Abstract
Haemophilus influenzae type b (Hib) was a major cause of pediatric disease in the United Kingdom prior to the introduction of routine Hib immunization in 1992. An unexpected resurgence of cases of vaccine failure was observed with fully vaccinated children from 1999 onward. We investigated whether Hib isolates causing vaccine failures in the United Kingdom could have undergone a change in their population structure to elude the protective effect of Hib vaccine. Molecular epidemiology studies were carried out with 376 isolates from invasive infections (164 vaccine failures and 212 controls). Genetic variability was higher in controls than in vaccine failures. Of the four major clusters obtained, cluster I comprised 92.2% of the total isolates: 156 vaccine failures (95%) and 193 (91%) controls. Cluster IV was specific for vaccine failures but included only four isolates. The increased number of cases of invasive Hib in fully vaccinated children in the United Kingdom was caused by isolates belonging not to a particular or new genotype but to genotypes already circulating in the prevaccination era, before conjugate Hib vaccines were available.
Prior to the introduction of Haemophilus influenzae type b (Hib) conjugate vaccination, this pathogen was an important cause of pediatric morbidity and mortality. However, in several countries, including the United States, the United Kingdom, and The Netherlands, a reemergence of Hib invasive disease in well-vaccinated children has been reported (6, 8, 15, 17).
In the United Kingdom, the annual incidence was approximately 30 per 100,000 for children under 5 years of age (1) but declined by 90% following the inclusion of Hib conjugate vaccine in the routine infant immunization schedule in October 1992 (9).
However, from 1999 onward, the incidence of invasive Hib disease in the United Kingdom began to increase again, rising from 0.66 in 1998 to 2.96 per 100,000 children aged less than 5 years in 2001 (17), with most of the cases occurring in children who had been fully immunized according to the United Kingdom's primary infant immunization schedule for Hib vaccine at 2, 3, and 4 months (14). An increase in cases was also seen with nonvaccinated older children and adults.
In the United Kingdom, possible reasons for the increase included a lower-than-expected direct protection from infant immunization, the wearing off of the initial impact of the “catch-up” campaign, and the use, in 2000 and 2001, of a less immunogenic combination vaccine with an acellular pertussis component (17). It is, however, possible that some of the increase may have been due to adaptive changes in the population structure of Haemophilus influenzae type b following the widespread use of Hib vaccine. Such adaptations in the population structure of Bordetella pertussis, leading to an increase in cases of pertussis, have been observed (12). Little information is available about the molecular epidemiology of Hib isolated from cases of vaccine failures (3).
Although the population structure of Hib isolates is highly clonal (13), a comprehensive evaluation of Hib invasive isolates from vaccine failure cases compared with isolates causing invasive infection in nonvaccinated children has not been carried out. Accordingly, we attempted to ascertain whether Hib isolates causing vaccine failures in the United Kingdom could have undergone a change in their population structure allowing them to elude the protective effect of Hib vaccine.
MATERIALS AND METHODS
Bacterial isolates.
The Health Protection Agency identified cases of invasive Hib disease through referral to the Haemophilus Reference Unit and reports of confirmed infections to the Communicable Disease Surveillance Centre from laboratories in England and Wales. Between 1992 and 2000, surveillance of children was enhanced by reporting of cases via the British Pediatric Surveillance Unit in collaboration with the Oxford Vaccine Group. A case was included if H. influenzae had been isolated from a normally sterile site. All cases confirmed as Hib by the Haemophilus Reference Unit were included. Strains of H. influenzae were confirmed as serotype b by standard microbiological procedures and capsular genotyping.
A case of “true” Hib vaccine failure was defined as a child between 0 and 16 years of age in whom H. influenzae was cultured from a normally sterile site, where the invasive infection occurred more than 1 week after three doses of Hib conjugate vaccine given in the first year of life or more than 2 weeks after a single dose given to a child over 12 months of age.
