Abstract
Staphylococcus aureus staphylococcal cassette chromosome mec type IV (SSCmec IV) is associated with virulent community-acquired methicillin-resistant Staphylococcus aureus (MRSA) and frequent horizontal transfer among staphylococci. To gain insight into the mechanism of transfer, we studied the ccrA/B type 2 recombinase-mediated excision of SCCmec IV (n = 5 strains) and SCCmec II (n = 2). In SCCmec IV- but not SCCmec II-containing strains, spontaneous excision of the cassette was observed. Introduction of ccrA/B type 2 recombinase genes under control of an S. aureus bacterial phage promoter in the different strains yielded excision of SCCmec II and multiple excision variants of SCCmec IV. Sequencing of the alternatively excised products in SCCmec IV strains identified a 100-bp shortened SCCmec′ variant and a 5,877-bp, conserved SCC-like element that lacks mecA and ccrA/B recombinases. Excision of the SCC-like element in wild-type S. aureus was dependent on the presence of SCCmec. The element could be excised separately or as part of a novel composite cassette together with SCCmec. The relative abundance of and variety in SCCmec IV excisions may contribute to the frequency of horizontal transfer and genetic plasticity in SCCmec IV MRSA strains.
Methicillin-resistant Staphylococcus aureus (MRSA) is a major nosocomial pathogen (2). Methicillin resistance is conferred by the mecA-encoded PBP2′, which has a low affinity for beta-lactam antibiotics. mecA is located on the staphylococcal cassette chromosome mec (SCCmec), a large 20- to 65-kb mobile element in S. aureus (9). Based on the class of mecA gene complex and the type of ccr gene complex present, SCCmec cassettes are classified as types I to V (6, 7, 12). SCCmec type II (SCCmec II) carries ccrA/B type 2 recombinases, and SCCmec IV carries ccrA/B type 2 or type 4 recombinases (depending on the classification scheme used), whereas SCCmec V carries a ccrC recombinase. After cloning of the ccrA/B type 2 and ccrC promoter-gene complex into SCCmec II and V MRSA strains, respectively, chromosomal excision and integration of the SCCmec cassette have been observed (6, 9). The ccr recombinases are anticipated to cleave DNA in the attB site directly upstream of the integration site sequence (ISS) for SCC. The ISS has a putative consensus sequence, GANGCNTATCANAANTNN, which is present at the 3′ end of orfX in S. aureus (6).
The relatively small SCCmec IV may be more frequently transferred among staphylococci and is associated with community-acquired MRSA (14). We have previously described a possible case of in vivo transfer of SCCmec IV from Staphylococcus epidermidis to S. aureus (16). In this study, three strains are described which were consecutively isolated from a single patient. Methicillin-susceptible Staphylococcus aureus (MSSA) strain wkz-1 shared among others a unique pulsed-field gel electrophoresis pattern with MRSA strain wkz-2, except for the presence of an SCCmec IV cassette. A cassette with the same restriction pattern was found in S. epidermidis strain O7.1, which may have served as the SCCmec donor strain (16). The mechanism of SCCmec transfer, however, is still unknown.
To gain insight into the mechanism of transfer, we further characterized these strains and studied the excision of SCCmec IV in these and other MRSA strains. Our data show that chromosomal excision mediated by ccrA/B type 2-containing recombinases is a dynamic process that is certainly not confined to SCCmec IV alone. We provide evidence for the excision and circularization of SCCmec IV in wild-type S. aureus and S. epidermidis strains and the formation and excision of a new composite SCCmec.
MATERIALS AND METHODS
Strains.
