Abstract
Pseudomonas aeruginosa is inherently resistant to most conventional antibiotics. The mechanism of resistance of this bacterium is mainly associated with the low permeability of its outer membrane to these agents. We sought to assess the bactericidal efficacy of liposome-entrapped aminoglycosides against resistant clinical strains of P. aeruginosa and to define the mechanism of liposome-bacterium interactions. Aminoglycosides were incorporated into liposomes, and the bactericidal efficacies of both free and liposomal drugs were evaluated. To define the mechanism of liposome-bacterium interactions, transmission electron microscopy (TEM), flow cytometry, lipid mixing assay, and immunocytochemistry were employed. Encapsulation of aminoglycosides into liposomes significantly increased their antibacterial activity against the resistant strains used in this study (MICs of ≥32 versus ≤8 μg/ml). TEM observations showed that liposomes interact intimately with the outer membrane of P. aeruginosa, leading to the membrane deformation. The flow cytometry and lipid mixing assays confirmed liposome-bacterial membrane fusion, which increased as a function of incubation time. The maximum fusion rate was 54.3% ± 1.5% for an antibiotic-sensitive strain of P. aeruginosa and 57.8% ± 1.9% for a drug-resistant strain. The fusion between liposomes and P. aeruginosa significantly enhanced the antibiotics' penetration into the bacterial cells (3.2 ± 2.3 versus 24.2 ± 6.2 gold particles/bacterium, P ≤ 0.001). Our data suggest that liposome-entrapped antibiotics could successfully resolve infections caused by antibiotic-resistant P. aeruginosa through an enhanced mechanism of drug entry into the bacterial cells.
Cystic fibrosis (CF) is the most common inherited lethal genetic disorder in Caucasian populations. CF is the result of mutations in the cystic fibrosis transmembrane conductance regulator gene leading to a series of cellular dysfunctions (21). Although many organs are affected, the colonization of the lungs and recurrent infections with Pseudomonas aeruginosa are the major cause of morbidity and mortality in CF patients (13). In addition, as an opportunistic microbe, P. aeruginosa causes acute pneumonia in individuals with underdeveloped or impaired immune defense systems (7, 34, 39).
P. aeruginosa exhibits several antibiotic resistance mechanisms including enzymatic inactivation of drugs, target site alteration, and antibiotic efflux systems (11, 12, 30, 31, 35). However, the low outer-membrane permeability of P. aeruginosa is the major factor contributing to antibiotic resistance (10, 11, 18, 30). Reports indicate that as many as 90% of P. aeruginosa strains isolated from CF patient lungs are multidrug resistant with low outer-membrane permeability (37, 43). Furthermore, P. aeruginosa in CF patient lungs undergoes a phenotypic change from nonmucoid to mucoid and adopts a biofilm mode of growth that is more resistant to antibiotics (20, 39, 41).
Pseudomonal lung infections are commonly treated with aminoglycosides alone or in combination with β-lactams (5, 14). Aminoglycosides are potent antibiotics that inhibit protein synthesis by binding to the bacterial ribosomes (19). Some of the major problems associated with aminoglycosides, however, include serious ototoxicity and nephrotoxicity as well as the emergence of low-permeability drug-resistant P. aeruginosa strains (4, 30, 32).
A delivery system that reduces the drugs' toxicity while increasing their therapeutic index is of great interest, and liposomes can provide these benefits. Liposomes are colloidal vesicles ranging from a few nanometers to several micrometers in diameter (1, 16). Hydrophilic drugs such as aminoglycosides can be entrapped in aqueous compartments of liposomes, whereas hydrophobic drugs are incorporated in their lipid bilayers (17).
The objectives of this study were (i) to investigate the antibacterial activity of liposome-entrapped aminoglycosides (amikacin, gentamicin, and tobramycin), (ii) to assess their bactericidal efficacy by performing killing curve assays, and (iii) to investigate their mechanism of action.
MATERIALS AND METHODS
Chemicals.
Amikacin, gentamicin, and tobramycin were obtained from Fisher Scientific (Ottawa, Ontario, Canada). 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) was from Northern Lipids (Vancouver, British Columbia, Canada). Cholesterol, Triton X-100, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (NBD-PE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (Rh-PE), and glutaraldehyde solution grade 1 were purchased from Sigma-Aldrich Canada Ltd. (Oakville, Ontario, Canada). All other chemicals were obtained from Fisher Scientific.
Organisms.
Nonmucoid strains (PA-1 and PA-48912-2) and mucoid strains (PA-48912-1 and PA-48913) of P. aeruginosa used in this study were isolated from sputum of CF patients with pulmonary infections at the Memorial Hospital (Sudbury, Ontario, Canada) and were maintained as described elsewhere (29). PA-48912-2 and PA-48913 were resistant to all aminoglycosides, with MICs ranging from 32 to 1,024 μg/ml. Although we do not know the mechanism of antibiotic resistance in these isolates, we infer that membrane impermeability is the culprit because their probe results were negative for all 14 aminoglycoside resistance genes by the DNA hybridization technique.
We also used laboratory strains of Staphylococcus aureus (ATCC 29213) and P. aeruginosa (ATCC 27853 and ATCC 10145) as test organisms as well as reference strains as quality control measures.