In order to determine whether or not the population structure of Hib strains causing vaccine failures in the United Kingdom could be differentiated from that of Hib isolates in the prevaccination era, a representative sample of 164 Hib strains isolated in England and Wales from 1991 to 2003 from cases of true vaccine failures was genetically compared with 212 Hib control strains recovered from invasive infections in unvaccinated children and adults. The controls included both historical (Hib isolated before 1992, when routine Hib immunization was introduced in the United Kingdom) and contemporary (from 1992 onward) controls (Table 1).
TABLE 1.
Distribution of the Haemophilus influenzae type b isolates included in the study
| Sample groupa | No. (%) of strains in group |
|---|---|
| Vaccine failure | 164 (43.5) |
| First period (1993-1999) | 134 (35.5) |
| Second period (2000-2003) | 30 (8.0) |
| Control | 212 (56.5) |
| Children, historic control (before 1993)* | 144 (38.3) |
| Children, contemporary control (2000-2003)** | 29 (7.8) |
| Adult, historic control (before 1993)* | 20 (5.3) |
| Adult, contemporary control (2000-2003) | 19 (5.1) |
| Total | 376 (100) |
*, no vaccine available; **, vaccine available but children were not vaccinated.
A total of 376 Hib isolates from invasive infections in the United Kingdom constituted the study collection. These Hib isolates were isolated during three time periods. Strains from the first period, 1991 to 1992, included invasive Hib isolates from children (n = 144) and adults (n = 20) from the prevaccination era and were considered historical controls, reflecting the Hib isolates causing invasive infections in England and Wales before conjugate Hib vaccines were introduced in the United Kingdom. Strains from the second period, from 1993 to 1999, included 134 invasive Hib isolates from true vaccine failures. Strains from the third period, from 2000 to 2003, included Hib isolates from the period of resurgence in vaccine failures in children (n = 30), invasive Hib isolates from nonvaccinated children (n = 29), and isolates from invasive cases in adults (n = 19). The last two groups were considered to be contemporary controls. The Eagan reference strain was used as a known Hib positive control.
Molecular epidemiology studies.
Genetic molecular epidemiology studies were carried out by pulsed-field gel electrophoresis (PFGE), a highly discriminatory and reproducible method widely used in H. influenzae molecular epidemiology studies (3, 4, 19). The PFGE procedure was performed as described previously (4). The restriction endonuclease SmaI (MBI Fermentas, Vilnius, Lithuania) was used at the manufacturer's suggested temperature. Restriction fragments were separated by PFGE in 1% agarose gel (Boehringer Mannheim, Germany) in 0.5× Tris-borate-EDTA buffer (45 mM Tris, 45 mM boric acid, 1.0 mM EDTA, pH 8.0) with a Bio-Rad contour-clamped homogeneous electric field mapper apparatus. The initial pulse time of 5 s was increased linearly to 50 s over 23.3 h at 6 V at 13°C. Gels were then stained with ethidium bromide and photographed under UV light.
Genetic analysis.
A similarity matrix was created based on a binary system according to the presence or absence of bands at 21 molecular size positions in PFGE gels. This information was first analyzed by the Multivariate Statistical Package (London, United Kingdom). Simpson's genetic diversity index (×100) (10, 15) was used for calculation of the discriminative power of the PFGE procedure applied to the collection of Hib isolates and analysis of the genetic diversity between vaccine failures and control isolates.
As the previous procedure did not fully inform about the genetic relatedness among isolates in the study collection, the similarity matrix was further analyzed by a second genetic software system (Fingerprinting II Informatix software; Bio-Rad, CA) specifically designed for the analysis of the genetic relatedness among isolates based on band PFGE patterns. In this case, the genetic similarity was calculated by the unweighted-pair group method using average linkages and shown in a dendrogram. Similarity was adjusted by Dice's coefficients, with a tolerance of 0.4%. Multivariate analysis of variance (mANOVA) was carried out to determine the statistical significance of the major fingerprinting groups according to the Fingerprinting II software.
