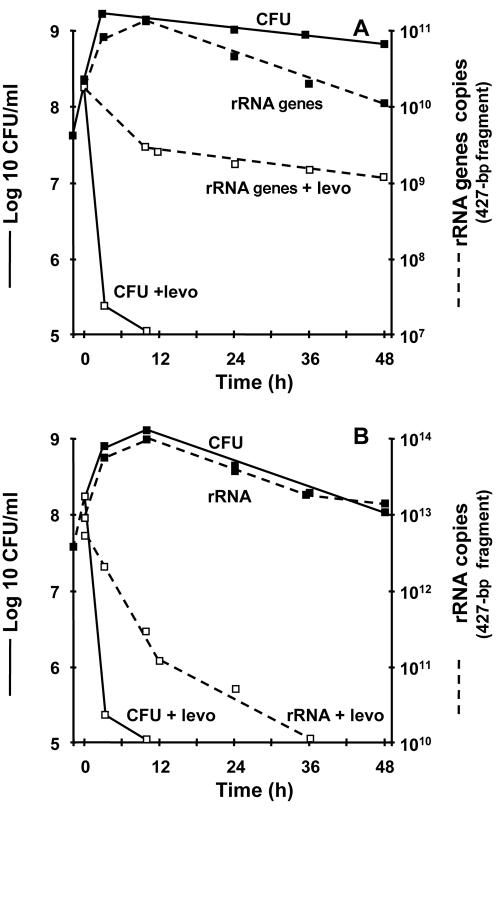
To assess whether degradation of 16S rRNA might also apply to killing by a mechanistically different antibiotic, we repeated the experiments with the quinolone levofloxacin. Quinolones generate breaks in the chromosomal ladder (17, 33) but are not expected to perturb rRNA. The stability of both the 16S rRNA genes (Fig. 4A) and the 16S rRNA (Fig. 4B) was tested by quantitative amplification of the 427-bp fragment described above. Killing by levofloxacin was fast, as previously reported (11). Amplification of both rRNA genes and rRNA paralleled bacterial viability during the logarithmic and stationary growth phases. After addition of levofloxacin, the quantities of rRNA gene amplicons remained remarkably stable, in spite of rapid drug-induced killing (>3 log10 CFU in 6 h) (Fig. 4A). In contrast, the quantities of 16S rRNA copies dropped by >3 log10, thus reflecting the bactericidal effect of the drug (Fig. 4B). Therefore, quantitative amplification of the 427-bp fragment of 16S rRNA correlated with killing by both cell wall inhibitors and DNA gyrase inhibitors.
FIG. 4.
Bacterial growth and levofloxacin (levo)-induced killing monitored by viable counts and quantification of either rRNA genes (A) or rRNA (B). (A) The number of copies of rRNA genes paralleled viable counts during bacterial growth but remained stable during levofloxacin treatment in spite of extensive killing. (B) In contrast, the number of copies of rRNA paralleled viable counts during both growth and drug-induced killing. Details and experimental reproducibility are described in the legend for Fig. 1.
Large fragments of rRNA are better markers of drug-induced killing than smaller fragments.
Since large polynucleotide fragments might be degraded faster than smaller ones (23), we repeated the 16S rRNA experiment by amplifying a shorter (119-bp) segment internal to the 427-bp fragment described above (Fig. 5). The quantities of 119-bp amplicons correlated with viable counts during logarithmic growth and stationary phase but decreased by only 1 order of magnitude (instead of ≥3 orders of magnitude) during penicillin-induced killing (Fig. 5A). Nevertheless, they still revealed a difference between the kill-susceptible parent and the Tol1 mutant (Fig. 5B). Thus, the shorter 119-bp fragment was less optimal than the longer 427-bp fragment for assessment of penicillin-induced killing and tolerance.
FIG. 5.
