Abstract
We developed a multiplex PCR-based reverse line blot assay to identify 23 pneumococcal serotypes represented in the polysaccharide vaccine, using 334 well-characterized isolates, representing all 90 serotypes, and 268 “unknowns.” The assay identified all target serotypes, but 11, which cross-react with 1 to 4 nonvaccine serotypes, could be distinguished using serotype-specific antisera.
Previously, we developed a molecular capsule type prediction system for 90 Streptococcus pneumoniae serotypes, based on a combination of partial cpsA-cpsB sequencing and serotype- and/or serogroup-specific PCR (3, 6). While this system is useful, it is too slow, expensive, and labor-intensive for routine use.
Multiplex PCR-based reverse line blot hybridization (mPCR/RLB) is a promising method for simultaneous detection and genotyping of microorganisms, which we have used previously in several applications (5, 9, 10). In the present study we applied it to identification of the 23 S. pneumoniae serotypes represented in the polysaccharide vaccine (Pneumovax 23; Merck & Co, Inc.).
The design of primers and probes was based on recently published full cps gene cluster sequences of all 90 pneumococcal serotypes (http://www.sanger.ac.uk/Projects/S_pneumoniae/CPS/) and others available in GenBank (3, 6; F. Kong, G. L. Gilbert, L. Wang, D. Liu, and J. Tao, 13 April 2004, Australian Patent Office). For the mPCR, we modified the primers we had used previously for serotype-specific PCR for the 23 vaccine serotypes (3, 6; Kong et al., Australian Patent Office) and a published S. pneumoniae-specific primer (7). For the RLB, we designed two probes for each of the 23 serotypes, as well as two S. pneumoniae-specific control probes (Table 1). Probes and primers were designed to have similar physical characteristics so as to allow simultaneous amplification and hybridization in a multiplex reaction system (5); they were synthesized by Sigma-Aldrich (Sydney, Australia). Primers were biotinylated at the 5′ end and probes had a 5′-amine group (5).
TABLE 1.
Oligonucleotide primers and probes for multiplex PCR-based reverse line blot assay used in this study
| Primera | Target | Specificityb | Tm (°C)c | GenBank accession no. | Sequence (5′ to 3′)d |
|---|---|---|---|---|---|
| SpIaSbe | Pneumolysin | S. pneumoniae | 61.43 | M17717 | 384ATT TCT GTA ACA GCT ACC AAC GA406 |
| SpSp | Pneumolysin | S. pneumoniae | 65.49 | M17717 | 531AGC GAT AGC TTT CTC CAA GTG G552 |
| SpAp | Pneumolysin | S. pneumoniae | 75.10 | M17717 | 556ACC CCA GCA ATT CAA GTG TTC GCG579 |
| SpIbAbe | Pneumolysin | S. pneumoniae | 61.69 | M17717 | 730GAA TTC CCT GTC TTT TCA AAG TC708 |
| 1Sb | wzy | Serotype 1 | 59.03 | Z83335 | 10307GGG ACT TTA ATT TTA TGC AGT G10328 |
| 1Ap | wzy | Serotype 1 | 59.22 | Z83335 | 10402AAA TTT CAC AAT TAT CAT TGC C10381 |
| 1Sp | wzy | Serotype 1 | 62.00 | Z83335 | 10529CTG GCT TTG GCA ACT TTG10546 |
| 1Ab | wzy | Serotype 1 | 60.67 | Z83335 | 10576CAC AAT GGC TTT AGA AGG TAG AG10554 |
| 2Sb | wzy | Serotype 2 | 59.86 | AF026471 | 9730CGG CAT TGT ATT CTT TAT ATC G9751 |