Additional statistical analyses were carried out by calculations of Fisher's exact test and the 95% confidence interval (95% CI) and odds ratio (OR) for the analysis of contingency tables (GraphPad Prism; San Diego, CA).
RESULTS
The initial overview of the genetic distribution of the 376 Hib study strains was carried out using the Multivariate Statistical Package software. Seventy-seven diverse groups of isolates were obtained: 19 of these groups (24.6%) contained 310 Hib isolates (82.4%), all of them common to both vaccine failures and control isolates, 40 groups (47 Hib strains) contained control isolates only, and 18 groups (19 Hib strains) had vaccine failure isolates only.
The results of Simpson's diversity index were as follows: 78% for Hib isolates causing vaccine failures and 81% for controls; 79% for children's isolates and 90% for isolates from adults; 80% for children's controls and 90% for adult controls; 83% for prevaccination children's isolates; and 76% for postvaccination isolates. Among the Hib control isolates, 50 diversity groups were obtained from 173 children's isolates in comparison with 23 groups among 39 adult isolates (P = 0.0006; OR = 0.28; 95% CI = 0.13 to 0.57), suggesting that more genetic variability was present in Hib controls than in Hib vaccine failure cases and that this variability was due mainly to adult Hib controls. In addition, 46 diversity groups were found among 144 Hib strains obtained from children in the prevaccination era compared to 40 groups from 193 strains from the postvaccination era (P = 0.02; OR = 1.8; 95% CI = 1.1 to 2.9). The same comparison between adult isolates did not reach statistical significance.
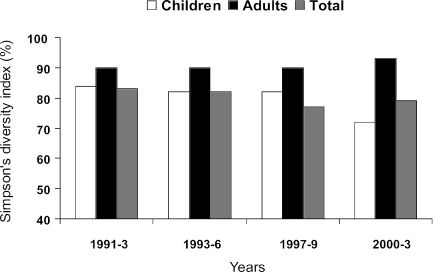
Figure 1 shows the genetic diversity of Hib isolates, as measured by Simpson's genetic diversity index, stratified by time period in 3-year intervals from 1991 to 2003. From 1991 to 1999, the genetic diversity indices for children's isolates were similar, whereas they decreased in the last period of 2000 to 2003. For adult Hib isolates, this indices (although higher than those for children) also were similar between 1991 and 1999 but increased in isolates of the last period (2000 to 2003) (Fig. 1).
FIG. 1.
Genetic diversity of Haemophilus influenzae type b strains isolated from patients with invasive disease, stratified by time period. Simpson's diversity index based on the PFGE is displayed.
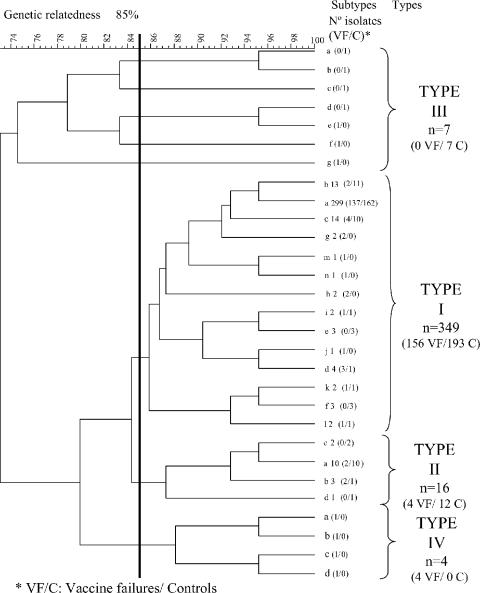
Additional analysis of the study collection to determine genetic relatedness was carried out using the Fingerprinting II software (see Material and Methods). At a genetic distance of 85 to 100%, which would include all identical isolates or those that are highly related (16), four major PFGE clusters were observed, referred to as type I to type IV (Fig. 2). Type I included 349 Hib (92.2%) isolates (156 vaccine failures [95%] and 193 controls [91%]) and the Eagan positive Hib control; type II, type III, and type IV had 16 (4.2%), 7 (1.8%), and 4 (1.1%) isolates, respectively (Fig. 2). Types III and IV were composed of individual isolates. Type I was separated further into 14 subtypes (“a” to “n”) (Fig. 2), where the majority of type I isolates were subtypes a (79.4%), b (3.4%), and c (3.4%); the remaining 11 subtypes contained one to four isolates each. The Eagan reference strain belonged to type I, subtype j. The most important group was type I, subtype a, with 299 isolates that consisted of 137 isolates from cases of vaccine failure and 162 from controls (with 123 isolated from the prevaccination era and 176 from the postvaccination era).
