Abstract
Daptomycin is a lipopeptide antibiotic with potent activity against gram-positive bacteria. Complete-genome comparisons of laboratory-derived Staphylococcus aureus with decreased susceptibility to daptomycin and their susceptible parent were used to identify genes that contribute to reduced susceptibility to daptomycin. Selective pressure of growth in sublethal concentrations of daptomycin resulted in the accumulation of mutations over time correlating with incremental decreases in susceptibility. Single point mutations resulting in amino acid substitutions occurred in three distinct proteins: MprF, a lysylphosphatidylglycerol synthetase; YycG, a histidine kinase; and RpoB and RpoC, the β and β′ subunits of RNA polymerase. Sequence analysis of mprF, yycF, yycG, rpoB, and rpoC in clinical isolates that showed treatment-emergent increases in daptomycin MICs revealed point mutations in mprF and a nucleotide insertion in yycG, suggesting a role for these genes in decreased susceptibility to daptomycin in the hospital setting.
Staphylococcus aureus can evolve drug resistance by altering one or more nucleotides within its 2.8-Mbp genome. Single nucleotide changes confer resistance to a wide range of antimicrobials, including rifampin, fluoroquinolones, and quinupristin-dalfopristin (8, 9, 15, 24, 42). Multiple mutations are required for increased resistance to other drugs, such as linezolid (32, 36). Identification of the mutation(s) that confers resistance is essential to understanding the genetic mechanism(s) of drug resistance and often elucidates the mechanics behind drug action.
Standard genetic methods and sequence analysis of predicted resistance-determining regions are traditionally used to identify genomic regions involved in drug resistance. These methods are limited to defined genetic loci and often fail to detect mutations scattered throughout the genome. Complete-genome comparisons of resistant and sensitive S. aureus strains provide a new, unbiased approach to identifying regions involved in drug resistance. S. aureus is a prime candidate for this new approach since there are six published genomes (3, 12, 10, 14, 23). Whole-genome comparisons have identified genes involved in virulence and resistance in S. aureus (14, 19). Nevertheless, the multitude of synonymous nucleotide alterations between different strains of S. aureus complicates the identification of single nucleotide changes that may be responsible for specific drug resistance (14, 17). Wong et al. showed that a DNA hybridization-based custom microarray service called comparative genome sequencing (CGS) can be used to detect nucleotide differences between viral genomes (43). In this study, we used CGS to identify mutations that correlate with increases in the daptomycin MIC for laboratory-derived S. aureus strains derived from S. aureus MW2.
Daptomycin is a lipopeptide antibiotic active against a wide range of gram-positive organisms, including methicillin-resistant and vancomycin-resistant S. aureus (2, 37). The current susceptibility criteria for daptomycin, based on the normal unimodal population distribution, are as follows: susceptible, MIC ≤ 1; nonsusceptible, MIC ≥ 2 (28). There is not enough clinical experience yet to provide a resistance designation. The MIC for 90% of the S. aureus strains tested is 0.5 μg/ml, and surveillance studies have not reported any strains for which the MIC is ≥4 μg/ml (5, 34). Rare treatment-emergent S. aureus strains for which the MICs are 2 to 4 μg/ml have been reported (13, 41). This study demonstrates that an accumulation of mutations over time correlates with incremental increases in the daptomycin MIC. Two of the genes identified by CGS as correlating with low-level MIC increases were found to contain mutations in daptomycin-nonsusceptible clinical isolates, supporting the validity of this approach in identifying clinically relevant resistance mechanisms.
MATERIALS AND METHODS
Strains and growth conditions.
S. aureus strains were routinely propagated on tryptic soy agar with 5% sheep blood (bioMérieux, Lombard, IL) or in Mueller-Hinton broth (Difco, Detroit, MI) supplemented with calcium chloride (final Ca2+ concentration, 50 mg/liter) (MHBc) at 37°C. All of the strains used in this study are listed in Table 1.
TABLE 1.
Bacterial strains used in this study
| Strain or pair | Cubist strain designation(s) | Description | Reference or sourcea |
|---|---|---|---|
| S. aureus laboratory strains | |||
| MW2 | CB1118 | Community-acquired MRSAb; sequenced genome, GenBank accession no. BA000033 | 3; B. N. Kreiswirth |
| MW2-CB1616 | CB1616-d1 to CB1616-d20 | Daily representatives from serial passage expt CB1616 | This study |
| MW2-CB1617 | CB1617-d1 to CB1617-d20 | Daily representatives from serial passage expt CB1617 | This study |
| MW2-CB1618 | CB1618-d1 to CB1618-d20 | Daily representatives from serial passage expt CB1618 | This study |
| MW2-CiproR | grlA mutant from fifth day of ciprofloxacin serial passage | This study | |
| S. aureus clinical isolates | |||
| Pair 1 | |||
| CB1482 | MRSA, 9 December 2003 blood specimen; daptomycin MIC = 0.5 μg/ml | 13; LSI | |
| CB184 | MRSA, 11 May 2004 blood specimen; daptomycin MIC = 4 μg/ml | 13; LSI | |
| Pair 2 | |||
| CB1483 | MRSA, 31 December 2003 tibial tissue specimen; daptomycin MIC = 0.25 μg/ml | 13; LSI | |
| CB185 | MRSA, 18 February 2004 epidural tissue specimen; daptomycin MIC = 4 μg/ml | 13; LSI | |
| Pair 3 | |||
| CB1631 | MRSA, June 2004 blood specimen; daptomycin MIC = 0.5 μg/ml | 41; LSI | |
| CB1634 | MRSA, August 2004 T10-11; disk specimen; daptomycin MIC = 4 μg/ml | 41; LSI |
B. N. Kreiswirth, Public Health Research Institute, Newark, NJ; LSI, Laboratory Specialist, Inc., Westlake, OH.
MRSA, methicillin-resistant S. aureus.