| 2Ap | wzy | Serotype 2 | 60.40 | AF026471 | 9849CCA ATA AAT CTT GTG TGA ATA TAA CTG9823 |
| 2Sp | wzy | Serotype 2 | 60.76 | AF026471 | 9989GCA ACA TTT CAA TCT TAT GGT G10010 |
| 2Ab | wzy | Serotype 2 | 63.05 | AF026471 | 10049CGT TTG TAT CCA TTT AAC TGC ATC10026 |
| 3Sb | wze | Serotype 3 | 59.32 | Z47210 | 5807TTG ATA TTC CCC TTG ACA ATA G5828 |
| 3Sp | wze | Serotype 3 | 59.62 | Z47210 | 5910TTT ACT ACA GTC CTC TTT CTC TGC5887 |
| 3Ap | wze | Serotype 3 | 58.63 | Z47210 | 6056GCC TCA TCC TTA ATT GGA G6038 |
| 3Ab | wze | Serotype 3 | 61.69 | Z47210 | 6102GGA GGC TTC AAG ATT CAA CTC6082 |
| 4Sb | wzy | Serotype 4 | 61.09 | AF316639 | 9619CCG TCT ATA TTT ATA TGG GTT TGC9642 |
| 4Ap | wzy | Serotype 4 | 60.08 | AF316639 | 9774TTG AAA CCC CAT ATA CTC ATT G9753 |
| 4Sp | wzy | Serotype 4 | 60.49 | AF316639 | 9848GGA TTT TGT TTT GTT ATT CTG TAG G9872 |
| 4Ab | wzy | Serotype 4 | 59.48 | AF316639 | 9934CCT GAT AAT TTT GTA CTT CTG AAT G9910 |
| 5Sb | wzy | Serotype 5 | 61.23 | AY336008 | 6052GTT TTC CCA ATA GTA GTT TGC G6073 |
| 5Ap | wzy | Serotype 5 | 61.86 | AY336008 | 6118TGC GAT AAA ATA GAT AAG GCA ATC6095 |
| 5Sp | wzy | Serotype 5 | 62.20 | AY336008 | 6275TGC TTT GTT AGA TAT AGT TAC GGG AG6300 |
| 5Ab | wzy | Serotype 5 | 62.04 | AY336008 | 6349CCC ACA GCC AAA TAG AGT TG6330 |
| 6B6ASb | wzy | Serogroup 6 (6A, 6B) | 58.77 | AF316640 | 9263TCA ACC TGC AGT AAT TTT AAC A9284 |
| 6B6AAp | wzy | Serogroup 6 (6A, 6B) | 60.95 | AF316640 | 9322TTA ACT AGA GCA CTT GCA ATC G9301 |
| 6B6Asp | wzy | Serogroup 6 (6A, 6B) | 59.01 | AF316640 | 9461GAA AGA AAT AAA TCC TTC AAA GAT AAT9487 |
| 6B6AAb | wzy | Serogroup 6 (6A, 6B) | 60.19 | AF316640 | 9555CTA CTT TCT GAA TTT CAC GGA TAT AAA G9528 |
| 7F7ASb | wzy | Serotypes 7F, 7A | 61.48 | CR931643 | 14476GCA AGT GTT TCA ATG GGA GTA14496 |
| 7F7AAp | wzy | Serotypes 7F, 7A | 58.71 | CR931643 | 14537AAA TTC CAC AAT ATT GGT AAT ACT G14513 |
| 7F7Asp | wzy | Serotypes 7F, 7A | 58.58 | CR931643 | 14776TTT TTG TAT GAT TTA AAT ATT GCG14799 |
| 7F7AAb | wzy | Serotypes 7F, 7A | 61.51 | CR931643 | 14820ACG GAG GGA CCA TAC AAT AAG14800 |
| 8Sb | wzy | Serotype 8 | 60.50 | AF316641 | 10827GAT GTT AGT TTC TTC GAT TCC AG10849 |
| 8Ap | wzy | Serotype 8 | 59.15 | AF316641 | 10870GAG GAA ACC CAC AAA GTC AAA AAA C10849 |
| 8Sp | wzy | Serotype 8 | 59.38 | AF316641 | 11012AAA ATT ATG TTT ACT TTA CGA GTT GG11037 |
| 8Ab | wzy | Serotype 8 | 61.32 | AF316641 | 11066TCA TAA TGA ATC GTA CCA ATC AAC11043 |
| 9N9LSb | wzy | Serotypes 9N, 9L | 62.46 | CR931647 | 9818TCA ATG GCG ACT TTA TTT GC9837 |
| 9N9LAp | wzy | Serotypes 9N, 9L | 58.53 | CR931647 | 9893GAA CTT TGG GAA TAT AAT CAA AAG9870 |
| 9N9LSp | wzy | Serotypes 9N, 9L | 62.15 | CR931647 | 10139GTC GGT TTC GAT TCT TTG C10157 |
| 9N9LAb | wzy | Serotypes 9N, 9L | 60.41 | CR931647 | 10179AGT CTA TTA TCT CCT GTA GGG TGC10156 |
| 9V9ASb | wzy | Serotypes 9V, 9A | 58.70 | AF402095 | 8546AAC TAT ATT TAC CCT ACT CTC CAC AG8571 |
| 9V9AAp | wzy | Serotypes 9V, 9A | 58.76 | AF402095 | 8639AAT CAG CGT TAC CTT ATA CTG TG8617 |
| 9V9ASp | wzy | Serotypes 9V, 9A | 59.62 | AF402095 | 8797GCC TCT TTT TAA CCT TTA TCT TGT8820 |
