Abstract
The performance of the new Abbott real-time human immunodeficiency virus type 1 (HIV-1) assay for HIV-1 RNA load determination in plasma was compared to that of the Abbott LCx HIV-1 RNA quantitative assay following automated RNA isolation by the Abbott m1000 extractor. The measured viral loads of 89 clinical specimens differed by mean 0.19 log10 copies/ml (95% confidence interval, 0.12 to 0.26 log10 copies/ml). Although the difference in viral load determinations was positively skewed in favor of the LCx assay, it did not reach statistical significance (P = 0.42). Results were linearly associated (R2 = 0.94) and strongly correlated (R = 0.96). Good performance was observed with HIV-1 subtypes other than B and circulating recombinant forms, although results obtained with two subtype G specimens and one H specimen showed a more substantial difference.
Since its introduction in the 1990s, measurement of plasma viral load (PVL) has become a cornerstone in the clinical management of human immunodeficiency virus type 1 (HIV-1) infection. The central role played by PVL has considerably increased laboratory workload, making necessary an improvement in technology. A variety of commercial assays are available for the measurement of HIV-1 PVL. Among them, real-time PCR is the latest development, offering many advantages over traditional molecular methods (3, 7, 11). These include (i) decreased performance time attributed to reduced cycle time, reduced amplicon size, and the elimination of an additional step needed for product detection; (ii) increased sensitivity as a result of the employment of fluorescence detection methods; (iii) decreased carryover contamination due to the use of a close system for the amplification and detection; and (iv) wider dynamic range. Further enhancements have been achieved by the introduction of automated nucleic acid extractions, resulting in completely automated assays with an average turnaround time of 2 h.
The objective of this study was to evaluate the performance of the new Abbott real-time HIV-1 assay (referred to as the real-time assay) for PVL quantitation in comparison with the Abbott LCx HIV-1 RNA quantitative assay (referred to as the LCx assay). Both assays target the integrase region of the HIV-1 genome. In the LCx assay, 24 samples can be processed in one run, including 21 clinical specimens and 3 controls. The range of quantification is from 50 to 1 million copies/ml. In the real-time assay, 48 samples can be processed in one run. The range of quantification is 40 to 10 million copies/ml. Previous studies have assessed the performance of the LCx assay and found the test suitable for the management of patients infected by HIV-1 group M subtypes (1, 4, 8, 9).
MATERIALS AND METHODS
Patient and samples.
Samples were randomly collected from 92 HIV-1-seropositive patients attending the Ian Charleson Day Centre clinic at Royal Free Hospital in London. Plasma was prepared from EDTA-anticoagulated blood and frozen at −80°C within 6 h of collection.
Viral load determination.
Plasma RNA was extracted and concentrated using the Abbott m1000 instrument, which uses magnetic particle technology to capture nucleic acid extracted from 1 ml of plasma. Each sample was examined in parallel using the LCx HIV-1 quantitative assay and the Abbott real-time HIV-1 assay according to the manufacturer's specification.
HIV-1 subtyping.
In 51 patients, with PVLs of >1,000 copies/ml and for which sufficient sample was available, HIV-1 subtype was determined from pol gene sequences obtained by the Viroseq kit (Celera Diagnostics). Briefly, total RNA was manually extracted from plasma, and the protease region and two-thirds of the reverse transcriptase region were amplified by reverse transcription-PCR and sequenced before being analyzed on a 3100-Avant genetic analyzer (Applied Biosystems, United Kingdom). Subtypes were determined by phylogenetic analysis employing Clustal X for sequence alignment and PAUP version 4 (Sinauer Associates) to generate a phylogenetic tree. Reference sequences were derived from the Los Alamos database (www.lanl.gov).
Statistical analysis.
Plasma HIV-1 RNA levels were log10 transformed before being subjected to statistical analysis. Averages were statistically compared by using the paired z test. Linear regression and correlation analysis were employed to determine assay relationship. The method of Bland and Altman was applied in order to assess the agreement between the two assays.