FIG. 2.
Genetic relatedness of Haemophilus influenzae type b isolated in the United Kingdom (from 1991 to 2003) from cases of vaccine failures and controls.
Multivariate analysis showed that 11 of the 21 bands initially recorded were critical for the segregation of the total collection of isolates into four clusters, although with different discriminative powers; for instance, one band of 146 kb was present in types I and II but not in the other two types, and another band, of 20 kb, was present only in type III. Clusters within types III and IV were clearly differentiated from types I and II by mANOVA (Lawley-Hotelling coefficient [L] = 0.06 and L = 0.18, respectively; P < 0.001), but clusters within types I and II could not be differentiated significantly (L = 0.46; P = 10.4), suggesting the possibility of one unique big cluster.
Further comparison between cluster types I, II, III, and IV showed that isolates belonging to the two most prevalent cluster types, types I and II, were distributed evenly into either the vaccine failure or the control group (Table 2). However, the minority clusters, types III and IV, were more prevalent in the control group (type III, P = 0.02) or the vaccine failure group (type IV, P = 0.03) (Table 2). The seven isolates of the type III cluster belonged to adult (n = 3) and children's (n = 4) control groups. Four isolates of the type IV cluster, 5616, 8106, 8272, and 9240, came from laboratories in southwestern England, as well as Midlands and London.
TABLE 2.
Distribution of invasive Haemophilus influenzae type b isolates from cases of vaccine failures and controls in the United Kingdom (1991 to 2003) into PFGE clusters
| PFGE cluster | No. (%) of strains
|
OR (95% CI) | Pa | |
|---|---|---|---|---|
| Vaccine failure | Control | |||
| Type I | 156 (41.3) | 193 (51.3) | 1.90 (0.8-4.4) | 0.16 |
| Type II | 4 (1.1) | 12 (3.1) | 0.42 (0.13-1.3) | 0.19 |
| Type III | 0 (0) | 7 (2.1) | 0.08 (0.004-1.4) | 0.02 |
| Type IV | 4 (1.1) | 0 (0) | 11.92 (0.63-223.1) | 0.03 |
| Total | 164 (43.5) | 212 (56.5) | ||
By Fisher's exact test.
DISCUSSION
From the data presented in this study, it appears that invasive Hib infections in the United Kingdom, in both pre- and postvaccination eras, were caused by a strongly clonal pathogen showing limited genetic diversity. Moreover, the increased number of invasive Hib cases in fully vaccinated children in the United Kingdom was not caused by isolates belonging to a particular or new genotype but by genotypes already circulating in the prevaccination era, before conjugate Hib vaccines were available. Although we found that cluster type IV was specific for Hib vaccine failures (Table 2), this should be interpreted with caution, as this cluster of highly related but not identical strains contained only four isolates (1.1%). We therefore conclude that this association appears to be very weak and may have occurred by chance.
Although the epidemiologies of vaccine failures seem very different in the United Kingdom and The Netherlands, a similar conclusion was obtained in a recent study carried out with Dutch invasive Hib isolates by using the molecular epidemiology techniques of multilocus sequence typing (MLST) and multiple-locus variable-number tandem repeat analysis (15). These authors noticed an increase in the genetic diversity of Hib in The Netherlands after vaccination was introduced. We also found an increase in the overall genetic diversity of Hib isolates in the United Kingdom in the last two postvaccination periods analyzed (from 1997 to 1999 and 2000 to 2003) (Fig. 1), but this was due to an increase of the genetic diversity in adult isolates not found to occur in children with true vaccine failure, so this finding is probably independent of Hib vaccine failure.