Antimicrobial testing.
MICs for S. aureus were determined according to Clinical Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) guidelines for broth microdilution (27, 28). MIC assays were performed in a 96-well plate format (100 μl/well). In order to detect incremental changes in drug susceptibility, an extended gradient was created by combining three sets of twofold serial dilutions from three different starting concentrations (20, 24, and 32 μg/ml). The extended gradient included daptomycin concentrations of 0, 0.020, 0.023, 0.031, 0.039, 0.047, 0.063, 0.078, 0.094, 0.125, 0.156, 0.188, 0.25, 0.313, 0.375, 0.5, 0.625, 0.75, 1, 1.25, 1.5, 2, 2.5, 3, 4, 5, 6, 8, 10, 12, 16, 20, 24, and 32 μg/ml, which extended over three rows of the 96-well plate. Control experiments utilized the same concentration gradient with ciprofloxacin. For the time line shown in Fig. 2, MIC determinations were repeated on a minimum of six colonies from each strain. All cultures were incubated for approximately 24 h. Growth was determined by measuring optical density at 600 nm on a SpectraMax plate reader (Molecular Devices, Sunnyvale, CA). Growth was defined as an optical density reading of ≥0.1.
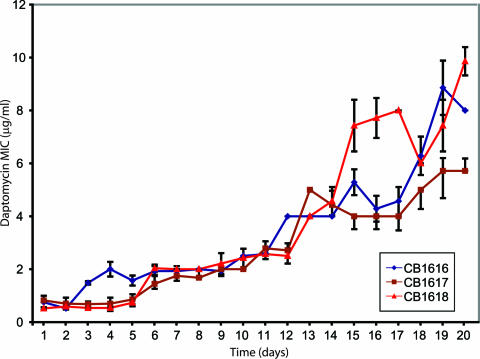
FIG. 2.
MIC analysis of daily archived colonies from three specific serial-passage experiments, CB1616, CB1617, and CB1618. Each spot on the graph represents the average MIC from more than six daptomycin MIC assays for an isolated representative of the daily population.
In vitro selection for decreased susceptibility to daptomycin or ciprofloxacin.
S. aureus MW2 was the drug-susceptible parental strain used for all serial-passage experiments. Similar protocols were used for daptomycin and ciprofloxacin; only the daptomycin protocol will be described in detail. Serial-passage experiments were performed in 96-well plates as a series of MIC experiments with the extended range of daptomycin concentrations described above. On day 1, each well was inoculated with approximately 104 CFU of exponentially growing S. aureus MW2. Following overnight growth, the MIC was determined and the highest drug concentration that permitted growth was diluted 1:1,000 in MHBc and used to inoculate the next day's MIC assay (a 50-μl diluted inoculum was added to 50 μl of medium), plates were incubated overnight as before, and the process was repeated for 20 days. Serial-passage experiments were given CB strain designations, i.e., CB1616, CB1617, and CB1618. Populations of bacteria from each day of the serial-passage experiment were stored at −80°C in MHBc supplemented with 20% glycerol. Individual colonies purified from these archived populations were given strain designations to indicate the serial passage from which they were isolated and the day on which they were taken from the serial-passage experiment. For example, CB1616-d3 was the designation of the strain of bacteria propagated from a single colony isolated from the population of bacteria on day 3 of the CB1616 serial-passage experiment. All colonies were propagated in MHBc. Genomic DNA was purified from cultures representing each day of the serial-passage experiment.
The stability of the daptomycin MICs was tested by passaging the CB1616-d20, CB1617-d20, and CB1618-d20 strains on antibiotic-free medium for 6 days. Genomic DNA from the day 20 isolates was analyzed by pulsed-field gel electrophoresis (after digestion with SmaI) and compared to S. aureus MW2 genomic DNA to confirm isogenicity (R. V. Goering, Creighton University School of Medicine, Omaha, NE).
Genomic DNA purification.
Genomic DNA was isolated with the Wizard Genomic DNA purification kit according to the standard protocol (Promega, Madison, WI). Bacteria were treated with lysostaphin (Sigma-Aldrich and/or AMBI [Lawrence, NY]) prior to lysis.
Complete-genome comparisons.
Complete-genome comparisons were performed with an array-based service (CGS) provided by NimbleGen Systems Inc. (Madison, WI) (1, 43). A complete description of the microarray design and the CGS comparison methodology used is provided in the supplementary-material sections of reference 1. The reference genome sequence used in the microarray was that of S. aureus subsp. aureus MW2 (GenBank accession number BA000033) (3). All CGS experiments compared the S. aureus MW2 reference strain to individual laboratory-derived strains with decreased susceptibility to the test drug. Daptomycin experiments utilized genomic DNAs from three nonsusceptible isolates, CB1616-d20, CB1617-d20, and CB1618-d20. CGS validation experiments utilized genomic DNA from a ciprofloxacin-resistant isolate (MW2-CiproR) with a known mutation in the topoisomerase IV gene, grlA. The microarray used for the validation experiment included bases 1 to 1543993 of the S. aureus MW2 genome.
PCR amplification and DNA sequencing.
CGS-identified single nucleotide changes were confirmed by sequencing of PCR-amplified regions surrounding each putative nucleotide alteration. The PCR and sequencing primers used are shown in Table 2. Single nucleotide changes identified by CGS that were not confirmed by sequencing were considered false positives and will not be described further. PCR amplification and sequencing were performed as a service for a fee (SeqWright, Inc., Houston, TX).