Bacterial growth and penicillin-induced killing of the S. gordonii parent (A) and Tol1 mutant (B) monitored by viable counts and quantification of a short (119-bp) fragment of 16S rRNA. The number of amplified 119-bp fragments increased proportionally to viable counts during bacterial growth but did not decrease proportionally to bacterial killing during penicillin treatment. Details and experimental reproducibility are described in the legend for Fig. 1.
Poor correlation between penicillin-induced killing and vital staining.
The parent S. gordonii strain and its Tol1 mutant were exposed to penicillin G as described above. Samples were removed just prior to and 24 h after drug addition and processed for both viable counts and vital staining. For each sample, 20 random areas containing 100 to 200 bacterial bodies were visually assessed by phase-contrast microscopy and counted for green (alive) and red (dead) staining. Before penicillin addition, all bacteria on the slides appeared in short chains of 2 to 10 cells at phase contrast. Accordingly, the number of green/red bacteria counted were 2,524/2,544 (98%) for the parent and 2,537/2,553 (99%) for the Tol1 mutant. After 24 h of penicillin treatment, the normal-looking organisms decreased by up to 300× in the penicillin-killed parent, as opposed to only 20× in the kill-tolerant mutant. Most bacteria appeared as single or double bulged cells, condensed ghosts, or intermediate forms. These forms were not stained by the BacLight system (dead bacteria should have appeared in red) and thus escaped the quantification by vital staining. Among the remaining forms, red/green counts were 670/1,165 (58%) for the parent (in spite of a killing of ≥3 log10 CFU/ml) and 2,166/2,924 (74%) for the Tol1 mutant, which had lost <1 log10 CFU/ml. As a control, heat-killed S. gordonii and Bacillus subtilis killed by membrane-active agents turned entirely red (data not shown). Thus, although combining phase-contrast microscopy and vital staining revealed a difference between live and antibiotic-killed bacteria, it was less accurate and more difficult to interpret than determining the numbers of copies of rRNA.
DISCUSSION
The present results disclosed a reproducibly good correlation between the loss of viable counts and the drop in the quantities of copies yielded by real-time PCR amplification of a 427-bp fragment of the 16S rRNA. Coherent results were obtained with both the parent strain and the tolerant mutant Tol1, as well as with mechanistically unrelated penicillin and levofloxacin, provided that a minimal (427-bp) fragment length of rRNA was used. A shorter (119-bp) template was much less reliable in differentiating between live and dead bacteria.
The experiments support the work by van der Vliet et al. (36), who observed that rRNA may be degraded during antibiotic-induced killing of Mycobacterium smegmatis treated with rifampin and ofloxacin. However, this pioneer report disclosed only a modest correlation between the viability loss and decay in rRNA, which underestimated drug-induced killing by >100× (36). The present molecular system was much more sensitive and predicted cell death at the same order of magnitude as actual killing. The principal difference between these two observations may rely on technical issues, with one experiment using nucleic acid sequence-based amplification (36) and one (presented herein) quantitative real-time PCR.
On the other hand, results from both the van der Vliet et al. report (36) and the present experiments are different from the results of Hellyer et al. (15), who studied the decrease in mRNA and 16S rRNA during treatment of M. tuberculosis with isoniazid and rifampin. These authors observed that cell death was accompanied by a sharp decrease in mRNA but found barely any decrease in 16S rRNA. However, rRNA was detected by sequential reverse transcription and nonquantitative PCR amplification of cDNA from a short (160-bp) rRNA fragment. This approach may have missed degradation of rRNA for at least two reasons. First, amplification was not quantitative, and second, the length of the rRNA fragment might have been suboptimal.
Both of these issues were critical in the present work. In particular, the inverse relation between polynucleotide length and lack of polynucleotide amplification is supported by earlier reports, for instance, during chlorine-induced killing of Legionella pneumophila (23). Long strands of nucleic acids have a greater chance than shorter strands to carry cleavage sites for endonucleases. In turn, cleaved strands cannot be amplified by PCR anymore. While DNA is very stable in spite of bacterial death (4, 7, 15, 16, 19-21, 29, 30, 34, 36, 37), rRNA may behave differently. In the present case, it is possible that antibiotic-induced killing allowed nonspecific deregulation of bacterial RNases (8, 9), leading to progressive RNA decay. Moreover, the kinetics of RNA decay were somewhat delayed compared to viability loss, thus complying with the assumption that RNA degradation was the consequence rather than the cause of cell death.