Preparation of liposomes.
Liposomes composed of DPPC and cholesterol (Northern Lipids, Vancouver, British Columbia, Canada) in a molar ratio of 2:1 (lipid to cholesterol) were prepared by a dehydration-rehydration method as previously reported (23).
Antimicrobial susceptibility testing.
The MICs of free and liposomal antibiotics for all strains were determined by the broth dilution technique as recommended by CLSI (formerly NCCLS) (26). Briefly, serial dilutions of free- or liposome-encapsulated antibiotics (1,024, 512, 256, 128, 64, 32, 16, 8, 4, 2, 1, and 0.5 μg/ml) in Mueller-Hinton broth were prepared. Bacterial suspensions were then added to each tube to achieve a final inoculum of 5 × 105 bacteria/ml. The lowest concentrations of antibiotic formulations that inhibited the visible bacterial growth were determined after 24 h. Two separate experiments in triplicate were performed for each formulation, and the procedure was validated by the quality control strains S. aureus and P. aeruginosa.
Time-kill method.
Killing curve assays were performed as previously described (28). Briefly, overnight cultures of P. aeruginosa in a final inoculum of 5 × 105/ml were incubated with either free- or liposome-encapsulated antibiotics at one, two, and four times their respective MICs. Control tubes contained no antibiotics. The tubes were then incubated at 37°C for 2, 4, 6, and 24 h. At the end of each time period, serial dilutions were prepared and the CFU on triplicate Muller-Hinton agar plates were determined.
Analysis of liposome-bacterium interactions. (i) Microscopic method.
Transmission electron microscopy (TEM) was utilized to monitor interactions between liposomes and bacteria. Briefly, overnight cultures (1.5 × 108 bacteria/ml) of the clinical isolates as well as the laboratory strain of P. aeruginosa were mixed with liposomes without antibiotic for 1 h at 37°C with agitation. Samples of bacterial suspensions on Formvar-coated copper grids were examined using the Hitachi HD-2000 transmission electron microscope.
(ii) Flow cytometry assays.
We chose a highly resistant clinical strain (PA-48912-2) and a sensitive laboratory strain (ATCC 10145) of P. aeruginosa to investigate the framework of liposome-bacterium interactions. Liposomes were labeled with a biological membrane probe, PKH2-GL (PKH2-GL labeling kit; Sigma, St. Louis, MO), as previously described (33). Briefly, PKH2-GL at a final concentration of 4 × 10−6 M was added to empty liposome suspensions in an isosmotic labeling vehicle. The reaction was stopped with bovine serum albumin, and the pellet was then washed twice before assessment of the liposomal labeling efficiency by fluorescence-activated cell sorting (FACS). Aliquots of overnight bacterial cultures in phosphate-buffered saline (PBS) were then incubated with either labeled liposomes, free PKH2-GL (positive control), or PBS alone (negative control). The mixtures (4 × 10−6 M) were incubated at 37°C with agitation, and samples were taken after 0.5-, 1-, 6-, and 10-h intervals. The aliquots were centrifuged through a sucrose cushion (21), washed twice, and fixed with 1% paraformaldehyde. Triplicate samples were then analyzed with an Epics Elite flow cytometer (Becton Dickinson, Mississauga, Canada).
(iii) Lipid mixing assay.
Integration of liposomes into bacterial membranes was assessed by the lipid mixing assay, which is based on the extent of resonance energy transfer between two fluorophores. We utilized NBD-PE or Rh-PE as the energy donor or acceptor, respectively (40). The labeled empty liposomes were mixed with bacterial suspensions (1.5 × 108 bacteria/ml) at 37°C with agitation. At different time intervals of 0.5, 1, 3, and 6 h, the aliquots (75 μl) were mixed with equal volumes of HEPES buffer and the fluorescence intensity was determined at 510 and 590 nm under steady-state excitation at 485 nm (NBD-PE maximum excitation level) using Fluostar Optima (BMG Labtech, Germany). Following each measurement, vesicles were disrupted with Triton X-100 to eliminate energy transfer, which allowed us to determine the NBD-PE and Rh-PE concentrations by their post-direct-excitation emission intensity. We calculated the percentage of liposomal fusion of three independent experiments as follows: fusion = (Ft − Fo)/(Ffinal − Fo), where Ft is the fluorescence intensity at each time point and Fo and Ffinal are the initial and final fluorescence intensities after vesicle disruption, respectively.
Antibiotic penetration assessment.