TABLE 2.
Oligonucleotide primers and PCR products used in this study
| Primer | 5′-3′ primer sequence | Product length (bp) |
|---|---|---|
| rpoB amplificationa | ||
| rpoB-1Fwd | AGCGGATCACATAATTTTTGAGGGG | 977 |
| rpoB-1Rev | TTTTACGACGATCAAGCACTGTACC | |
| rpoB-2Fwd | GTGAACCACCAACTGTTGAAAATGC | 1,047 |
| rpoB-2Rev | CAACTTCATCATCCATGAAACGACC | |
| rpoB-3Fwd | TGACGTTCACTACTCTCACTATGGC | 1,077 |
| rpoB-3Rev | GCATGTAACAATCTTTCTTCGGCAG | |
| rpoB-4Fwd | CTAAGTTAGGACCTGAAGAAATCAC | 953 |
| rpoB-4Rev | GTTTTCACCTTTAACAATAGCCTCG | |
| rpoB-5Fwd | GAAGAAGATATGCCTTACTTACCAG | 1,063 |
| rpoB-5Rev | GTATTTACCACAACTACATTCCCAG | |
| Additional rpoB primers for sequence determination | ||
| rpoB-1RevS | GCTTGTTCAGTGTTTTCAGTG | |
| rpoB-2RevS | GCCATAGTGAGAGTAGTGAACG | |
| rpoB-3RevS | CATGATAACGGCATCCTCATAG | |
| rpoB-4RevS | AATATGGTCCTGTTGAACGC | |
| rpoB-5FwdS | AAGAAGCTGGTATGGCTCG | |
| rpoC amplificationa | ||
| rpoC-1Fwd | CCAAGTGTTCCAGAATCATTCCG | 1,070 |
| rpoC-1Rev | ATCGTCCACCATCTAATTGAACC | |
| rpoC-2Fwd | CCAATCATCCCACCAGAAATTCG | 998 |
| rpoC-2Rev | TTACCTACTGACGTAGCAAGAATC | |
| rpoC-3Fwd | ATGCAGTAAATACAGGCGCAATC | 950 |
| rpoC-3Rev | AGTACCACAGTCTTCTTCACGAAC | |
| rpoC-4Fwd | CAGTTGTTGAAATTTGGACAGATGC | 985 |
| rpoC-4Rev | GAACCTTCAGTTAATACTTCACCAC | |
| rpoC-5Fwd | AACCAGGTACACAGCTTACAATG | 959 |
| rpoC-5Rev | TTATCCTCCAAAGTTCTGCTTGC | |
| rpoC-6Fwd | AACCTGAAACAATCAACTACCG | 820 |
| rpoC-6Rev | CGACCACGACGACCATTATC | |
| Additional rpoC primers for sequence determination | ||
| rpoC-1FwdS | ACGCAATTTACAAAACAGGC | |
| rpoC-2RevS | GTTTTGTGCTGCTAACATCAAC | |
| rpoC-3RevS | ACGGAATGAAGATGTGATTGG | |
| rpoC-4FwdS | TCATTCCGTGAAGGTTTAACAG | |
| rpoC-5RevS | CGTATTTTACGTCGCTATAACG | |
| yycF and yycG amplificationa | ||
| yycFG-1Fwd | CAAAAATAGTAAGCGACATCCTGTG | 1,201 |
| yycFG-1Rev | GTGATAGTGCTTTTTGGACAGAAC | |
| yycFG-2Fwd | AAGTGGCTAAAACAACTACAATCCC | 1,056 |
| yycFG-2Rev | TGTTTCCTGCACAATCGTACTAAAG | |
| yycFG-3Fwd | ACGTAGACTGGACTCAGTTATCACC | 1,098 |
| yycFG-3Rev | ATACGACACTCATCAAGACGAGTAG | |
| Additional yycF and yycG primers for sequence determination | ||
| yycFG-1FwdS | GCATACGATGGTAATGATGCAG | |
| yycFG-2FwdS | GCTCCGTAAATGCACAAAAAG | |
| yycFG-3FwdS | TGCAGGAAACAGGATTTGTAAC | |
| mprF-amplificationa | ||
| mprF-1Fwd | ATGGTGCTTATAATGTTGGGCAG | 1,046 |
| mprF-1Rev | CGACACTAAAGTGCAGTACAATC | |
| mprF-2Fwd | GATAAGATTACATGGGTAAGATGGG | 1,002 |
| mprF-2Rev | CCAGAAGTAATAGCGCAATACAG | |
| mprF-3Fwd | TATTGTAGCTTTCCGTAGAGCAC | 915 |
| mprF-3Rev | GGCAACCATCTAATTAAATCGACTG | |
| mprF-4Fwd | CAAGTTACAGATCAACACATGCC | 964 |
| mprF-4Rev | GCAGAAGAACAAAAAATACGCCAAG | |
| Additional mprF primers for sequence determination | ||
| mprF-1RevS | CCGAAACCTACAATCGCATTC | |
| mprF-2RevS | ACGGAAAGCTACAATAAGCAG | |
| mprF-3FwdS | GGTATGATTGCATGGTTGTTTG | |
| mprF-4RevS | TGTGACGTATTACACGCATTAC | |
| MW2528 amplification | ||
| MW2528-1Fwd | GCGTATCATCATACCTTACGCATTG | 1,072 |
| MW2528-1Rev | ATTGTCATGGATGGGGATTTGCAC | |
| MW2528-2Fwd | TGTTGCCCAAACTAAGTCATCTTC | 960 |
| MW2528-2Rev | CTGAGTACAAGCATACACACAAGC | |
| MW2528-3Fwd | TCCATCTTTTTGACCGGCAATAAC | 800 |
| MW2528-3Rev | CATTATTACGCTGTGGGAAAATAGG | |
| Intergenic regions | ||
| 1512174C-G Fwd | TGTAATGTCATCAAGGTGTCTC | 709 |
| 1512174C-G Rev | CGTAACCCTCAAACTGGTAAAG | |
| 1654504C-T Fwd | TTGTTCAACCCAAGTATCTCCAGC | 902 |
| 1654504C-T Rev | ACAATTTGAAGGTGACGAATCACTC | |
| 2041052C-G Fwd | GGCTTGCGCTTCATTAACCAATTC | 994 |
| 2041052C-G Rev | CACATAAATAACCCCAAGTCTGCTG | |
| 2705330T-C Fwd | AAGTAATTCGTCAACGCGGTCAAC | 769 |
| 2705330T-C Rev | TTTAGCAGAGGCAAACATGGGCAC | |
| grlA amplification and sequencing primers for ciprofloxacin resistance validation | ||
| LF-3 | GCATTGCCAGATGTTCGTG | 278 |
| LF-4 | GCTTCAGTGTAACGCATTGC |
The primers designed for PCR amplification were also used to sequence the PCR products.