Staphylococcal strains used in this study include MSSA wkz-1, SCCmec IV MRSA wkz-2, S. epidermidis O7.1 (16), MRSA MR108 (5, 8), MW2 (1), Ca05 (JCSC1968), and 8/6-3P (JCSC1978) (11), and SCCmec II MRSA Mu50 and N315 (10). We used Escherichia coli DH10β and S. aureus strain RN4220 for cloning of the different ccrA/B constructs in S. aureus. S. aureus strain WVW 189 was used for the amplification of an S. aureus phage repressor promoter. WVW 189 is RN6390 containing pACL 1484 with a phage repressor promoter cloned in its EcoRI/XbaI site, upstream of GFPuvr. This phage promoter yields a strong, constant expression of green fluorescent protein in S. aureus (15). The primers used in this study are listed in Table 1. Each primer is given a unique code and a number corresponding to the location of the primer as shown in Fig. 2. Depending on the genetic variation in the strains at different primer positions, primers with different codes may share the same primer location number (Table 1).
TABLE 1.
Primers used for cloning and excision experiments
| Application | Code | Position in Fig. 2 | Location | Orientationa | Strain(s)b | Sequencec | TA (°C)d |
|---|---|---|---|---|---|---|---|
| Cloning | C1 | ccr promoter | F | wkz-2, O7.1 | CGGAATTCTGTAGAGTTTGCATCTATCCTGG | 68 | |
| C2 | ccrA/B | R | wkz-2, O7.1 | ACGCGTCGACTCTGTTTCTTCGAATCTGCAAAT | 74 | ||
| C3 | ccrA/B | F | wkz-2, O7.1 | GCTCTAGAATGAAACAACAATCTCTTGC | 62 | ||
| C4 | Phage promoter | F | WVW 189 | CGGAATTCCTTGTTTTGAATCAAGTCA | 66 | ||
| C5 | Phage promoter | R | WVW 189 | GCTCTAGACCGTTTGATAACTTCATAAT | 56 | ||
| Excision | E1 | 1 | Upstream of SCC-like element | F | wkz-2, MR108 | GCATTCAGATTATTGACTGTTGG | 58 |
| E2 | 2 | SCC-like element | R | wkz-2, MR108 | CTACCAGCAATACCTCATACC | 52 | |
| E3 | 3 | Upstream of SCCmec | F | wkz-2, MR108 | TTTTGCTGTTTTTATCACCATATTGAA | 59 | |
| E4 | Ca05 | AATTTACCAGACAGCCTGGTGC | 61 | ||||
| E5 | JCSC1978, Mu50, N315 | ATTTAATGTCCACCATTTAACA | 53 | ||||
| E6 | 4 | SCCmec | R | wkz-2, O7.1, Ca05 | GTCCTAACAAGCGGTCAACACC | 62 | |
| E7 | JCSC1978 | CATCAAACTTTAAGGGAGAAGC | 57 | ||||
| E8 | MW2, Ca05, wkz 2 | CCACGTTATGGAGGTGCTCTG | 62 | ||||
| E9 | MR108 | AACGGTCTGGACGAAGTAAGG | 59 | ||||
| E10 | Mu50, N315 | GAATCTTCAGCATGTGATTTA | 52 | ||||
| E11 | 5 | SCCmec | F | All | ATGAAAGACTGCGGAGGCTAACT | 61 | |
| E12 | 6 | SCCmec | R | All | CAGCCGCTTCATAAAGGGATT | 61 | |
| E13 | 7 | SCCmec | F | wkz-2 | GGCTGAAAAAACCGCATCAT | 61 | |
| E14 | 8 | orfX | R | All | AAACGACATGAAAATCACCAT | 56 |
F, forward primer; R, reverse primer.
Strain(s) on which primers were used.
Primer sequences depicted in bold have been described previously Katayama et al. (9).
Annealing temperature was chosen 5°C below the melting temperature of the primer. For primer combinations in the PCR, the lowest TA was chosen.