We utilized the immunogold technique to assess liposomal antibiotic penetration into the bacterial cells (33). Briefly, the antibiotic-resistant clinical strain of P. aeruginosa (PA-48912-2) was mixed with free or liposomal tobramycin at a final concentration of 128 μg/ml (two times the MIC of free tobramycin). PBS was used as a negative control. Samples were taken after 0, 2, 4, and 6 h of incubation at 37°C and centrifuged to remove the nonpenetrating drugs. The pellets were then prefixed in glutaraldehyde (0.5%), washed twice after 30 min at room temperature, and resuspended in PBS. For embedding in Spurr resin, samples were washed with 0.1 M cacodylate buffer (pH 7.4) and the pellets were encapsulated in 2% Bacto Agar and cut into 1-mm cubes. Several pieces of each sample were embedded in gelatin capsules filled with Spurr resin (epoxy resin) and polymerized overnight at 60°C. Ultrathin sections (70 to 90 nm) were then collected onto uncoated 300-mesh nickel grids and previewed under a Philips 400T transmission electron microscope. Selected samples were then prepared for immunogold labeling using monoclonal antibody to tobramycin (Cedarlane, Hornby, Ontario, Canada) and colloidal gold (10 nm) coupled to protein A/G (Sigma-Aldrich, Oakville, Ontario, Canada) (3). Control samples contained PBS instead of antitobramycin antibody. Samples were analyzed using a JEOL STEM (2011) transmission electron microscope, and images were captured with a Gatan Ultrascan digital camera.
Data analysis.
Bacterial counts in killing curves are expressed as the mean CFU ± standard deviation obtained from triplicate plates per dilution. The data from the lipid mixing assay are expressed as means ± standard errors of the means of three independent experiments. Comparisons were made by paired Student's t test, and the P ≤ 0.05 values were considered significant. Analysis of variance with the two-tailed Dunnett posttest analysis was used for multiple comparisons within and between the groups.
RESULTS
Antimicrobial activity of free and liposomal aminoglycosides.
We used small vesicles with a mean diameter of 210 ± 25 nm and an aminoglycoside entrapment efficiency of 28.20% ± 1.15% in this project. The MICs of free and liposomal aminoglycosides against nonmucoid and mucoid strains of P. aeruginosa isolates are significantly (P ≤ 0.05) lower than those of the corresponding free antibiotics (Table 1). For instance, PA-48912-2, a nonmucoid strain, was highly resistant to amikacin (MIC, 256 μg/ml) and tobramycin (MIC, 64 μg/ml) but sensitive (MIC, ≤8 μg/ml) to the aforementioned antibiotics encapsulated in liposomes. Likewise, the highly resistant mucoid strain PA-48913 (MIC, ≥256 μg/ml) was sensitive to liposomal amikacin (MIC, 8 μg/ml) but intermediate to liposomal gentamicin and tobramycin (MIC, 8 μg/ml). The MICs for quality control laboratory strains were within the acceptable limits established by CLSI (formerly NCCLS) (27). The liposomes containing PBS had no antibacterial activity. Likewise, the combination of empty liposomes with free drug had no additive effect on the antibacterial activity of these aminoglycosides.
TABLE 1.
Antimicrobial activities of free and liposomal aminoglycosides against one laboratory strain and four clinical strains of P. aeruginosaa
| P. aeruginosa strain | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| F-AMK | L-AMK | F-GEN | L-GEN | F-TOB | L-TOB | |
| ATCC 10145 | 8 | 4 | 4 | 2 | 2 | 1 |
| PA-1 | 16 | 4 | 16 | 2 | 4 | 4 |
| PA-48912-2 | 256 | 8 | 32 | 4 | 64 | 2 |
| PA-48912-1 | 4 | 2 | 32 | 0.5 | 1 | 0.5 |
| PA-48913 | 512 | 8 | 256 | 8 | 1,024 | 8 |
MICs were determined by utilizing a standard broth dilution technique (CLSI document M7-A6). Twofold dilution series of free (F) and liposomal (L) amikacin (AMK), gentamicin (GEN), and tobramycin (TOB) were prepared in a broth medium and were mixed with bacterial suspension to achieve a final inoculum of 5 × 105 bacteria/ml. The MICs were determined as the lowest concentration of the antibiotics that inhibited visible bacterial growth.
Time-killing study.
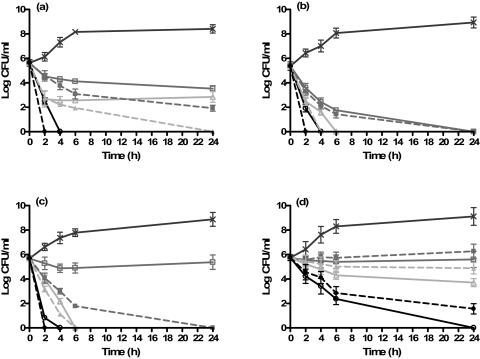
To confirm the MIC data and to evaluate the ability of liposomal aminoglycosides to eliminate P. aeruginosa, we performed killing curve assays on one laboratory strain (ATCC 10145) and three clinical isolates of P. aeruginosa. Figure 1a to d represents the killing curves of pseudomonal strains by free or liposomal aminoglycosides at one, two, or four times the MICs. Liposomal antibiotics were significantly (P ≤ 0.05) more effective in stopping the nonmucoid P. aeruginosa than the corresponding free drugs. For instance, liposomal amikacin and gentamicin at twice the MIC completely halted bacterial growth in 24 h. Although free drugs at twice the MIC had an initial bacterial killing power, they failed to eliminate bacterial growth. At four times the MIC, however, both liposomal and free drugs completely eradicated this strain in 2 and 4 h, respectively (Fig. 1a). The laboratory strain incubated with free tobramycin at one and two times the MIC demonstrated an initial drop in CFU after 6 h, but it regrew to its initial concentration after 24 h, while liposomal tobramycin at the MIC completely eradicated the bacteria in 6 h. At four times the MIC, both liposomal and free forms of tobramycin eliminated the laboratory strain of P. aeruginosa in 2 and 4 h, respectively (data not shown). Figure 1b demonstrates the killing curves of PA-48912-1, a mucoid clinical strain of P. aeruginosa. According to the MIC studies, this strain was sensitive to both forms of amikacin and tobramycin but their time-killing curve indicated a different efficacy pattern. For instance, at the MIC, free amikacin showed a modest initial drop in CFU that remained while the liposomal form led to a complete eradication in 4 h. At the MIC, both free and liposomal gentamicin led to a complete bacterial eradication in 24 h. However, we should point out that the concentration of gentamicin trapped in liposomes at this concentration (MIC, 0.5 μg/ml) was 64-fold lower than that of the free drug. Both liposomal and free gentamicin at two to four times the MIC led to complete bacterial eradication in 2 to 6 h, where free drugs lagged behind by 2 h.