To identify mutations present in clinical isolates, primers were designed to amplify and sequence the open reading frames containing mprF, yycF, yycG, rpoB, and rpoC (Table 2).
RESULTS
Isolation of mutants with decreased susceptibility to daptomycin.
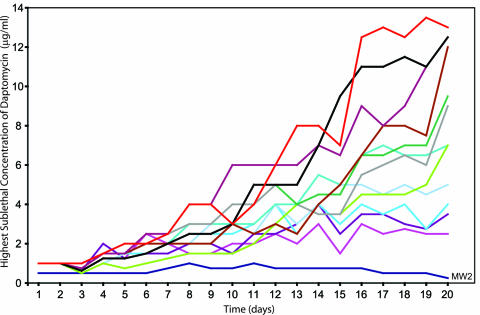
Isolates with decreased susceptibility to daptomycin were generated by serial passage of susceptible S. aureus MW2 (MIC, 0.75 μg/ml) through progressively increasing concentrations of daptomycin. The highest concentration of daptomycin in which the bacteria grew was plotted daily for 12 serial-passage experiments (Fig. 1). All experiments demonstrated the ability of S. aureus MW2 to grow in successively higher concentrations of daptomycin over time. After 20 days of serial passage, the MICs for individual experiments ranged from 3 to 20 μg/ml. The general trend toward growth in higher concentrations of daptomycin resulted from a series of small incremental changes over the course of 20 days, as opposed to ciprofloxacin or rifampin, where one or two mutations can account for the entire MIC shift (8, 42).
FIG. 1.
Serial-passage experiments with S. aureus MW2 selected for increasing resistance to daptomycin. Each line indicates a serial-passage experiment. The results for a control passage experiment with drug-free medium are included (the dark blue line labeled MW2).
Determination of the genetic changes that correlate with decreased susceptibility to daptomycin.
Genetic changes that correlate with increasing daptomycin MICs were identified by whole-genome comparisons between serial-passage-derived strains for which the daptomycin MIC increased and S. aureus MW2. Genomes were compared by using an array-based DNA hybridization and comparison service (CGS).
We validated CGS by analyzing a ciprofloxacin-resistant derivative of S. aureus MW2 from the fifth day of a serial passage in increasing concentrations of ciprofloxacin. Mutations in topoisomerase IV (grlA) are common first-step mutations in fluoroquinolone resistance (8). We designed primers to amplify and sequence a 278-bp region surrounding the fluoroquinolone resistance-determining region in topoisomerase IV (grlA) (Table 2) and identified a strain containing a C-to-T mutation resulting in a Ser80Phe substitution in GrlA. Genomic DNA from the mutant was compared with DNA from the parent. CGS identified the grlA mutation among 1.5 Mbp of genomic DNA (to contain costs during validation, we analyzed half of the complete genome sequence).
Three S. aureus strains with daptomycin MICs of 6, 8, and 10 μg/ml were selected from day 20 of independent serial-passage experiments for CGS analysis (CB1617-d20, CB1616-d20, and CB1618-d20). Figure 2 shows the daily daptomycin MICs for individual representatives of the archived populations. Multiple mutations were found in all three strains for which the daptomycin MIC increased. CGS-predicted nucleotide changes were confirmed by PCR amplification of 700 to 1,200 bp surrounding the putative mutation, followed by sequencing of the PCR product.
Confirmed alterations in strains for which the daptomycin MIC increased.
Single nucleotide changes that result in amino acid substitutions are listed in Table 3. Four proteins were identified by CGS: the lysylphosphatidylglycerol synthetase MprF (also known as FmtC), the sensor kinase YycG, and the β and β′ chains of RNA polymerase RpoB and RpoC. Each protein is altered by a single amino acid change. All three strains contain alterations in MprF and the RNA polymerase. Two out of three strains contain alterations in YycG.
TABLE 3.