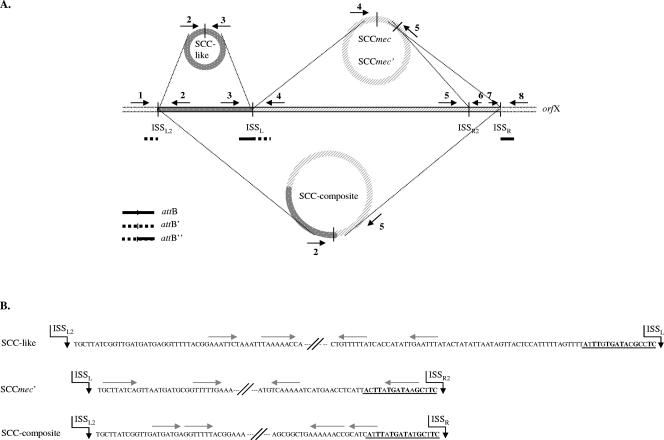
FIG. 2.
Diversity of SCC excision products in SCCmec IV MRSA. (A) Schematic presentation of different SCC excision products generated by type 2 ccrA/B recombinases in SCCmec IV strains. The arrows indicate the positions of the different primers used to detect excision. Numbers correspond to the primer position numbers in Table 1. Thick and dotted lines represent the chromosomal parts that reunite upon excision of the diverse SCC elements. For SCCmec, this site is known as attB. Upon excision of the SCC-like element and the composite cassette, alternative attB sites were formed that we named attB′ and attB", respectively. (B) ISS regions and indirect repeats of SCC excision products. The ISS consensus region is underlined, whereas consensus nucleotides are given in bold. Gray arrows denote indirect repeats. Vertical arrows indicate the positions at which the element is excised, at the 3′ end of the ISS.
Cloning.
Two different recombinant ccrA/B type 2 plasmids were constructed in which the ccrA/B genes were placed either under the control of their endogenous promoter or under the control of a phage repressor promoter. The endogenous −300-bp promoter region and adjacent ccrA/B genes were amplified by PCR from MRSA strain wkz-2, using primers C1 and C2 (Table 1). To generate the phage repressor ccrA/B construct, the wkz-2 ccrA/B genes were amplified using primers C2 and C3 with DNA from strain wkz-2. The phage repressor promoter was obtained from strain WVW 189 by using primers C4 and C5 (15). The endogenous promoter-ccrA/B fragment was cloned in the EcoRI-SalI site of shuttle vector pSK236, whereas the phage promoter and ccrA/B genes were cloned in the EcoRI-XbaI and XbaI-SalI sites of this vector, respectively. The pSK236 vector confers carbacillin resistance in E. coli and chloramphenicol resistance in S. aureus. The pSK236 recombinant plasmids were introduced in E. coli DH10β and subsequently introduced in the DNA restriction-deficient S. aureus strain RN4220 by electroporation. The endogenous ccrA/B-promoter construct was transduced into MRSA wkz-2, using phage 11 according to standard procedures. The phage repressor promoter-ccrA/B construct was transduced into MSSA wkz-1 and the MRSA strains wkz-2, Ca05, JCSC 1978, MW2, MR108, Mu50, and N315. The inserts of all bacterial clones were confirmed with PCR and sequencing.
PCR.
S. aureus was grown on Luria-Bertani agar plates (containing 10 mg/liter chloramphenicol when pSK236 derivatives were present) for 16 h at 37°C. DNA lysates were made using standard methods. Chromosomal DNA was purified using a QIAGEN purification kit (Hilden, Germany). Excision of SCCmec was measured by PCR, using divergent primers on both outer regions of the cassette that yield a PCR product only upon excision and circularization of the SCC element, analogous to the method described in reference 9. Excision was confirmed using two chromosomal primers flanking the SCC elements to detect the reformed attB site upon excision of the cassette (9). Except for PCRs to detect attB after SCC excision, which were performed on chromosomal purified DNA, all PCRs were performed on DNA lysates. This was necessary since the circular SCCmec was not efficiently purified by either plasmid or chromosomal DNA isolation protocols (data not shown). PCRs that did not yield products specific for SCC circularization, or the reformed attB site, were performed again with different template concentrations, using a touchdown PCR program. The touchdown PCR program was as follows: an initial denaturation step for 5 min at 94°C, followed by an initial 10 cycles of three steps consisting of 30 s at 94°C, 1 min at annealing temperature (TA) plus 10°C to TA (Table 1) decreasing 1°C per cycle, and 1 min 30 s at 72°C for extension. These first 10 cycles were followed by another 25 to 35 cycles of three steps consisting of 30 s at 94°C, 1 min at TA (Table 1), and 1 min 30 s at 72°C and a final 5-min extra extension. A PCR strategy according to Oliveira and de Lencastre (12) was performed for subtyping of SCCmec.