RESULTS
A total of 92 plasma samples were tested in parallel by the LCx assay and the real-time assay, including 67/92 (73%) samples from antiretroviral-drug-naïve patients and 25/92 (27%) samples from patients on highly active antiretroviral therapy (HAART). Overall, 63/92 (68%) samples were quantified by both assays, with viral loads ranging from 1.7 log10 to 6 log10 copies/ml for the LCx assay and 1.6 log10 to 6.4 log10 copies/ml for the real-time assay.
There were 14/92 (15%) samples undetectable by both assays. These included 13/14 samples from patients on stable HAART and 1/14 from a drug-naïve long-term nonprogressor. The latter sample also showed a viral load of <50 copies/ml when tested by the Cobas-Monitor 1.5 assay (Roche Diagnostics, Germany). Discrepant results at the lower limit of detection included 10/92 (11%) samples showing viral loads between 63 and 251 copies/ml by the LCx assay but <40 copies/ml by the real-time assay. Conversely, 2/92 (2%) samples showed viral loads of 1,200 and 158 copies/ml, respectively, by the real-time assay but <50 copies/ml by the LCx assay.
Finally, 3/92 samples (3%) were quantified by the LCx assay at levels of 284 −11,300 copies/ml but were inhibitory by the real-time assay. As there was insufficient sample for repeating testing, these samples were excluded from further analysis.
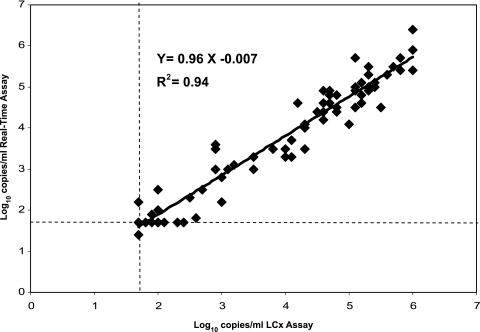
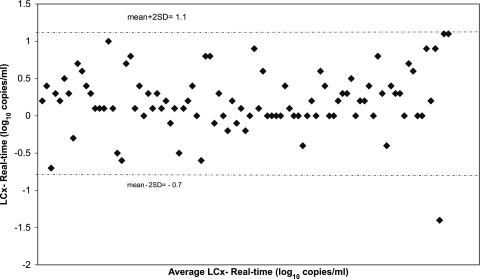
The two assays showed a high degree of correlation (Fig. 1). The linear regression equation for log10 HIV RNA copies per milliliter was y = 0.96x − 0.007 (R2 = 0.94). Viral loads obtained by the two assays differed on average by 0.19 log10 (95% confidence interval, 0.12 to 0.26 log10 copies/ml). The difference was not significant as determined by the paired z test (P = 0.42). Agreement between the two methods was calculated by the method of Bland and Altman by plotting differences against the average (Fig. 2). One of 92 (1%) samples was out of the 95% interval of agreement, with a difference of −1.4 log10 copies/ml (<50 copies/ml by LCx and 1,259 copies/ml by the real-time assay). The sample was from a patient on HAART infected with HIV-1 subtype B.
FIG. 1.
Correlation of viral load results obtained with 87 plasma samples tested in parallel by the Abbott LCx HIV-1 RNA quantitative assay and the Abbott real-time HIV-1 assay. The linear regression trend is shown. The lowest limit of detection was adjusted to 1.7 log10 copies/ml for both assays.
FIG. 2.
Agreement of plasma viral load determinations obtained by the Abbott LCx HIV-1 RNA quantitative assay and the Abbott real-time HIV-1 assay. The dotted horizontal lines correspond to the 95% limit of agreement, given by the mean difference plus or minus twice the standard deviation of the difference. The sample outside the 95% interval was obtained from a patient infected with HIV-1 subtype B. SD, standard deviation.
Table 1 summarizes results obtained by the two assays from 51 samples with known HIV-1 subtypes. The coefficient of correlation (R2) between the two assays was 0.94 for subtype B and 0.91 for genotypes other than B. The difference in PVL quantitation was not significant overall, although results obtained with two subtype G specimens and one H specimen showed a more substantial difference (Table 1). The amplification curved did not show any grade of inhibition for these three samples in the real-time assay. The samples were insufficient for further investigation.
TABLE 1.