RNA degradation was not formally demonstrated herein. However, other causes of impairment of amplification seem less likely. Secondary structures impeding amplification are one alternative, but such structures should not have sustained the PCR conditions. Another alternative could involve inhibitors of amplification present only in antibiotic-killed cells. However, such inhibitors should also affect amplification of the smaller 119-bp internal fragment, which was not the case with these observations. Ultimately, the precise limit of RNA size useful for differentiation between live and dead bacteria was not determined. This size could indeed vary between different RNA stretches and between different bacteria. Nevertheless, the experiments described herein provide clear evidence that size must be taken into account.
Killing of Staphylococcus aureus and E. coli by physical or chemical means indicated that 16S rRNA was unstable after autoclaving (24) but was more stable after killing by less drastic conditions, such as lower temperatures (60 or 80°C), ethanol, or UV irradiation (24, 32). In one study (24), the authors used both Northern blotting and reverse transcription followed by nonquantitative PCR to detect rRNA. Fragment length (ca. 400 bp or larger) was not an issue. However, since cDNA was amplified nonquantitatively, the PCR data are difficult to interpret. On the other hand, Northern blotting clearly indicated a hierarchy of rRNA degradation, with total disappearance of rRNA at harsh (120°C) conditions, partial disappearance at less harsh (80°C) conditions, and minimal disappearance after UV treatment. It was suggested that the less drastic conditions could respect the physical integrity of the ribosomes, thus preventing them from attack by RNases or other enzymes (24). If so, then penicillin and levofloxacin treatments appear more like harsh conditions than milder conditions. Indeed, real-time PCR precisely detected both drug-induced killing and survival of the parent S. gordonii strain and its Tol1 mutant.
Ultimately, quantitative assessment of rRNA was a better marker of drug-induced killing than vital staining. In this particular case, the poor performance of vital staining was due to physical alterations of dying bacteria, which became unstainable by the coloration system. Although the effect might be more pronounced with cell wall active drugs than with other compounds, this kind of limit was also reported for other systems, especially when the plasma membrane is not physically altered (37).
Taken together, the present results revive the perspective of using molecular markers to detect antibiotic-induced killing of bacteria. They clarify certain differences between various reports and help delineate a molecular system that might detect drug-induced killing. An interesting observation was the inverse relationship between rRNA amplification and the length of the amplicons. This raises the possibility of combining both short and long amplicons in the same assay. Short amplicons would detect both the presence and the nature of the organism, while long amplicons would assess its viability. Calculating a ratio between the short and long fragments could provide information on the total overkilled cells both in vitro and in clinical samples.
Molecular tools are increasingly applied for rapid diagnosis, detection of noncultivable pathogens, and detection of antibiotic resistance genes. In the future they will also include virulence genes such as toxins. Yet, an important limit of current molecular methods is that they do not differentiate between live bacteria and dead remnants. The present observations indicate that analyzing rRNA fragments could help solve the issue. At this stage, the technique using RT-PCR followed by real-time PCR is still too complicated for clinical or field application. However, the results identify at least one of the molecular approaches that might be worth pursuing into engineering development.
Acknowledgments
This work was supported by grants no. 3200-65371.01 and 3200-65371/2 from the Swiss National Fund for Scientific Research.
We thank Marlyse Giddey, Sylvie Rossier, and Stéphane Piu for outstanding technical assistance.