FIG. 1.
Killing curves for a laboratory strain and three clinical strains of P. aeruginosa. Bacteria were exposed to one (squares), two (triangles), or four (circles) times the MICs (Table 1) of free (solid lines) and liposomal (broken lines) antibiotics. Control samples (×) contained no antibiotics. (a) ATCC 10145 exposed to amikacin; (b) PA-48912-1 exposed to gentamicin; (c) PA-48912-2 exposed to amikacin; (d) PA-48913 exposed to tobramycin.
Figure 1c demonstrates the killing curves of PA-48912-2, a nonmucoid clinical strain that is highly resistant to free amikacin and tobramycin but sensitive to all liposomal aminoglycosides. At the MIC (32-fold less than that of the free drug), only liposomal amikacin eliminated this resistant strain in 24 h. At two to four times the MIC, however, both free and liposomal amikacin completely eradicated the bacteria in 2 to 6 h.
Unlike amikacin, neither free gentamicin nor tobramycin eradicated PA-48912-2 even at the concentration equal to four times the MIC. Liposomal drugs, however, showed improved killing rates, although complete eradication was demonstrated only at four times the MIC in 24 h.
Figure 1d displays the killing curves of PA-48913, a mucoid clinical strain of P. aeruginosa which was extremely resistant (MIC, ≥256 μg/ml) to all free aminoglycosides tested but sensitive or intermediate to their liposomal formulations. Liposomal amikacin at twice the MIC (16 μg/ml) abolished bacterial growth in 6 h. The same killing effect was achieved with 512 μg/ml of the free amikacin. Liposomal tobramycin demonstrated a killing pattern similar to that of the free drug but at much lower concentrations (8 versus 1,024 μg/ml). Although liposomal gentamicin displayed a better killing pattern than that of the free drug, it was unable to completely eradicate this strain.
Collectively, the MICs shown in Table 1 and the killing curves described above confirm the higher potency of liposomal aminoglycosides than of the free antibiotics against resistant strains of P. aeruginosa.
Analysis of liposome-bacterium interactions by TEM.
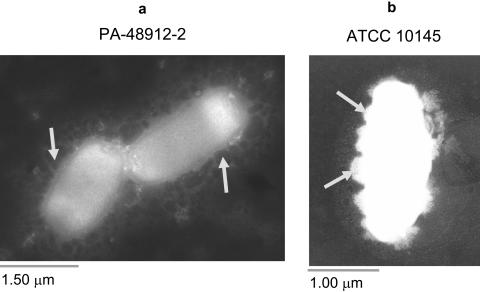
Figure 2a represents the interaction between antibiotic-free liposomes and PA-48912-2, a nonmucoid clinical strain that is highly resistant to free amikacin and tobramycin but sensitive to their liposomal formulations. The liposomes surrounded the cells, and some fused with the bacterial outer membrane (arrows) after 1 h of incubation at 37°C (Fig. 2b). Similar interactions were observed with other strains tested.
FIG. 2.
Analysis of liposome-bacterium interactions by transmission electron microscopy. Empty liposomes were incubated with overnight cultures of different pseudomonal strains at 37°C for 1 h and visualized by TEM. The liposomes (dark spheres) surrounded the bacterial cells (a; arrows), and some induced membrane deformation (b; arrows).
Flow cytometry analysis of liposome-bacterium interactions.
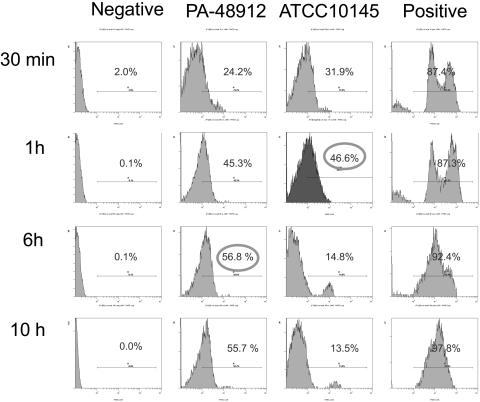
Integration of PKH2-GL-labeled empty vesicles into the membrane of P. aeruginosa is demonstrated by FACS analysis (Fig. 3). Bacteria incubated with PKH2-GL alone (positive control) or PBS (negative control) were used as controls. Liposomal PKH2-GL was incorporated into the membranes of antibiotic-sensitive as well as resistant strains of P. aeruginosa in a time-dependent fashion. The laboratory strain reached a maximum incorporation level of 46.6% in 1 h while the resistant clinical strain reached its peak of 56.8% in 6 h. Positive controls demonstrated that PKH2-GL is compatible with the membrane of P. aeruginosa, as it labeled nearly all bacterial membrane (97.8%) in 10 h. Samples were analyzed in triplicate, and data confirmed the fusion of liposomes with bacterial membranes as suggested by TEM observations.