Comparison of HIV-1 plasma viral loads obtained by the Abbott LCx HIV-1 RNA quantitative assay and the Abbott real-time HIV-1 assay, according to the infecting HIV-1 subtype
| Sample(s) with indicated genotype | No. of samples | Mean viral load (log10 copies/ ml) ± standard deviation by indicated assay
|
|
|---|---|---|---|
| LCx | Real-time | ||
| B | 21 | 4.1 ± 0.7 | 3.8 ± 0.6 |
| Non-B | 30 | 3.9 ± 0.4 | 3.8 ± 0.4 |
| A | 9 | 4.4 ± 1.3 | 4.1 ± 1.4 |
| C | 6 | 4.4 ± 1.6 | 4.4 ± 1.4 |
| D | 2 | 2.3 ± 0.9 | 2.6 ± 0.7 |
| G | 2 | 4.2 ± 0.1 | 3.4 ± 0.1 |
| H | 1 | 5.0 | 4.1 |
| CRF01_AE | 2 | 4.7 ± 0.2 | 4.8 ± 0.4 |
| CRF02_AG | 5 | 4.8 ± 0.6 | 4.9 ± 0.8 |
| CRF13 | 1 | 3.2 | 3.1 |
| CRF06 | 1 | 5.3 | 5.3 |
| CRF02_AG/Aa | 1 | 4.7 | 4.6 |
Mosaic pol sequence.
DISCUSSION
This study showed an excellent correlation and a high degree of concordance between the new assay, Abbott real-time HIV-1 assay and the Abbott LCx HIV RNA quantitative assay, with all except one differences falling within the 95% interval of agreement. The one divergent sample showed a PVL of 1,200 copies/ml by the real-time but <50 copies/ml by the LCx assay. The sample had been collected from a patient on HAART who was infected with HIV-1 subtype B. In both assays the sample met the quality control criteria to be considered and acceptable result, the negative, low and high positive controls were within the acceptable range and the internal controls were not inhibited.
The differences in PVL determination were positively skewed, indicating that the viral load estimations obtained by the LCx assay were overall higher than those obtained by the real-time assay. However, the difference did not reach statistical significance. These observations indicate that the real-time assay can be used for those patients previously managed by the LCx assay.
There were 14 samples that showed undetectable PVL by both assays. Of these, 13 were from patients on stable HAART with undetectable plasma viremia documented for at least one year. One drug-naïve black African patient with undetectable viral load was a long-term nonprogressor with a CD4 cell count consistently above 1,000 cells/mm3, suggesting that he may have low levels of virus replication. The patient also showed an undetectable viral load by the Roche Cobas-Monitor 1.5 assay, which targets a different region of the HIV-1 genome (gag gene).
Reliable performance at the lower limit of detection is a critical feature of PVL determination. The prevalence of discrepancies at this level was 13%, in most cases represented by samples that gave low-level PVL measurements in the LCx assay (61 to 251 copies/ml) but were <40 copies/ml in the real-time assay. Of note, all these were samples from patients on stable HAART and with longstanding viral suppression (<50 copies/ml). We conducted a reproducibility analysis of PVLs greater than 50 copies/ml but less than 400 copies/ml obtained by the LCx assay in patients with histories of suppressed viral loads while on stable HAART. Results showed that only 8/32 (25%) results of the PVL were reproduced in repeated testing (data not shown). This finding suggests that there may be a greater degree of error for the LCx assay at the lower limit of detection than for the real-time assay.