Single nucleotide changes located in open reading frames
| Strain and sitea | Base change | Amino acid change | Locus | Day of change |
|---|---|---|---|---|
| MW2-CiproR | ||||
| 1358553 | C→T | S80F | grlA | |
| CB1616-d20 | ||||
| 575764 | T→C | F632S | rpoC | 11 |
| 1366542 | C→T | P314L | mprF | 3 |
| CB1617-d20 | ||||
| 26304 | T→C | S221P | yycG | 19 |
| 573039 | T→G | I953S | rpoB | 14 |
| 1366635 | C→T | T345I | mprF | 5 |
| CB1618-d20 | ||||
| 26430 | C→T | R263C | yycG | 9 |
| 573435 | C→T | A1086V | rpoB | 13 |
| 576750 | C→A | Q961K | rpoC | 20 |
| 1366634 | A→G | T345A | mprF | 6 |
Location of mutations in the S. aureus strain MW2 sequence (GenBank accession no. BA000033). For MW2-CiproR, only the first 1.5 Mb of the genome sequence were tested.
Nucleotide changes found in intergenic regions are described in Table 4. All three strains contained the same G-to-A change 77 nucleotides (nt) upstream of MW2528 (acetate-coenzyme A [CoA] ligase [EC 6.2.1.1]/acetyl-CoA synthetase).
TABLE 4.
Single nucleotide changes located in intergenic regions
| Strain and sitea | Base change | Location |
|---|---|---|
| CB1616-d20 | ||
| 1512174 | C→G | 22 nt upstream of MW1361 (hypothetical protein) |
| 2694054 | G→A | 77 nt upstream of MW2528 (acetate-CoA ligase [EC 6.2.1.1]/acetyl-CoA synthetase) |
| CB1617-d20 | ||
| 1654504 | C→T | 14 nt upstream of rpsU, 30S ribosomal protein S21 |
| 2041052 | C→G | 41 nt upstream of MW1875, hypothetical protein, putative transcription regulator, GntR family |
| 2694054 | G→A | 77 nt upstream of MW2528 (acetate-CoA ligase [EC 6.2.1.1]/acetyl-CoA synthetase) |
| CB1618-d20 | ||
| 2694054 | G→A | 77 nt upstream of MW2528 (acetate-CoA ligase [EC 6.2.1.1]/acetyl-CoA synthetase) |
| 2705330 | T→C | 13 nt upstream of nrdD, anaerobic ribonucleoside triphosphate reductase |
Location of mutations in the S. aureus strain MW2 sequence (GenBank accession no. BA000033).
Multiple mutations acquired over time.
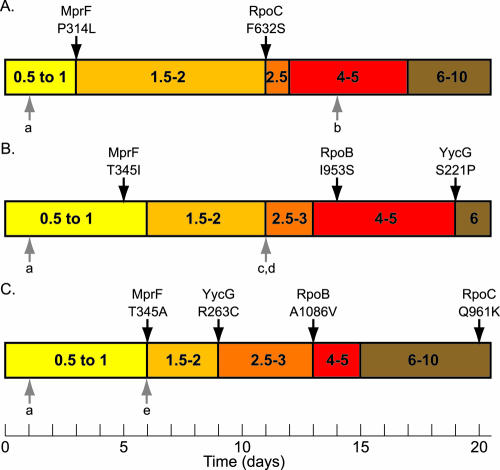
To determine the order of mutation acquisition, we sequenced the mutated regions in archived strains representing the 20 days of daptomycin serial passage. Figure 3 outlines the three serial-passage experiments and indicates the first day on which each mutation was identified. In all three serial-passage experiments, mutations accumulated over time. Of note, the nucleotide change at position 2694054, 77 nt upstream of MW2528, was the first mutation to appear in all three serial-passage experiments, although it did not correlate directly with an increase in the daptomycin MIC. The second change found in all three serial-passage experiments occurred in mprF. In two out of three strains, the mprF mutation correlated with the first increase in the daptomycin MIC; in CB1617, the mutation occurred 1 day before the MIC shift. Changes in yycG, rpoB, and rpoC also correlated with increases in daptomycin MICs, although the order of acquisition of these mutations is strain specific. Multiple mutations also correlated with decreased fitness, as evidenced by smaller colonies and a decreased growth rate for CB1618-d20 (data not shown).
FIG. 3.
Time lines representing three serial-passage experiments. Panels: A, CB1616; B, CB1617; C, CB1618. The daptomycin MICs are divided into five ranges, as indicted by colored bars, with daptomycin MIC in micrograms per milliliter. Black arrows above the bars indicated the first day that a mutation resulting in an amino acid change was identified. The protein name and the amino acid change found in each protein are indicated directly above the respective arrow. Gray arrows below the bars represent the first day that a mutation was identified in the intergenic regions (a, 2694054; b, 1512174; c, 1654504; d, 2041052; e, 2705330; see Table 4 for additional details). Once present, the mutation was found on all subsequent days of the experiment.
CGS-identified genes are altered in posttreatment clinical isolates for which the daptomycin MIC increased.
The genes identified in serial-passage strains, mprF, yycF, yycG, rpoB, and rpoC, were analyzed in three pairs of clinical isolates. Each pair consisted of a susceptible baseline isolate (MICs, ≤1 μg/ml) and a daptomycin-nonsusceptible, clonal, posttherapy isolate (MIC of 4 μg/ml) (Table 1 and Table 5). All daptomycin-nonsusceptible isolates had mutations in mprF leading to a single amino acid change in the protein. One of the three strains had a nucleotide insertion resulting in a frameshift in yycG. Since yycG is part of an essential two-component response regulator, we were surprised to find a frameshift mutation early in the yycG open reading frame; this result was confirmed multiple times. No changes were identified in yycF, rpoB, or rpoC in clinical isolates.
TABLE 5.