Cluster type I fingerprinting accounted for the vast majority of Hib isolates whether causing vaccine failure or not; in addition, type II (16 isolates) was in fact closely related to type I according to mANOVA, being in practice one unique huge cluster. Accordingly, so far it cannot be concluded that the selective pressure posed by the widespread use of conjugate Hib vaccine after 1992 has substantially modified the population structure of Hib in the United Kingdom.
The discriminative power of the PFGE technique as determined in this study was very high (78% in Hib producing vaccine failures and 81% in controls) according to Simpson's discriminative index. It was even higher than that of other molecular epidemiology methods, like multiple-locus variable-number tandem repeat analysis (75.9%) and MLST (41.4%) (15). However, PFGE has potential limitations, as bands of the same size may or may not be the same DNA fragment and as a change in one restriction site can mean more than one band change. To overcome these limitations, MLST has been proposed as the definitive molecular epidemiological tool for several bacterial species, including H. influenzae (11). Although PFGE and MLST have not been directly compared for Hib, 77 capsulated isolates of types e and f of H. influenzae studied by both techniques gave almost identical strong clonal distributions (B. Aracil and J. Campos, unpublished observations).
According to the data presented in this study, we believe that other explanations for the recent increase in invasive Hib disease in the United Kingdom should be sought. Trotter et al. (17) concluded that the use of a combination Hib-diphtheria-tetanus vaccine with an acellular pertussis component was a major contributory factor in the increase in Hib disease observed in the United Kingdom from 1998 to 2003. Conjugate Hib vaccines have reduced immunogenicity when combined with diphtheria-tetanus-acellular pertussis compared to diphtheria-tetanus-whole-cell pertussis (2). This effect is strongly dose dependent (7) and is more marked with accelerated schedules of the type adopted in the United Kingdom (18).
The Department of Health in the United Kingdom responded rapidly to the increase in Hib disease in 2003 by withdrawing preparations of the combination Hib-diphtheria-tetanus-acellular pertussis vaccine and by conducting a second catch-up campaign, offering a further dose of Hib vaccine to all children aged 6 months to 4 years (5). This campaign has proved extremely successful, and the number of cases in the age group targeted for the vaccine has declined dramatically.
Acknowledgments
We are grateful to the Oxford Vaccine Group.
This study was supported by grants from the Instituto de Salud Carlos III, Programa Intramural (references 02/16 and 03/32), and Fondo de Investigaciones Sanitarias (reference PI 040899). B.A. is a recipient of a fellowship from the Instituto de Salud Carlos III (reference 01/0037).
REFERENCES
- 1.Anderson, E. C., N. T. Begg, S. C. Crawshaw, R. M. Hargreaves, A. J. Howard, and M. P. E. Slack. 1995. Epidemiology of invasive Haemophilus influenzae infections in England and Wales in the pre-vaccination era (1990-2). Epidemiol. Infect. 115:89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, F., P. Heath, F. Shackley, N. Maclennan, and L. Diggle. 1998. Effect of combination with an acellular pertussis, diphtheria, tetanus vaccine on antibody response to Hib vaccine (PRP-T). Vaccine 16:637-642. [DOI] [PubMed] [Google Scholar]
- 3.Campos, J., B. Aracil, F. Román, and M. Pérez-Vázquez. 2003. Molecular epidemiology of Haemophilus influenzae type b isolated from children with clinical cases of conjugate vaccine failures. J. Clin. Microbiol. 41:3915-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos, J., M. Hernando, F. Román, M. Pérez-Vázquez, B. Aracil, J. Oteo, E. Lázaro, and F. de Abajo. 2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 42:524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chief Medical Officer, Chief Nursing Officer, and Chief Pharmaceutical Officer. 17. February 2003. Planned Hib vaccination catch-up campaign. PL/CMO/2003/1. http://www.dh.gov.uk/assetRoot/04/01/34/87/04013487.pdf.