As HIV-1 infection continues to spread, its genetic diversification increases worldwide. Subtype-related variability in the performance of viral load assays has been widely reported (2, 6, 8, 10). Previous studies have demonstrated that the high degree of nucleotide conservation within the targeted pol region enables the LCx assay to quantify HIV-1 subtypes A through G efficiently (4, 8, 9). In this study, subtype-related variability in assay performance was evaluated by using 51 specimens from patients infected with subtype B, non-B subtypes A through H, and various circulating recombinant forms. Most specimens were quantified by the two assays with comparable PVL measurements. A more substantial difference was observed with samples from two patients infected with subtype G and one patient infected with subtype H which showed lower PVL levels by the real-time assay than by the LCx assay. Given the limited number of samples, the significance of this observation is unclear. In London, 42% of newly diagnosed HIV-1-infected patients carry non-B subtypes or recombinant forms, including 2.5% with subtype G and approximately 0.5% with subtype H. Although the prevalence of subtypes G and H is generally low in Europe, with both subtypes being found mainly in Central Africa, subtype G is among the most prevalent non-B subtypes found in some European countries, such as Spain and Portugal (5). Using a reference HIV-1 subtype panel, including subtypes G and H, Braun et al. reported excellent performance of the real-time assay with PVLs higher than those measured by the Bayer bDNA and Roche Amplicor Monitor v1.5 assays (P. Braun, R. Ehret, F. Zabbai, S. Thamm, S. Schaffer, and H. Knechten, Eur. Meet. Mol. Diagn.). Thus, while this finding indicates that the real-time assay can be considered appropriate for the managements of patients infected in many parts of the world, ongoing vigilance is recommended to evaluate assay performance with existing and emerging divergent strains.
REFERENCES
- 1.Amendola, A., L. Bordi, C. Angeletti, E. Girardi, G. Ippolito, and M. R. Capobianchi. 2004. Comparison of LCx with other current viral load assays for detecting and quantifying human immunodeficiency virus type 1 RNA in patients infected with circulating recombinant form A/G (CRF02). J. Clin. Microbiol. 42:811-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annette, A., L. Knut, S. Anders, and A. Jan. 1997. Subtype-specific problems with quantification of plasma HIV-1 RNA. AIDS 11:859-865. [DOI] [PubMed] [Google Scholar]
- 3.Bankowski, M. J., and S. M. Anderson. 2004. Real-time nucleic acid amplification in clinical microbiology. Clin. Microbiol. Newsletter 26:9-15. [Google Scholar]
- 4.De Mendoza, C., J. Alcami, M. Sainz, D. Folgueira, and V. Soriano. 2002. Evaluation of the Abbott LCx quantitative assay for the measurement of human immunodeficiency virus RNA in plasma. J. Clin. Microbiol. 40:1518-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lospitao, E., A. Alvarez, V. Soriano, and A. Holguin. 2005. HIV-1 subtypes in Spain: a retrospective analysis from 1995 to 2003. HIV Med. 6:313-320. [DOI] [PubMed] [Google Scholar]
- 6.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Christopherson, J. P. Spadoro, K. K. Y. Young, V. Polonis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of and improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith, T. F., J. R. Uhl, M. J. Espy, M. L. Sloan, E. A. Vetter, M. F. Jones, J. E. Rosenblatt, and F. R. Cockerill. 2004. Development, implementation, and trend analysis of real-time PCR tests fro the clinical microbiology laboratory. Clin. Microbiol. Newsletter 26:145-153. [Google Scholar]
- 8.Swanson, P., J. B. Harris, V. Holzmayer, S. G. Devare, G. Schochetman, and J. Hackett, Jr. 2000. Quantification of HIV-1 group M (subtypes A-G) and group O by the LCx HIV RNA quantitative assay. J. Virol. Methods 89:97-108. [DOI] [PubMed] [Google Scholar]
- 9.Swanson, P, C. De Mendoza, Y. Joshi, A. Golden, R. L. Hodinka, V. Soriano, S. G. Devare, and J. Hackett, Jr. 2005. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v 1.5, VERSANT HIV-1 RNA 3.0 and NucliSens HIV-1 QT. J. Clin. Microbiol. 43:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Triques, K., J. Coste, J. L. Perret, C. Segarra, E. Mpoudi, J. Reynes, E. Delaporte, A. Butcher, K. Dreyer, S. Herman, J. Spadoro, and M. Peeters. 1999. Efficiency of four version of the AMPLICOR HIV-1 MONITOR test for quantification of different subtypes of human immunodeficiency virus type 1. J. Clin. Microbiol. 37:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, S., and R. E. Rothman. 2004. PCR-based diagnostics for infectious diseases: uses, limitations and future applications in acute-settings. Lancet Infect. Dis. 4:337-348. [DOI] [PMC free article] [PubMed] [Google Scholar]