| 9V9AAb | wzy | Serotypes 9V, 9A | 61.56 | AF402095 | 8860ACC GGA AAA AGC AAT TGA G8842 |
| 10A10BSb | wzy | Serotypes 10A, 10B | 61.86 | CR931649 | 7172TGA GCT ATT TAA GGA CCT GGG7192 |
| 10A10BAp | wzy | Serotypes 10A, 10B | 62.30 | CR931649 | 7206GTT TAG AAA CCT TGC CCA GG7187 |
| 10A10BSp | wzy | Serotypes 10A, 10B | 62.25 | CR931649 | 7353ACC ATA TGG TAT TTG TTG CCT G7374 |
| 10A10BAb | wzy | Serotypes 10A, 10B | 61.76 | CR931649 | 7450GCA AGC GTC ACT TTC TTG A7432 |
| 11A11DSb | wzy | Serotypes 11A 11D | 59.48 | CR931653 | 11368GAA ATA TCG CCA TTC ATC AG11387 |
| 11A11DAp | wzy | Serotypes 11A, 11D | 59.34 | CR931653 | 11407TGT AAG ATG AAA AAA GCA TGC11387 |
| 11A11DSp | wzy | Serotypes 11A, 11D | 59.42 | CR931653 | 11676GGA TTT CTC GTA TCA GCA TAT TAC11699 |
| 11A11DAb | wzy | Serotypes 11A, 11D | 62.68 | CR931653 | 11744CAA CAG CAA CTG TGC CAC T11726 |
| 124446Sb | wzy | Serotypes 12F, 12A, 12B, 44, 46 | 59.41 | CR931660 | 8914TGA ATA TGG ACG GTG GAG8931 |
| 124446Ap | wzy | Serotypes 12F, 12A, 12B, 44, 46 | 58.48 | CR931660 | 8951GAA GAA GTT CAA CAA TCG CT8932 |
| 124446Sp | wzy | Serotypes 12F, 12A, 12B, 44, 46 | 59.37 | CR931660 | 9104GCA TGA TAT GGG AAG TTT TG9123 |
| 124446Ab | wzy | Serotypes 12F, 12A, 12B, 44, 46 | 59.69 | CR931660 | 9155AGC AAA GAA AGC CGA AAG9138 |
| 14Sb | wzy | Serotype 14 | 60.41 | X85787 | 7376CCT ACT TCC AAA ACA GTT TAT GC7398 |
| 14Ap | wzy | Serotype 14 | 59.77 | X85787 | 7494CCA TAC AAA AAG ACG GTG TAT C7473 |
| 14Sp | wzy | Serotype 14 | 59.30 | X85787 | 7587GCA TTT AAT TGG CTA ATA GCA G7608 |
| 14Ab | wzy | Serotype 14 | 59.69 | X85787 | 7652GTC AAT ATT GAT TGG CAT TTT C7631 |
| 15B15CSb | wzy | Serotypes 15B, 15C | 62.63 | CR931664 | 7797TAA TAA GCG GAT GAT TGT AGC G7818 |
| 15B15CAp | wzy | Serotypes 15B, 15C | 58.19 | CR931664 | 7837GAG GTA TAG TTG GAT AAA ACG C7816 |
| 15B15CSp | wzy | Serotypes 15B, 15C | 62.54 | CR931664 | 7976GAG CAG GAA TCA GAA CAC AAT C7997 |
| 15B15CAb | wzy | Serotypes 15B, 15C | 60.91 | CR931664 | 8148TAT ACT GAT TAA CTT TCC AGA TGG G8124 |
| 17FSb | wzy | Serotype 17F | 59.84 | CR931670 | 13988AGA GGG ATT GTT GAA GGT ATT C14009 |
| 17FAp | wzy | Serotype 17F | 60.89 | CR931670 | 14035GGA AGT GAA CGT CAA ATC TTT T14014 |
| 17FSp | wzy | Serotype 17F | 61.75 | CR931670 | 14250CAA TGC TAT GTC GCA AAT ATT G14271 |
| 17FAb | wzy | Serotype 17F | 61.34 | CR931670 | 14295AGT AGT CTC GCA TTT CTA TCA TCC14272 |
| 18Sb | wzy | Serogroup 18 (18F, 18A, 18B, 18C) | 60.45 | AF316642 | 12208AAT TGT TCT TTT CCT GTA CTC AGT C12232 |
| 18Ap | wzy | Serogroup 18 (18F, 18A, 18B, 18C) | 58.72 | AF316642 | 12260ATT CAA C/TTG GGT TCA TTA CG12241 |
| 18Sp | wzy | Serogroup 18 (18F, 18A, 18B, 18C) | 58.74 | AF316642 | 12425GGA GGA CTT AGT CAA TTT ATC TTG12448 |
| 18Ab | wzy | Serogroup 18 (18F, 18A, 18B, 18C) | 62.13 | AF316642 | 12478CGA ACC ATT GAA ACT ATC ATC TG12456 |
| 19ASb | wzy | Serotype 19A | 61.40 | AF094575 | 9260TGT ATT TGC CCT TAT TAA TGT GC9282 |
| 19AAp | wzy | Serotype 19A | 61.12 | AF094575 | 9336TGC CAC TAA TAA TCA AAA GAT AAG C9312 |
| 19ASp | wzy | Serotype 19A | 60.81 | AF094575 | 9422CCA ATT CTG GAA AAT AGC TCT TAC9445 |