FIG. 3.
Flow cytometric analysis of liposome-bacterium interactions. A sensitive laboratory strain, ATCC 10145, and a resistant clinical strain, PA-48912-2, were incubated with PBS (left panels), labeled empty liposomes (middle panels), or free PKH2-GL (right panels) for 0.5, 1, 6, and 10 h at 37°C with agitation. Percentages of the labeled bacterial cells are indicated.
Analysis of fusion by lipid mixing assay.
The fusion between labeled empty liposomes and bacterial membranes resulted in a decreased efficiency of resonance energy transfer as measured by a fluorescence spectrophotometer. The percentage of fusion between liposomes and a nonresistant laboratory strain reached a maximum fusion of 54.3% ± 1.5% in 1 h. On the other hand, the maximum fusion degree of a resistant clinical strain (57.8% ± 1.9%) was reached in 6 h. Overall, there was no significant difference in fusion rates obtained by FACS analysis and the by lipid mixing assay (Table 2). Further, both techniques demonstrated that maximum fusion occurs sooner in the antibiotic-sensitive strain, 1 h compared to 6 h in the antibiotic-resistant strain.
TABLE 2.
Liposomal fusion rates of a sensitive laboratory strain (ATCC 10145) and a resistant clinical strain (PA-48912-2) determined by flow cytometry and lipid mixing assay (as detailed in the text)
| Time (h) | Bacterial strain | % Fusion
|
|
|---|---|---|---|
| FACS | Lipid mixing | ||
| 0.5 | ATCC 10145 | 31.9 | 32.6 ± 1.4 |
| PA-48912-2 | 24.2 | 29.3 ± 1.0 | |
| 1 | ATCC 10145 | 46.6 | 54.3 ± 2.6 |
| PA-48912-2 | 45.3 | 51.4 ± 0.7 | |
| 6 | ATCC 10145 | 14.8 | 11.4 ± 5.0 |
| PA-48912-2 | 56.8 | 57.8 ± 3.3 | |
Determination of antibiotic penetration by immunocytochemistry.
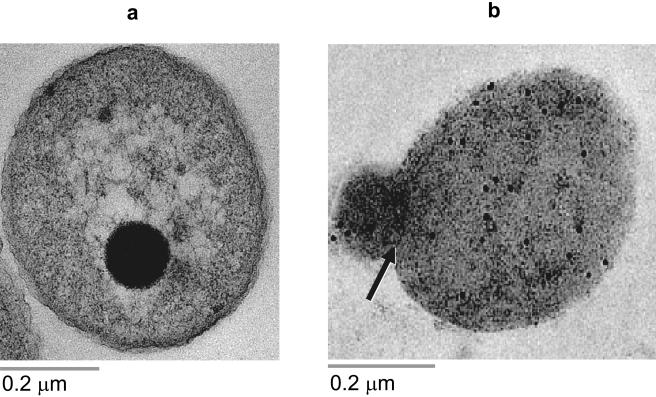
The amount of encapsulated or free immunogold-labeled tobramycin within a resistant clinical strain of P. aeruginosa (PA-48912-2) increased as a function of incubation time, regardless of the antibiotic formulation (Fig. 4). No significant differences were observed in the penetration of antibiotic between free and liposomal tobramycin in 2 h. After 6 h of incubation, however, the amount of colloidal gold in the cytoplasm of bacteria exposed to liposomal tobramycin was significantly higher than that in bacteria that received free antibiotics (24.2 ± 6.2 versus 3.2 ± 2.3 gold particles/bacterium; P ≤ 0.001). Control samples receiving PBS or antitobramycin alone were negative for gold labeling inside the bacterial cells. The background labeling, gold particles not associated with bacteria, was negligible.
FIG. 4.
Determination of antibiotic penetration by immunocytochemistry. A resistant clinical strain (PA-48912-2) was incubated with free (a) or liposomal (b) tobramycin at a final concentration of 128 μg/ml for 6 h at 37°C with agitation. Colloidal gold-labeled antibiotics in the bacterial cytoplasm are shown as dark dots (10 nm).
DISCUSSION
Chronic lung infections caused by P. aeruginosa are the leading cause of death in CF patients (8, 9, 13). Despite the use of aggressive antibiotic therapy, complete eradication remains virtually impossible (12, 13). This, in part, is due to the high intrinsic resistance of P. aeruginosa to antimicrobial agents, owing primarily to its low outer-membrane permeability (10, 11). In this communication, we report the mechanism of enhanced antibacterial activity of aminoglycosides enclosed in our newly developed DPPC-cholesterol formulation.