REFERENCES
- 1.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birch, L., C. E. Dawson, J. H. Cornett, and J. T. Keer. 2001. A comparison of nucleic acid amplification techniques for the assessment of bacterial viability. Lett. Appl. Microbiol. 33:296-301. [DOI] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Ruef, E. C. Bottger, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin. Infect. Dis. 37:167-172. [DOI] [PubMed] [Google Scholar]
- 4.Branger, S., J. P. Casalta, G. Habib, F. Collard, and D. Raoult. 2003. Streptococcus pneumoniae endocarditis: persistence of DNA on heart valve material 7 years after infectious episode. J. Clin. Microbiol. 41:4435-4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, D. F. 2001. Detection of methicillin/oxacillin resistance in staphylococci. J. Antimicrob. Chemother. 48(Suppl. 1):65-70. [DOI] [PubMed] [Google Scholar]
- 6.Caldelari, I., B. Loeliger, H. Langen, M. P. Glauser, and P. Moreillon. 2000. Deregulation of the arginine deiminase (arc) operon in penicillin-tolerant mutants of Streptococcus gordonii. Antimicrob. Agents Chemother. 44:2802-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canvin, J. M., S. C. Goutcher, M. Hagig, C. G. Gemmell, and R. D. Sturrock. 1997. Persistence of Staphylococcus aureus as detected by polymerase chain reaction in the synovial fluid of a patient with septic arthritis. Br. J. Rheumatol. 36:203-206. [DOI] [PubMed] [Google Scholar]
- 8.Carpousis, A. J. 2002. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30:150-155. [PubMed] [Google Scholar]
- 9.Carpousis, A. J., A. Leroy, N. Vanzo, and V. Khemici. 2001. Escherichia coli RNA degradosome. Methods Enzymol. 342:333-345. [DOI] [PubMed] [Google Scholar]
- 10.Entenza, J. M., I. Caldelari, M. P. Glauser, P. Francioli, and P. Moreillon. 1997. Importance of genotypic and phenotypic tolerance in the treatment of experimental endocarditis due to Streptococcus gordonii. J. Infect. Dis. 175:70-76. [DOI] [PubMed] [Google Scholar]
- 11.Entenza, J. M., I. Caldelari, M. P. Glauser, and P. Moreillon. 1999. Efficacy of levofloxacin in the treatment of experimental endocarditis caused by viridans group streptococci. J. Antimicrob. Chemother. 44:775-786. [DOI] [PubMed] [Google Scholar]
- 12.Francois, P., D. Pittet, M. Bento, B. Pepey, P. Vaudaux, D. Lew, and J. Schrenzel. 2003. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauduchon, V., L. Chalabreysse, J. Etienne, M. Celard, Y. Benito, H. Lepidi, F. Thivolet-Bejui, and F. Vandenesch. 2003. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valve tissue. J. Clin. Microbiol. 41:763-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldenberger, D., A. Kunzli, P. Vogt, R. Zbinden, and M. Altwegg. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 35:2733-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellyer, T. J., L. E. DesJardin, G. L. Hehman, M. D. Cave, and K. D. Eisenach. 1999. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J. Clin. Microbiol. 37:290-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellyer, T. J., T. W. Fletcher, J. H. Bates, W. W. Stead, G. L. Templeton, M. D. Cave, and K. D. Eisenach. 1996. Strand displacement amplification and the polymerase chain reaction for monitoring response to treatment in patients with pulmonary tuberculosis. J. Infect. Dis. 173:934-941. [DOI] [PubMed] [Google Scholar]
- 17.Hiasa, H., D. O. Yousef, and K. J. Marians. 1996. DNA strand cleavage is required for replication fork arrest by a frozen topoisomerase-quinolone-DNA ternary complex. J. Biol. Chem. 271:26424-26429. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, W. R., Jr., R. G. Barletta, R. Udani, J. Chan, G. Kalkut, G. Sosne, T. Kieser, G. J. Sarkis, G. F. Hatfull, and B. R. Bloom. 1993. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science 260:819-822. [DOI] [PubMed] [Google Scholar]
- 19.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keer, J. T., and L. Birch. 2003. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53:175-183. [DOI] [PubMed] [Google Scholar]