Sequencing.
All PCR-amplified fragments were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and sequenced. DNA regions upstream of SCCmec were sequenced using an LA PCR in vitro cloning kit (Takara Mirus Bio Corporate Information, Madison, WI) according to the manufacturer's protocol. This kit enables nested PCR amplification of unknown sequences upstream of known sequences. Sequences were analyzed against all available sequences by using the BLAST algorithm. Multilocus sequence typing (MLST) was performed according to Enright et al. (4).
RESULTS
Genetic characterization of strains associated with horizontal transfer of SCCmec.
MRSA wkz-2 may originate from the transfer of SCCmec IV from S. epidermidis O7.1 to MSSA wkz-1 (16). Therefore, excision of SCCmec was studied primarily in these strains. To this end, first these strains and the SCCmec elements carried by them were further genetically characterized. Both S. aureus strain wkz-1 and S. aureus strain wkz-2 have MLST sequence type 30. The SCCmec types of strains wkz-2 and O7.1 were identical according to the multiplex PCR developed by Oliveira and de Lencastre (12). Both cassettes carry mec complex B, type 2 ccrA/B, and loci A and D. We sequenced the left (400 bp) and right (1,800 bp; in the proximity of orfX) extremities of SCCmec in both the wkz-2 and O7.1 strains. The DNA sequences were 100% identical between the two strains. These sequences completely matched the outer ends of SCCmec IV carried by community-acquired MRSA strain Ca05 (GenBank accession no. AB063172). Subsequently, the ccrA and ccrB genes of strains wkz-2 and O7.1 were sequenced. As expected, the complete ccrA/B DNA sequences were also identical between the strains. On their turn, these ccrA/B sequences were indistinguishable from the ccrA/B of strain MR108 (GenBank accession no. AB096217) but differed from that of Ca05 (data not shown). When we used the methods described above, the SCCmec of S. epidermidis O7.1 and MRSA wkz-2 were indistinguishable.
Excision of SCCmec in SCCmec II and IV MRSA.
To address the first step in horizontal transfer of SCCmec, we examined SCCmec excision in SCCmec IV MRSA (n = 5), the SCCmec IV-positive S. epidermidis strain O7.1, and SCCmec II MRSA strains (n = 2). All these strains carry ccrA/B type 2 recombinases. Excision results for the MSSA wkz-1, MRSA wkz-2, and S. epidermidis O7.1 strains are displayed in Fig. 1, whereas the excision data for all other strains are summarized in Table 2. A schematic of the different excision products and the location of the different excision primers is given in Fig. 2.
FIG. 1.