Data reported here indicate significant differences in MICs between free and liposomal formulations. These findings agree with our previous observations on enhanced susceptibility of antibiotic-resistant P. aeruginosa to liposomal gentamicin, despite the difference in method of preparation (22). The dehydration-rehydration method used in this study yielded small vesicles (210 ± 25 nm in diameter) with high entrapment efficiency (28.20% ± 1.15%), due to the reduced hydrophobic forces (45).
The time-kill assays further confirmed the higher potency of liposomal aminoglycosides than of the free antibiotics. For instance, liposomal amikacin eradicated a nonmucoid clinical strain at the MIC (8 μg/ml), whereas the free drug at 256 μg/ml was inactive. Considering the narrow therapeutic range of amikacin (16 to 32 μg/ml), this formulation could be the drug of choice against this bacterium. Other studies have shown improved efficacy of liposomal antibiotics of different formulations (2, 24, 25). For instance, tobramycin encapsulated into negatively charged fluid liposomes displayed stronger bactericidal activity than the free drug (2). Nacucchio et al. demonstrated that encapsulation of piperacillin in liposomes prepared with phosphatidylcholine and cholesterol (molar ratio, 1:1) protected the drug from hydrolysis by staphylococcal β-lactamase as well (24).
Several hypotheses including reduced electrostatic repulsion of liposomal antibiotics or protection of the drugs from bacterial enzymes may explain the mechanism of enhanced antimicrobial activities of liposomal formulations (36-38). We hypothesized that the enhanced antimicrobial activity of these formulations is due to their fusion with the bacterial outer membrane. The TEM data show that liposomes interact intimately with the outer membrane of P. aeruginosa. This was evident from membrane deformation and signs of membrane swelling. Integration of the PKH2-GL-labeled liposomes with bacterial membranes was confirmed by flow cytometry as well. This association indicates a direct incorporation of liposomal phospholipids into the bacterial membranes because (i) the probe inserts its aliphatic carbon tails into membranes (15, 38) and (ii) dissociation of the probe from the liposomes to the bacterial membrane via an aqueous environment is unlikely due to the strong hydrophobic nature of this probe. Further, we applied a centrifugation step with a sucrose cushion as well as successive PBS washes to rule out the possibility of liposomal adsorption to bacteria.
An apparent reduction in incorporation of the tracer with ATCC 10145 after 1 h could be due to the tracer's degradation as well as bacterial growth since the labeled liposomes had no antibiotics. The latter phenomenon seems to be the main reason, however, because the reduction was more pronounced with ATCC 10145 than with the clinical strains with lower growth rates.
The lipid mixing assay confirmed that the interaction between liposomes and bacteria is fusion while ruling out the possibility of adsorption or aggregation (6, 44). Based on the assay's principle, fusion of fluorescent vesicles to nonfluorescent lipids, i.e., bacterial cells, results in probe dilution and a decrease in resonance energy transfer efficiency due to an increase in distance between NDB-PE and Rh-PE (40). However, aggregation or adsorption of liposomes to cells does not induce any change in resonance energy transfer. The time needed to reach the maximum fusion rate for a sensitive laboratory strain (54.3% ± 1.5%) and a resistant clinical strain (57.8% ± 1.9%) of P. aeruginosa was 1 and 6 h, respectively. This variation in fusion rate agrees with both TEM and FACS data and could be due to bacterial permeability to antibiotics. Although liposomal fusion with the resistant strain was significantly delayed, the overall level of fusion was not affected (Table 2).
Finally, we employed immunogold techniques to assess whether liposomal fusion with bacterial membrane resulted in increased antibiotic penetration inside the cell. In this assay, we chose tobramycin because of the availability of antibodies and the clinical strain PA-48912-2 due to its antibiotic susceptibility profile. Bacteria exposed to liposomal tobramycin revealed more cytosolic antibiotics than those exposed to free drug. Enhanced antibiotic penetration through liposomal fusion suggests that pseudomonal membrane permeability plays a major role in the mechanism of antibiotic resistance (Table 1).
The phenomenon of enhanced liposomal antibiotic penetration into bacterial cells has been reported by other investigators as well. Sekeri-Pataryas et al. showed that negatively charged liposomes containing penicillin could overcome the cell wall barrier of P. aeruginosa and deliver the antibiotic to the cells (36). Furthermore, these authors demonstrated that nonpermeable substances such as albumin can be introduced into the cells by the use of liposomes. This particular report, however, did not reveal the activity of entrapped drugs nor the mechanism by which liposomes interacted with the cells. Liposome-bacterium fusion, however, has been demonstrated with the delivery of entrapped horseradish peroxidase to the periplasmic space of Escherichia coli and Salmonella enterica serovar Minnesota (42).
In conclusion, liposomal formulations reported here could deliver a sufficient amount of aminoglycosides into antibiotic-impermeable bacteria. Application of several methods confirmed liposome-bacterial membrane fusion as the molecular mechanism of this phenomenon. Our research is now aimed at evaluating the efficacy of these liposomal formulations in animal models of pulmonary infections.