- 21.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 22.Loeliger, B., I. Caldelari, A. Bizzini, P. Stutzmann Meier, P. A. Majcherczyk, and P. Moreillon. 2003. Antibiotic-dependent correlation between drug-induced killing and loss of luminescence in Streptococcus gordonii expressing luciferase. Microb. Drug Resist. 9:123-131. [DOI] [PubMed] [Google Scholar]
- 23.McCarty, S. C., and R. M. Atlas. 1993. Effect of amplicon size on PCR detection of bacteria exposed to chlorine. PCR Methods Appl. 3:181-185. [DOI] [PubMed] [Google Scholar]
- 24.McKillip, J. L., L. A. Jaykus, and M. Drake. 1998. rRNA stability in heat-killed and UV-irradiated enterotoxigenic Staphylococcus aureus and Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4264-4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatomi, Y., and J. Sugiyama. 1998. A rapid latex agglutination assay for the detection of penicillin-binding protein 2′. Microbiol. Immunol. 42:739-743. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 1996. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-T. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.Oliveira, K., G. W. Procop, D. Wilson, J. Coull, and H. Stender. 2002. Rapid identification of Staphylococcus aureus directly from blood cultures by fluorescence in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 40:247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel, J. B. 2001. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 6:313-321. [DOI] [PubMed] [Google Scholar]
- 29.Recorbet, G., C. Picard, P. Normand, and P. Simonet. 1993. Kinetics of the persistence of chromosomal DNA from genetically engineered Escherichia coli introduced into soil. Appl. Environ. Microbiol. 59:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanowski, G., M. G. Lorenz, and W. Wackernagel. 1993. Use of polymerase chain reaction and electroporation of Escherichia coli to monitor the persistence of extracellular plasmid DNA introduced into natural soils. Appl. Environ. Microbiol. 59:3438-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shepard, C. C. 1969. Further experience with the kinetic method for the study of drugs against Mycobacterium leprae in mice. Activities of DDS, DFD, ethionamide, capreomycin and PAM 1392. Int. J. Lepr. Other Mycobact. Dis. 37:389-397. [PubMed] [Google Scholar]
- 32.Sheridan, G. E., C. I. Masters, J. A. Shallcross, and B. M. MacKey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka, M., Y. Onodera, Y. Uchida, K. Sato, and I. Hayakawa. 1997. Inhibitory activities of quinolones against DNA gyrase and topoisomerase IV purified from Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2362-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Heijden, I. M., B. Wilbrink, A. E. Vije, L. M. Schouls, F. C. Breedveld, and P. P. Tak. 1999. Detection of bacterial DNA in serial synovial samples obtained during antibiotic treatment from patients with septic arthritis. Arthritis Rheum. 42:2198-2203. [DOI] [PubMed] [Google Scholar]
- 35.van der Vliet, G. M., S. N. Cho, K. Kampirapap, J. van Leeuwen, R. A. Schukkink, B. van Gemen, P. K. Das, W. R. Faber, G. P. Walsh, and P. R. Klatser. 1996. Use of NASBA RNA amplification for detection of Mycobacterium leprae in skin biopsies from untreated and treated leprosy patients. Int. J. Lepr. Other Mycobact. Dis. 64:396-403. [PubMed] [Google Scholar]
- 36.van der Vliet, G. M., P. Schepers, R. A. Schukkink, B. van Gemen, and P. R. Klatser. 1994. Assessment of mycobacterial viability by RNA amplification. Antimicrob. Agents Chemother. 38:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Villarino, A., O. M. Bouvet, B. Regnault, S. Martin-Delautre, and P. A. D. Grimont. 2000. Exploring the frontier between life and death in Escherichia coli: evaluation of different viability markers in live and heat- or UV-killed cells. Res. Microbiol. 151:755-768. [DOI] [PubMed] [Google Scholar]
- 38.Williams, D. L., T. L. Pittman, T. P. Gillis, M. Matsuoka, and Y. Kashiwabara. 2001. Simultaneous detection of Mycobacterium leprae and its susceptibility to dapsone using DNA heteroduplex analysis. J. Clin. Microbiol. 39:2083-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]