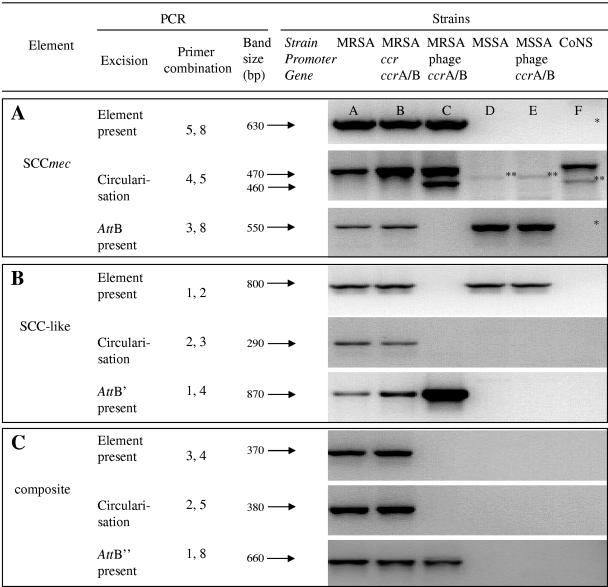
Excision of three different SCC elements in MRSA strain wkz-2, MSSA strain wkz-1, and coagulase-negative staphylococcus (CoNS) strain O7.1. These strains were possibly involved in the in vivo horizontal transfer of SCCmec IV from S. epidermidis to S. aureus (16). Excision of the SCC elements was measured in wild-type strains and in strains complemented with the ccrA/B type 2 recombinase genes under the control of either the endogenous ccr promoter or a phage repressor promoter. Excision of the SCC elements was measured by PCR: first, the presence of the element was detected, subsequently the circularization of the element, and finally the reformed attB site upon excision of the element. attB′ and attB" are alternative attB sites reformed after excision of the SCC-like element and the SCC-like/SCCmec composite element, respectively (Fig. 2). All PCR products were confirmed by sequencing. The primer combinations used correspond to the primer position numbers depicted in Fig. 2. Note that the wkz-2 strain complemented with the phage promoter-ccrA/B construct lost the SCC-like element and consequently was negative in any PCR using primers located on this element (primers E2 and E3, location numbers 2 and 3 [Fig. 2]). *, No product due to the absence of CoNS O7.1-specific primers outside of SCCmec. **, Aspecific bands, as confirmed by sequencing.
TABLE 2.
Excision of SCC elements in different S. aureus strains
| Strain | Country | Year | Location | SCCmec type | Excisiona
|
|||
|---|---|---|---|---|---|---|---|---|
| SCCmec | SCCmec′ | SCC-like element | Composite | |||||
| wkz-1 | Netherlands | 2000 | Unknown | — | ||||
| O7.1 | Netherlands | 2000 | Unknown | IV | + | — | ||
| MW2 | United States | 1998 | Community | IVa | + | — | ||
| Ca05 | United States | 1998 | Community | IVa | + | +/− | ||
| JCSC1978 | United States | 1998 | Community | IVb | +/− | I | ||
| MR108 | Japan | 1982 | Community | IVc | +/− | I | + | + |
| wkz-2 | Netherlands | 2000 | Hospital | IV | + | I | + | + |
| Mu50 | Japan | 1996 | Hospital | II | I | I | ||
| N315 | Japan | 1982 | Hospital | II | I | I | ||
Excision of four different SCC elements in MRSA SCCmec II and SCCmec IV strains. Analogous to the excision results shown in more detail in Fig 1, excision was determined by PCRs specific for the presence and circularization of the SCC element and on the reformed attB site upon excision of the element. An empty box indicates that the element is not present in the strain. −, No excision. +, Excision in wild-type strains. Both the excision and attB PCRs are positive. +/−, Excision in wild-type strains is partly demonstrated. The excision PCR is below the detection limit, and the attB-specific PCR is positive. In addition, excision is induced after introduction of the phage promoter-ccrA/B construct in the strain. I, Inducible excision. Excision is induced after introduction of the phage promoter-ccrA/B construct in the strain.
Spontaneous excision of the SCCmec IV cassette was detected in MRSA wkz-2 and S. epidermidis strain O7.1 (Fig. 1A). In these strains, both the circularization PCR and the attB PCR yielded PCR products with the predicted sequence (Fig. 1A, lanes A and F). Excision frequencies in MRSA wkz-2 (1:5,000) and S. epidermidis strain O7.1 (1:1,000) were estimated by semiquantitative excision and mecA PCR (data not shown).
Excision of SCCmec IV was also observed for strains MW2 and Ca05. For the remaining SCCmec IV MRSA strains, JCSC 1978 and MR108, circularization of SCCmec was not detected whereas the attB PCR was positive. In contrast, excision of SCCmec II in strains Mu50 and N315 was not observed either by circularization or by reformation of the attB site (Table 2).
We constructed several clones with enhanced ccrA/B activity as a tool to explore ccrA/B-mediated excision of SCCmec in all S. aureus strains. In these clones, ccrA/B type 2 recombinases were placed under the control of either the endogenous ccr promoter or a phage promoter.