Single nucleotide changes found in daptomycin-nonsusceptible posttherapy clinical isolates
| Strain | Sitea | Base change | Amino acid change | Locus |
|---|---|---|---|---|
| CB184 | 1366486 | C→T | S295L | mprF |
| CB185 | 1368078 | C→T | L826F | mprF |
| CB1634 | 1368078 | C→T | L826F | mprF |
| CB1634 | 26121 | Inserted Ab | Frameshift | yycG |
Location of mutations S. aureus strain MW2 sequence (GenBank accession no. BA000033).
Confirmed in multiple independent PCRs and sequencing reactions.
DISCUSSION
Development of increases in the daptomycin MIC.
The daily passage of S. aureus MW2 in increasing concentrations of daptomycin generated isogenic strains for which the daptomycin MIC increased. These strains were then used in complete-genome comparisons with their daptomycin-susceptible parent strain to reveal mutations that correlate with increases in the daptomycin MIC. The serial-passage experiments revealed information about the acquisition of decreased susceptibility to daptomycin. By comparing 12 serial-passage experiments from a single strain background, we observed a gradual increase in the daptomycin MIC over the 20 days of the experiment (Fig. 1). All serial-passage experiments gave rise to an increase in the daptomycin MIC from 0.75 μg/ml to between 3 and 20 μg/ml after 20 days. The largest increase in the daptomycin MIC was relatively small (30-fold increase) compared with the greater-than-140-fold increase that resulted when ciprofloxacin resistance was selected for in a similar serial-passage experiment (unpublished observation). On a day-to-day basis, decreases in daptomycin susceptibility resulted in less-than-fourfold changes in the MIC and no two experiments followed the same path to the final MIC. Since single nucleotide changes can correlate with specific decreases in drug susceptibility, we propose that the series of small elevations in the MIC may result from a number of mutational changes.
Genes involved with decreased susceptibility to daptomycin.
Four open reading frames representing three distinct proteins are implicated in daptomycin MIC increases: MprF, a lysylphosphatidylglycerol synthetase; YycG, a sensor histidine kinase; and RpoB and RpoC, the β and β′ subunits of the RNA polymerase. These proteins were altered in at least two out of the three strains tested.
Mutations in mprF were found in all three strains, as well as in the three nonsusceptible clinical isolates. The mprF mutations correlated with the first increase in the daptomycin MIC. Genetic and biochemical evidence implicates MprF in the addition of a lysine to membrane phosphatidylglycerol to produce lysylphosphatidylglycerol (18, 30). The composition of the cell membrane influences the antimicrobial activity of cationic antimicrobial peptides and peptides from the innate immune system (22, 26, 31, 40). MprF knockout mutants have increased susceptibility to cationic antimicrobial peptides, methicillin in methicillin-resistant S. aureus, and altered vancomycin resistance levels (21, 29, 33). The impact of the observed mutations on MprF activity is under investigation. Nonetheless, the influence of the phospholipid content of the cell membrane on daptomycin activity should be expected. Previous studies have shown that daptomycin enters phospholipid bilayers and causes loss of membrane potential and potassium leakage (20, 38). Calcium and phospholipid content influences the amount of daptomycin that enters liposomes (20). Further studies of how the amino acid changes in MprF alter protein function may lead to a better understanding of how daptomycin interacts with the S. aureus cell membrane.
The second protein implicated in decreased susceptibility to daptomycin is the histidine kinase YycG of the essential two-component response regulator YycF/YycG (25). This protein was altered in two out of three of the serial-passage strains and one of the nonsusceptible clinical isolates. Studies with Bacillus subtilis and S. aureus have identified numerous genes predicted to be regulated by YycF/YycG (4, 7, 11, 16). The exact mechanism by which the changes in YycG alter the protein's function is not known. Martin et al. suggested that yycF and yycG are involved in cell permeability and showed that loss of yycF activity results in increased susceptibility to macrolide and lincosamide antibiotics and unsaturated long-chain fatty acids (25). Since YycF/YycG is essential, we predict that the alterations resulting in daptomycin resistance do not shut down the protein's function. Interestingly, the alteration in the daptomycin-nonsusceptible clinical isolate is the result of the insertion of a single nucleotide. This insertion should create a frameshift early in the protein sequence. We might have predicted that early frameshift mutations in yycG would result in cell death. Possible explanations include an alternative sensor kinase for YycF or the production of a functional protein even with the nucleotide insertion.
The third enzyme implicated in daptomycin MIC increases is RNA polymerase. Nucleotide changes conferring amino acid alterations in both the β and β′ subunits of RNA polymerase (RpoB and RpoC) were observed; these tended to appear later in the serial-passage experiment. The amino acid changes found in association with increases in the daptomycin MIC have not been described previously. They do not lie within the regions of rpoB known to confer resistance to the RNA polymerase inhibitor rifampin, nor do they overlap with mutations in rpoB and rpoC that confer resistance to other antibiotics, e.g., zwittermicin, streptolydigin, and microcin J25 (6, 35, 39, 42, 44, 45). Mutations in rpoB and rpoC could reflect direct interaction between daptomycin and the polymerase or could alter global gene expression in ways that alter susceptibility.
Five additional mutations were identified in intergenic regions. One of these, a mutation 77 bp upstream of MW2528, a putative acetate CoA ligase/acetyl-CoA synthetase, was found in all three strains. Although this mutation did not correlate with an elevation of the daptomycin MIC, it may be required in combination with other mutations for daptomycin MIC increases. Alternatively, selective pressures of growth under the serial-passage experimental conditions, separate from drug pressure, may immediately select for this nucleotide alteration. The G-to-A nucleotide change was not found in the three nonsusceptible posttherapy clinical isolates.