Acknowledgments
We thank Eric Tsang for his technical assistance. All clinical isolates of P. aeruginosa were kindly provided by the Department of Microbiology, Memorial Hospital, Sudbury, Ontario, Canada.
This work was partly supported by a research grant from LURF (Laurentian University Research Funds).
REFERENCES
- 1.Barratt, G. 2003. Colloidal drug carriers: achievements and perspectives. Cell. Mol. Life Sci. 60:21-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaulac, C., S. Sachetelli, and J. Lagace. 1998. In-vitro bactericidal efficacy of sub-MIC concentrations of liposome-encapsulated antibiotic against gram-negative and gram-positive bacteria. J. Antimicrob. Chemother. 41:35-41. [DOI] [PubMed] [Google Scholar]
- 3.Bendayan, M. 1995. Colloidal gold post-embedding immunocytochemistry. Prog. Histochem. Cytochem. 29:1-159. [DOI] [PubMed] [Google Scholar]
- 4.Black, F. O., S. Pesznecker, and V. Stallings. 2004. Permanent gentamicin vestibulotoxicity. Otol. Neurotol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 5.Conway, S. P., K. G. Brownlee, M. Denton, and D. G. Peckham. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321-332. [DOI] [PubMed] [Google Scholar]
- 6.Deeba, F., H. N. Tahseen, K. S. Sharad, N. Ahmad, S. Akhtar, M. Saleemuddin, and O. Mohammad. 2005. Phospholipid diversity: correlation with membrane-membrane fusion events. Biochim. Biophys. Acta 1669:170-181. [DOI] [PubMed] [Google Scholar]
- 7.Elkin, S., and D. Geddes. 2003. Pseudomonal infection in cystic fibrosis: the battle continues. Expert Rev. Anti-Infect. Ther. 1:609-618. [DOI] [PubMed] [Google Scholar]
- 8.Geller, D. E., W. H. Pitlick, P. A. Nardella, W. G. Tracewell, and B. W. Ramsey. 2002. Pharmacokinetics and bioavailability of aerosolized tobramycin in cystic fibrosis. Chest 122:219-226. [DOI] [PubMed] [Google Scholar]
- 9.Geller, D. E., M. Rosenfeld, D. A. Waltz, and R. W. Wilmott. 2003. Efficiency of pulmonary administration of tobramycin solution for inhalation in cystic fibrosis using an improved drug delivery system. Chest 123:28-36. [DOI] [PubMed] [Google Scholar]
- 10.Hancock, R. E. 1997. The bacterial outer membrane as a drug barrier. Trends Microbiol. 5:37-42. [DOI] [PubMed] [Google Scholar]
- 11.Hancock, R. E. 1998. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin. Infect. Dis. 27:93-99. [DOI] [PubMed] [Google Scholar]
- 12.Hauser, A. R., and P. Sriram. 2005. Severe Pseudomonas aeruginosa infections. Tackling the conundrum of drug resistance. Postgrad. Med. 117:41-48. [DOI] [PubMed] [Google Scholar]
- 13.Heijerman, H. 2005. Infection and inflammation in cystic fibrosis: a short review. J. Cyst. Fibros. 4:3-5. [DOI] [PubMed] [Google Scholar]
- 14.Hill, D., B. Rose, A. Pajkos, M. Robinson, P. Bye, S. Bell, M. Elkins, B. Thompson, C. Macleod, S. D. Aaron, and C. Harbour. 2005. Antibiotic susceptibilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J. Clin. Microbiol. 43:5085-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horan, P. K., M. J. Melnicoff, B. D. Jensen, and S. E. Slezak. 1990. Fluorescent cell labeling for in vivo and in vitro cell tracking. Methods Cell Biol. 33:469-490. [DOI] [PubMed] [Google Scholar]
- 16.Langner, M., and T. E. Kral. 1999. Liposome-based drug delivery systems. Pol. J. Pharmacol. 51:211-222. [PubMed] [Google Scholar]
- 17.Lian, T., and R. J. Ho. 2001. Trends and developments in liposome drug delivery systems. J. Pharm. Sci. 90:667-680. [DOI] [PubMed] [Google Scholar]
- 18.MacLeod, D. L., L. E. Nelson, R. M. Shawar, B. B. Lin, L. G. Lockwood, J. E. Dirk, G. H. Miller, J. L. Burns, and R. L. Garber. 2000. Aminoglycoside-resistance mechanisms for cystic fibrosis Pseudomonas aeruginosa isolates are unchanged by long-term, intermittent, inhaled tobramycin treatment. J. Infect. Dis. 181:1180-1184. [DOI] [PubMed] [Google Scholar]
- 19.Magnet, S., and J. S. Blanchard. 2005. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 105:477-498. [DOI] [PubMed] [Google Scholar]
- 20.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 21.Mall, M., and K. Kunzelmann. 2005. Correction of the CF defect by curcumin: hypes and disappointments. Bioessays 27:9-13. [DOI] [PubMed] [Google Scholar]
- 22.Mugabe, C., A. O. Azghani, and A. Omri. 2005. Liposome-mediated gentamicin delivery: development and activity against resistant strains of Pseudomonas aeruginosa isolated from cystic fibrosis patients. J. Antimicrob. Chemother. 55:269-271. [DOI] [PubMed] [Google Scholar]
- 23.Mugabe, C., A. O. Azghani, and A. Omri. 2006. Preparation and characterization of dehydration-rehydration vesicles loaded with aminoglycoside and macrolide antibiotics. Int. J. Pharm. 307:244-250. [DOI] [PubMed] [Google Scholar]