In SCCmec II strains Mu50 and N315, SCCmec excision could be induced after introduction of the phage promoter-ccrA/B construct (Table 2). Induced SCCmec II excision in strain N315 is in agreement with that described by Katayama et al. (9). Interestingly, two SCCmec excision products were observed in strain wkz-2 (Fig. 1A, lane C) and all other MRSA strains complemented with the phage promoter-ccrA/B construct, except for strain MW2 (Table 2). Sequencing of excision products and chromosomal remnants in these strains showed that SCCmec was cleaved either at the “normal” ISSR next to orfX, or 100 bp upstream at an alternative consensus sequence called ISSR2, yielding a shortened SCCmec called SCCmec′ (Fig. 2). In wild-type strains, excision of SCCmec′ was detected only in strain Ca05, as determined with primers E4 and E12 (Table 2). Circularization of the 100-bp fragment between the target sites ISSR2 and ISSR was not detected in any of the clones (data not shown).
SCC-like element.
Two of the phage promoter-ccrA/B clones (MRSA wkz-2 and MRSA MR108) lost their capability to bind primer 3 upstream of SCCmec (see Fig. 2 for primer location). The introduction of the ccrA/B construct in these strains was accompanied by the absence of amplification when primer 3 was used (Fig. 1A, lane C, Fig. 1C, lane C, and Table 2). We therefore hypothesized that these clones lost the SCCmec upstream region due to aberrant ccr recombinase-mediated excision. Using an LA PCR in vitro cloning kit, we identified a 5,877-bp SCC-like element present in MRSA wkz-2 and MRSA MR108 wild-type strains, which was lost in these strains upon introduction of the phage promoter-ccrA/B construct. The SCC-like element contains ISSs at its extremes, two unknown open reading frames, three transposases, and an element that matches parts of prophage PV83 (7).
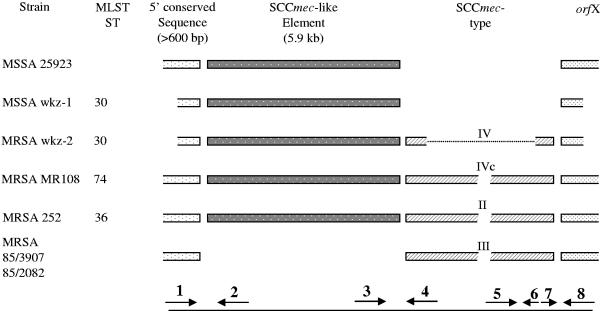
Using the BLAST algorithm, we examined the presence of SCC-like elements among published staphylococcal sequences deposited in GenBank. Besides MSSA strain wkz-1 and MRSA strain wkz-2, the SCC-like element is present in the MSSA strain 25923, an SCCmec II MRSA strain (MRSA 252), and an SCCmec IV MRSA strain (MR108). In all of these strains, a conserved region of at least 600 bp was identified upstream of the SCC-like element. Two SCCmec III MRSA strains (85/3907 and 85/2082) lacked the SCC-like element but did contain this 600-bp upstream region (Fig. 3).
FIG. 3.
Schematic presentation of the chromosomal organization of the SCC-like element in different S. aureus strains. Arrows indicate the positions of the different primers used to detect excision of the various elements. Numbers correspond to the primer position numbers in Table 1.
Excision of SCC-like element.
Is the SCC-like element mobile? To study this, we examined the excision of the SCC-like element separately or in combination with SCCmec in different strains. The SCC-like element was excised in MRSA wkz-2 (Fig. 1B, lane A) and MRSA MR108 wild-type strains (Table 2), as determined by a PCR specific for circularization and a PCR on the alternative reformed attB site that we called attB′ (Fig. 2). Interestingly, SCC-like excision was observed neither in the MSSA wkz-1 wild-type strain nor in MSSA wkz-1 strains complemented with the phage promoter-ccrA/B construct, although they contained this element (Fig. 1B, lanes D and E). Finally, excision and circularization of a composite element consisting of the SCC-like element and SCCmec were observed in MRSA wkz-2 (Fig. 1C, lane A) and MRSA 108 (Table 2), as measured by a PCR specific for circularization and a PCR on the chromosomal remnants called attB′ (Fig. 2). Excision of the composite cassette at the alternative ISSR2 was not detected (data not shown).