- 6.Dargan, J. M., P. M. Coplan, K. M. Kaplan, and A. Nikas. 2000. Reemergence of invasive Haemophilus influenzae type b disease in Alaska: is it because of vaccination with polyribosylribitol phosphate outer membrane protein complex (PRP-OMPC) or failure to vaccinate with PRP-OMPC? J. Infect. Dis. 181:806-809. [DOI] [PubMed] [Google Scholar]
- 7.Daum, R. S., C. E. Zenko, G. Z. Given, G. A. Ballanco, H. Parikh, and K. Germino. 2001. Magnitude of interference after diphtheria-tetanus toxoids-acellular pertussis /Haemophilus influenzae type b capsular polysaccharide-tetanus vaccination is related to the number of doses administered. J. Infect. Dis. 184:1293-1299. [DOI] [PubMed] [Google Scholar]
- 8.Galil, K., R. Singleton, O. S. Levine, M. A. Fitzgerald, L. Bulkow, M. Getty, B. A. Perkins, and A. Parkinson. 1999. Reemergence of invasive Haemophilus influenzae type b disease in a well-vaccinated population in remote Alaska. J. Infect. Dis. 179:101-106. [DOI] [PubMed] [Google Scholar]
- 9.Hargreaves, R. M., M. P. E. Slack, A. J. Howard, E. C. Anderson, and M. E. Ramsay. 1996. Changing patterns of invasive Haemophilus influenzae disease following introduction of the Hib vaccination programme. BMJ 312:160-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 41:1623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mooi, F. R., V. M. van Loo, and A. J. King. 2001. Adaptation of Bordetella pertussis to vaccination: a cause for its reemergence? Emerg. Infect. Dis. 7(Suppl. 3):526-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musser, J. M., J. S. Kroll, D. M. Granoff, E. R. Moxon, B. R. Brodeur, J. Campos, et al. 1990. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev. Infect. Dis. 12:75-111. [DOI] [PubMed] [Google Scholar]
- 14.Ramsay, M. E., J. McVernon, N. J. Andrews, P. T. Heath, and M. P. Slack. 2003. Estimating Haemophilus influenzae type b vaccine effectiveness in England and Wales by use of the screening method. J. Infect. Dis. 188:481-485. [DOI] [PubMed] [Google Scholar]
- 15.Schouls, L. M., A. van der Ende, I. van de Pol, C. Schot, L. Spanjaard, P. Vauterin, D. Wilderbeek, and S. Witteveen. 2005. Increase in genetic diversity of Haemophilus influenzae serotype b (Hib) strains after introduction of Hib vaccination in The Netherlands. J. Clin. Microbiol. 43:2741-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trotter, C. L., M. E. Ramsay, and M. P. Slack. 2003. Rising incidence of Haemophilus influenzae type b disease in England and Wales indicates a need for a second catch-up vaccination campaign. Commun. Dis. Public Health 6:55-58. [PubMed] [Google Scholar]
- 18.Vidor, E., A. Hoffenbach, and M. A. Fletcher. 2001. Haemophilus influenzae type b vaccine: reconstitution of lyophilised PRP-T vaccine with a pertussis-containing paediatric combination vaccine, or a change in the primary series immunisation schedule may modify the serum anti-PRP antibody response. Curr. Med. Res. Opin. 17:197-209. [DOI] [PubMed] [Google Scholar]
- 19.Wang, C. C., L. K. Siu, M. K. Chen, Y. L. Yu, F. M. Lin, M. Ho, and M. L. Chu. 2001. Use of automated riboprinter and pulsed-field gel electrophoresis for epidemiological studies of invasive Haemophilus influenzae in Taiwan. J. Med. Microbiol. 50:277-283. [DOI] [PubMed] [Google Scholar]