- 24.Nacucchio, M. C., M. J. Bellora, D. O. Sordelli, and M. D'Aquino. 1985. Enhanced liposome-mediated activity of piperacillin against staphylococci. Antimicrob. Agents Chemother. 27:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nacucchio, M. C., M. J. Gatto Bellora, D. O. Sordelli, and M. D'Aquino. 1988. Enhanced liposome-mediated antibacterial activity of piperacillin and gentamicin against gram-negative bacilli in vitro. J. Microencapsul. 5:303-309. [DOI] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard—6th ed. M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 27.National Committee for Clinical Laboratory Standards. 2003. MIC testing supplemental tables. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 28.Omri, A., and M. Ravaoarinoro. 1996. Comparison of the bactericidal action of amikacin, netilmicin and tobramycin in free and liposomal formulation against Pseudomonas aeruginosa. Chemotherapy 42:170-176. [DOI] [PubMed] [Google Scholar]
- 29.Omri, A., Z. E. Suntres, and P. N. Shek. 2002. Enhanced activity of liposomal polymyxin B against Pseudomonas aeruginosa in a rat model of lung infection. Biochem. Pharmacol. 64:1407-1413. [DOI] [PubMed] [Google Scholar]
- 30.Poole, K. 2005. Aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 49:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole, K., and R. Srikumar. 2001. Multidrug efflux in Pseudomonas aeruginosa: components, mechanisms and clinical significance. Curr. Top. Med. Chem. 1:59-71. [DOI] [PubMed] [Google Scholar]
- 32.Rougier, F., D. Claude, M. Maurin, A. Sedoglavic, M. Ducher, S. Corvaisier, R. Jelliffe, and P. Maire. 2003. Aminoglycoside nephrotoxicity: modeling, simulation, and control. Antimicrob. Agents Chemother. 47:1010-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sachetelli, S., H. Khalil, T. Chen, C. Beaulac, S. Senechal, and J. Lagace. 2000. Demonstration of a fusion mechanism between a fluid bactericidal liposomal formulation and bacterial cells. Biochim. Biophys. Acta 1463:254-266. [DOI] [PubMed] [Google Scholar]
- 34.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweizer, H. P. 2003. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: unanswered questions. Genet. Mol. Res. 2:48-62. [PubMed] [Google Scholar]
- 36.Sekeri-Pataryas, K. H., C. Vakirtzi-Lemonias, H. A. Pataryas, and J. N. Legakis. 1985. Liposomes as carriers of14C-labelled penicillin and 125I-labelled albumin through the cell wall of Pseudomonas aeruginosa. Int. J. Biol. Macromol. 7:379-381. [Google Scholar]
- 37.Shawar, R. M., D. L. MacLeod, R. L. Garber, J. L. Burns, J. R. Stapp, C. R. Clausen, and S. K. Tanaka. 1999. Activities of tobramycin and six other antibiotics against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob. Agents Chemother. 43:2877-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spotl, L., A. Sarti, M. P. Dierich, and J. Most. 1995. Cell membrane labeling with fluorescent dyes for the demonstration of cytokine-induced fusion between monocytes and tumor cells. Cytometry 21:160-169. [DOI] [PubMed] [Google Scholar]
- 39.Sriramulu, D. D., H. Lunsdorf, J. S. Lam, and U. Romling. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667-676. [DOI] [PubMed] [Google Scholar]
- 40.Struck, D. K., D. Hoekstra, and R. E. Pagano. 1981. Use of resonance energy transfer to monitor membrane fusion. Biochemistry 20:4093-4099. [DOI] [PubMed] [Google Scholar]
- 41.Theilacker, C., F. T. Coleman, S. Mueschenborn, N. Llosa, M. Grout, and G. B. Pier. 2003. Construction and characterization of a Pseudomonas aeruginosa mucoid exopolysaccharide-alginate conjugate vaccine. Infect. Immun. 71:3875-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomlinson, S., P. W. Taylor, and J. P. Luzio. 1989. Transfer of phospholipid and protein into the envelope of gram-negative bacteria by liposome fusion. Biochemistry 28:8303-8311. [DOI] [PubMed] [Google Scholar]
- 43.Westbrock-Wadman, S., D. R. Sherman, M. J. Hickey, S. N. Coulter, Y. Q. Zhu, P. Warrener, L. Y. Nguyen, R. M. Shawar, K. R. Folger, and C. K. Stover. 1999. Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob. Agents Chemother. 43:2975-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilschut, J., and D. Hoekstra. 1986. Membrane fusion: lipid vesicles as a model system. Chem. Phys. Lipids 40:145-166. [DOI] [PubMed] [Google Scholar]
- 45.Zadi, B., and G. Gregoriadis. 2000. A novel method for high-yield entrapment of solutes into small liposomes. J. Liposome Res. 10:73-80. [Google Scholar]