In summary, excision of SCCmec IV occurred among wild-type MRSA and MRSE strains, whereas SCCmec excision could be induced in SCCmec II MRSA strains. In addition, four different mobile SCC elements were detected in S. aureus. Two excision products, SCCmec and SCCmec′, were observed in MRSA strain Ca05. Three excision products, the SCC-like element, SCCmec, and a novel composite cassette, were observed in SCCmec IV MRSA strains wkz-2 and MR 108 (Fig. 2).
DISCUSSION
Here, we report for the first time ccrA/B type 2 mediated excision and circularization of SCC elements among wild-type SCCmec type IV strains under normal laboratory conditions. In addition, we observed the excision of several ISS-flanked elements within one MRSA strain, either individually or as part of a composite cassette, which is suggestive of a high genetic plasticity among these strains.
Interestingly, the excision results suggest an optimum length for cassettes to become excised and circularized. Circularization could not be detected for the small, 100-bp fragment at the orfX extremity of SCCmec, and excision of the 52-kb SCCmec II cassette in Mu50 and N315 was inducible only. In contrast, excision of the 6-kb SCC-like element, the 20-kb SCCmec IV cassette, and the 26-kb composite cassette could be readily detected in wild-type strains (Fig. 2). Since the SCCmec II and SCCmec IV strains carried the same ccrA/B type and ISSs, this difference in excision efficiency may be due to differences in cassette size rather than differences in recombinase activity or recombinase target sequences. Spontaneous excision of SCCmec′, however, is probably a rare phenomenon. In contrast to strains complemented with the phage promoter-ccrA/B construct, concomitant excision of SCCmec and SCCmec′ could not be detected in the Ca05 wild-type strain (determined with primer combination 4 and 5) (data not shown). This may be explained by the much lower excision frequency of SCCmec′ than that of SCCmec in the Ca05 wild-type strain, resulting in detectable PCR amplification of the latter element only. Comparison of all ISSs involved in excision in our experiments confirms the putative ISS reported earlier by Ito et al. (6).
The SCC-like element does not contain any known virulence or resistance genes. Nevertheless, its presumed mobility (in the presence of SCCmec), together with the observation that an ancient MSSA strain and several virulent epidemic MRSA strains possess this element, suggests that its presence may be beneficial for the bacterial host. We hypothesize that similar to the “train-wagon”-like organization of gene cassettes in integrons (13), several ISS-flanked elements may be coupled to SCCmec and get mobilized either as a composite cassette or alone by using the recombinases of the adjacent SCCmec cassette. The same mobilization mechanism may apply for the newly described arginine catabolic mobile element in the virulent, community-acquired MRSA strain USA300 (3). Similar to the SCC-like element, this element is likely to be excised by ccrA/B recombinases contained in the adjacent SCC element. In addition, it may be possible that SCC excision and reintegration at another ISS position will lead to shuffling of SCC elements, thereby conferring genetic plasticity without the need for horizontal transfer. We could not, however, detect shuffling of SCC elements in our strains (data not shown).
Several lines of evidence suggest that the SCC-like element is subject to horizontal transfer. First, it is flanked by ISS consensus elements. Second, the element is well conserved among several MSSA and MRSA strains of different MLST sequence types and MRSA strains carrying different SCCmec cassettes, whereas it is absent in other strains that share the same sequences upstream of the SCC-like element (Fig. 3). Finally, our data show that the SCC-like element can be excised separately from the chromosome. Our data strongly suggest that the excision of the SCC-like element is dependent on the presence of SCCmec, as SCC-like excision was undetectable in the MSSA wkz-1 strain. Although it is likely that SCC-like excision depends on SCCmec containing ccrA/B recombinases, we cannot exclude other mechanisms, as excision of the SCC-like element could not be induced in the MSSA wkz-1 strain after introduction of a phage promoter-ccrA/B construct.