The identification of the same three proteins in at least two out of the three CGS-analyzed strains supports the involvement of these proteins in daptomycin MIC increases. Mutations in mprF and yycG are further implicated by their identification in nonsusceptible isolates that have emerged in the clinic after weeks of daptomycin therapy (13, 41). These results suggest that selective pressures found in laboratory serial-passage experiments parallel the selective pressures encountered in clinical settings, particularly in difficult infections where organisms might not be exposed to adequate levels of antibiotic (e.g., undrained abscess, dead bone). Further understanding of disease states may help in establishing proper treatment protocols and avoiding resistance development.
Laboratory-derived S. aureus strains for which the daptomycin MICs are greater than or equal to 6 μg/ml accumulated three or more mutations in three distinct genetic regions. Since the accumulation of multiple mutations over the course of the serial-passage experiment appears to be required for the acquisition of daptomycin MICs of 6 to 10 μg/ml, this may explain the lack of clinically derived S. aureus isolates for which the daptomycin MICs are high (≥16 μg/ml).
CGS analysis identified between four and six mutations per strain for which the daptomycin MIC was elevated (Tables 3 and 4 and Fig. 3). Most of the mutations correlate with elevations in the daptomycin MIC. Nevertheless, some increases in the daptomycin MIC are not associated with known mutations. Our experiments may not have identified all of the mutations that correlate with increases in the daptomycin MIC because of limitations in the CGS technology and because of the experimental design. CGS is based on DNA hybridization and can only identify single nucleotide changes and deletions in nonredundant DNA sequences. Single nucleotide insertions, certain rearrangements, and mutations in repetitive sequences might not be detected. Our experimental design provided CGS analysis only of strains from the 20th day of the serial-passage experiment on the basis of the assumption that the majority of the mutations acquired over time will be fixed in the genome. Although this assumption appears to hold true for the mutations isolated on day 20, we did not perform CGS on daily isolates, so we may have missed a number of mutations. Mutations acquired early in the experiment that decrease daptomycin susceptibility could be lost after selection for subsequent mutations that supersede the need for the previous mutation. CGS analysis on each daily isolate would be necessary to determine the presence or absence of such transitional genetic alterations.
The ability to detect multiple mutations scattered throughout the genome that correlate with resistance is a significant advance over traditional genetic approaches and can be applied to any sequenced genome. After confirmation of success with S. aureus, Albert et al. went on to identify mutations that correlate with metronidazole resistance in Helicobacter pylori (1). The present study expands our understanding of daptomycin susceptibility. Future study of these genes will expand our knowledge of how daptomycin functions and might provide guidance leading to better treatment of patients.
Acknowledgments
We thank the employees of Cubist Pharmaceuticals, Inc., who helped edit the manuscript.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 2.Alder, J. D. 2005. Daptomycin: a new drug class for the treatment of gram-positive infections. Drugs Today 41:81-90. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 4.Clausen, V. A., W. Bae, J. Throup, M. K. Burnham, M. Rosenberg, and N. G. Wallis. 2003. Biochemical characterization of the first essential two-component signal transduction system from Staphylococcus aureus and Streptococcus pneumoniae. J. Mol. Microbiol. Biotechnol. 5:252-260. [DOI] [PubMed] [Google Scholar]
- 5.Critchley, I. A., D. C. Draghi, D. F. Sahm, C. Thornsberry, M. E. Jones, and J. A. Karlowsky. 2003. Activity of daptomycin against susceptible and multidrug-resistant gram-positive pathogens collected in the SECURE study (Europe) during 2000-2001. J. Antimicrob. Chemother. 51:639-649. [DOI] [PubMed] [Google Scholar]
- 6.Delgado, M. A., M. R. Rintoul, R. N. Farias, and R. A. Salomon. 2001. Escherichia coli RNA polymerase is the target of the cyclopeptide antibiotic microcin J25. J. Bacteriol. 183:4543-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubrac, S., and T. Msadek. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J. Bacteriol. 186:1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrero, L., B. Cameron, and J. Crouzet. 1995. Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrero, L., B. Cameron, B. Manse, D. Lagneaux, J. Crouzet, A. Famechon, and F. Blanche. 1994. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol. Microbiol. 13:641-653. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuchi, K., Y. Kasahara, K. Asai, K. Kobayashi, S. Moriya, and N. Ogasawara. 2000. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology 146:1573-1583. [DOI] [PubMed] [Google Scholar]
- 12.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayden, M. K., K. Rezai, R. A. Hayes, K. Lolans, J. P. Quinn, and R. A. Weinstein. 2005. Development of daptomycin resistance in vivo in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5285-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holden, M. T. G., E. J. Feil, J. A. Linsay, S. J. Peacock, N. P. J. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggestt, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper, D. C. 2002. Fluoroquinolone resistance among gram-positive cocci. Lancet Infect. Dis. 2:530-538. [DOI] [PubMed] [Google Scholar]
- 16.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 49:1639-1655. [DOI] [PubMed] [Google Scholar]
- 17.Hughes, A. L., and R. Friedman. 2005. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J. Bacteriol. 187:2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichihashi, N., K. Kurokawa, M. Matsuo, C. Kaito, and K. Sekimizu. 2003. Inhibitory effects of basic or neutral phospholipid on acidic phospholipid-mediated dissociation of adenine nucleotide bound to DnaA protein, the initiator of chromosomal DNA replication. J. Biol. Chem. 278:28778-28786. [DOI] [PubMed] [Google Scholar]
- 19.Ito, T., K. Okuma, X. X. Ma, H. Yuzawa, and K. Hiramatsu. 2003. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist. Updates 6:41-52. [DOI] [PubMed] [Google Scholar]
- 20.Jung, D., A. Rozak, M. Okon, and R. E. W. Hancock. 2004. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem. Biol. 11:949-957. [DOI] [PubMed] [Google Scholar]
- 21.Komatsuzawa, H., K. Ohta, T. Fujiwara, G. H. Choi, H. Labischinski, and M. Sugai. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 203:49-54. [DOI] [PubMed] [Google Scholar]
- 22.Kristian, S. A., M. Durr, J. A. Van Strijp, B. Neumeister, and A. Peschel. 2003. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect. Immun. 71:546-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 24.Malbrunhy, B., A. Cunu, B. Bozdogan, B. Fantin, V. Zarrouk, S. Dutka-Malen, C. Feger, and R. Leclercq. 2002. Resistance to quinupristin-dalfopristin due to mutations of L22 ribosomal protein in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2200-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, P. K., T. Li, D. Sun, D. P. Biek, and M. B. Schmid. 1999. Role in cell permeability of an essential two-component system in Staphylococcus aureus. J. Bacteriol. 181:3666-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Midorikawa, K., K. Ouhara, H. Komatsuzawa, T. Kawai, S. Yamada, T. Fujiwara, K. Yamazaki, K. Sayama, M. A. Taubman, H. Kurihara, K. Hashimoto, and M. Sugai. 2003. Staphylococcus aureus susceptibility to innate antimicrobial peptides, β-defensins and CAP18, expressed by human keratinocytes. Infect. Immun. 71:3730-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard. NCCLS publication no. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.National Committee for Clinical Laboratory Standards. 2004. Standards for antimicrobial susceptibility testing, 13th informational supplement. NCCLS publication no. M100-S14. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 29.Nishi, H., H. Komatsuzawa, T. Fujiwara, N. McCallum, and M. Sugai. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:4800-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oku, Y., K. Kurokawa, N. Ichihashi, and K. Sekimizu. 2004. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150:45-51. [DOI] [PubMed] [Google Scholar]
- 31.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pillai, S. K., G. Sakoulas, C. Wennersten, G. M. Eliopoulos, R. C. Moellering, Jr., M. J. Ferraro, and H. S. Gold. 2002. Linezolid resistance in Staphylococcus aureus: characterization and stability of resistant phenotype. J. Infect. Dis. 186:1603-1607. [DOI] [PubMed] [Google Scholar]
- 33.Ruzin, A., A. Severin, S. L. Moghazeh, J. Etienne, P. A. Bradford, S. J. Projan, and D. M. Shlaes. 2003. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim. Biophys. Acta 1621:117-121. [DOI] [PubMed] [Google Scholar]
- 34.Sader, H. S., T. R. Fritsche, J. M. Streit, and R. N. Jones. 2005. Daptomycin in vitro activity tested against gram-positive strains collected from European and Latin American medical centers in 2003. J. Chemother. 17:477-483. [DOI] [PubMed] [Google Scholar]
- 35.Severinov, K., D. Markov, E. Severinova, V. Nikiforov, R. Landick, S. A. Darst, and A. Goldfarb. 1995. Streptolydigin-resistant mutants in an evolutionarily conserved region of the β′ subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 270:23926-23929. [DOI] [PubMed] [Google Scholar]
- 36.Shinabarger, D. L. 1999. Mechanism of action of the oxazolidinone antibacterial agents. Expert Opin. Investig. Drugs 8:1195-1202. [DOI] [PubMed] [Google Scholar]
- 37.Silverman, J. A., N. Oliver, T. Andrew, and T. Li. 2001. Resistance studies with daptomycin. Antimicrob. Agents Chemother. 45:1799-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silverman, J. A., N. G. Perlmutter, and H. M. Shapiro. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stabb, E. V., and J. Handelsman. 1998. Genetic analysis of zwittermicin A resistance in Escherichia coli: effects on membrane potential and RNA polymerase. Mol. Microbiol. 27:311-322. [DOI] [PubMed] [Google Scholar]
- 40.Staubitz, P., H. Neumann, T. Schneider, I. Wiedemann, and A. Peschel. 2004. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 231:67-71. [DOI] [PubMed] [Google Scholar]
- 41.Vikram, H. R., N. L. Havill, L. M. Koeth, and J. M. Boyce. 2005. Clinical progression of methicillin-resistant Staphylococcus aureus vertebral osteomyelitis associated with reduced susceptibility to daptomycin. J. Clin. Microbiol. 43:5384-5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wichelhaus, T. A., B. Boddinghaus, S. Besier, V. Schafer, V. Brade, and A. Ludwig. 2002. Biological cost of rifampin resistance from the perspective of Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong, C. W., T. J. Albert, V. B. Vega, J. E. Norton, D. J. Cutler, T. A. Richmond, L. W. Stanton, E. T. Liu, and L. D. Miller. 2004. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 14:398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, X., and C. W. Price. 1995. Streptolydigin resistance can be conferred by alterations to either the β or β′ subunits of Bacillus subtilis RNA polymerase. J. Biol. Chem. 270:23930-23933. [DOI] [PubMed] [Google Scholar]
- 45.Yuzenkova, J., M. Delgado, S. Nechaev, D. Savalia, V. Epshtein, I. Artsimovitch, R. A. Mooney, R. Landick, R. N. Farias, R. Salomon, and K. Severinov. 2002. Mutations of bacterial RNA polymerase leading to resistance to microcin J25. J. Biol. Chem. 277:50867-50875. [DOI] [PubMed] [Google Scholar]