Ito et al. already suggested the presence of this SCC-like element in an S. aureus MSSA strain 25923 isolated in 1945 (DDBJ/EMBL/GenBank accession no. AB047239) (7). They hypothesized that this element may be an immobile remnant of a functional SCCmec that lost its recombinases (and mec region) (9). Our data suggest a second possibility: the element may have belonged to a composite SCC cassette or requires trans functions from an SCC with recombinases. After loss of the SCC cassette carrying the recombinase genes, the SCC-like element became immobilized in the genome. However, acquisition of an SCC element containing recombinases may mobilize this element again.
Such an event may have occurred in the S. aureus wkz-1 strain. We previously showed that wkz-1 and wkz-2 shared a unique genotype among 312 MRSA isolates, as measured by phage typing, their antibiotic resistance profiles, pulsed-field gel electrophoresis, and ribotyping. In addition, it was shown that the wkz-2 and O7.1 strains showed the same rare SCCmec restriction pattern (16). We now show that MSSA strain wkz-1 and MRSA strain wkz-2 belong to the same MLST genotype and that the MSSA wkz-1 strain and S. epidermidis strain O7.1 shared identical (partial) SCCmec sequences. These findings are in line with our earlier observation (16) of the putative in vivo transfer of SCCmec from S. epidermidis O7.1 to MSSA strain wkz-1, yielding MRSA strain wkz-2. It is therefore likely that the SCCmec element is mobile and may have been acquired from S. epidermidis O7.1 or another unknown source at some point in time. Excision of the SCC-like element was not detected in the wkz-1 strain, but upon introduction of SCCmec in the wkz-2 strain, the SCC-like element became mobile. In addition to its separate excision, the SCC-like element could be coexcised together with SCCmec in this strain. Thus, the SCC-like element may also hijack SCCmec for its mobilization. Taken together, these findings suggest that horizontal transfer of SCCmec created a novel mobile element composed of the SCC-like element and SCCmec.
In conclusion, this study identified four different SCC excision variants in SCCmec IV strains. The relative abundance of and variety in SCCmec IV excisions, exemplified by the formation and excision of a new SCCmec IV composite, may contribute to the frequency of horizontal transfer and genetic plasticity in SCCmec IV MRSA strains.
Acknowledgments
We kindly thank T. Ito for providing us with the MRSA strains MW2, MR108, Ca05, and JCSC 1978.
REFERENCES
- 1.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 2.Chopra, I. 2003. Antibiotic resistance in Staphylococcus aureus: concerns, causes and cures. Expert Rev. Anti. Infect. Ther. 1:45-55. [DOI] [PubMed] [Google Scholar]
- 3.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 4.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiramatsu, K., E. Suzuki, H. Takayama, Y. Katayama, and T. Yokota. 1990. Role of penicillinase plasmids in the stability of the mecA gene in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:600-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updates 6:41-52. [DOI] [PubMed] [Google Scholar]
- 9.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 11.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ploy, M. C., T. Lambert, J. P. Couty, and F. Denis. 2000. Integrons: an antibiotic resistance gene capture and expression system. Clin. Chem. Lab. Med. 38:483-487. [DOI] [PubMed] [Google Scholar]
- 14.Robinson, D. A., and M. C. Enright. 2004. Multilocus sequence typing and the evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 10:92-97. [DOI] [PubMed] [Google Scholar]
- 15.Rooijakkers, S. H., W. J. Van Wamel, M. Ruyken, K. P. van Kessel, and J. A. van Strijp. 2005. Anti-opsonic properties of staphylokinase. Microbes Infect. 7:476-484. [DOI] [PubMed] [Google Scholar]
- 16.Wielders, C. L., M. R. Vriens, S. Brisse, L. A. Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2001. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet 357:1674-1675. [DOI] [PubMed] [Google Scholar]