| 19AAb | wzy | Serotype 19A | 60.63 | AF094575 | 9506AAG TGC AAG ATT ATG AAT CTC TCT C9482 |
| 19FSb | wzy | Serotype 19F | 59.55 | U09239 | 7693TCA GTA TTT GCA CTG GTT AAT TC7715 |
| 19FAp | wzy | Serotype 19F | 61.85 | U09239 | 7800TGC CAT TAA AGG AAT CGA AA7781 |
| 19FSp | wzy | Serotype 19F | 60.35 | U09239 | 7858CAA TTT TGG AAA ATT GCT CTA AC7880 |
| 19FAb | wzy | Serotype 19F | 59.39 | U09239 | 7941AAG AAC AAG GTT GTA TAT TTC CTT C7917 |
| 20Sb | wzy | Serotype 20 | 61.18 | CR931679 | 7711CTT TAT CAG GAA TAC GCC AAT C7732 |
| 20Ap | wzy | Serotype 20 | 60.16 | CR931679 | 7759GCA TAA AAA ACA ATC GCT GTA G7738 |
| 20Sp | wzy | Serotype 20 | 62.31 | CR931679 | 7933CTG GGT CTG AAT TTG TAT CTC G7954 |
| 20Ab | wzy | Serotype 20 | 59.11 | CR931679 | 8010CTG TAT AAT AAC GAG AAC CAA CG7988 |
| 22F22ASb | wzy | Serotypes 22F, 22A | 61.86 | CR931682 | 13190AGG ATG CAG TAG ATA CCA GTG G13211 |
| 22F22AAp | wzy | Serotypes 22F, 22A | 59.68 | CR931682 | 13263TAA TAC CAT GGC ACT AGG AAT AAC13240 |
| 22F22ASp | wzy | Serotypes 22F, 22A | 58.30 | CR931682 | 13503ATG GCT ATC AAC TTT ATC TAG GAC13526 |
| 22F22AAb | wzy | Serotypes 22F, 22A | 60.25 | CR931682 | 13543TAT AAA CGG AGG TTG TTG TCC13523 |
| 23FSb | wzy | Serotype 23F | 60.39 | AF057294 | 8583TGA TAG TGA ACT TGG GAT TGT C8604 |
| 23FAp | wzy | Serotype 23F | 61.24 | AF057294 | 8694CTA TTT GCA AAC ACG TTG AGA G8673 |
| 23FSp | wzy | Serotype 23F | 58.36 | AF057294 | 8735GGG ATT AAT TTA CAA AAT CTT CC8757 |
| 23FAb | wzy | Serotype 23F | 58.19 | AF057294 | 8827CTT TAT CGG TAA GGT GGA TAA G8806 |
| 33F33A37Sb | wzy | Serotypes 33F, 33A, 37 | 61.77 | AJ006986 | 11362TCA ACT AGT CAA GGA TTT GAT GG11384 |
| 33F33A37Ap | wzy | Serotypes 33F, 33A, 37 | 60.62 | AJ006986 | 11421TGA TAC CAC AAG TAA CAG AGT CG11399 |
| 33F33A37Sp | wzy | Serotypes 33F, 33A, 37 | 59.52 | AJ006986 | 11589GAT GAT TTT GCA GAT GTA CTA TGA11612 |
| 33F33A37Ab | wzy | Serotypes 33F, 33A, 37 | 59.67 | AJ006986 | 11639CGT ATC AGA TTT GCG ATT TC11620 |
S, sense; A, antisense; b, biotin-labeled primer (primers were biotin labeled at the 5′ end); p, probe (probes were 5′ end C6 amine labeled).
Based on published sequence data for the whole cps gene cluster, it was not possible to design primers/probes that could distinguish some individual serotypes from one or four closely related ones (shown in boldface), usually (but not always) belonging to the same serogroup.
Tm values were provided by the primer synthesizer (Sigma-Aldrich).
Numbers represent the base positions at which primer/probe sequences start and finish (starting at “1” of the corresponding GenBank sequence).
Two primers published previously (8) were used as species-specific probes.
The DNA extraction method (4), the mPCR system, and the thermal profiles used were as previously described (5), except that we included 24 (rather than 10) primer pairs (Table 1) in a 25-μl mPCR system and used 1 U (rather than 0.5) of QIAGEN Hotstart Taq polymerase. For serotype 23F, 25 pM of each primer were used, whereas 12.5 pM of each primer was used for all other serotypes.
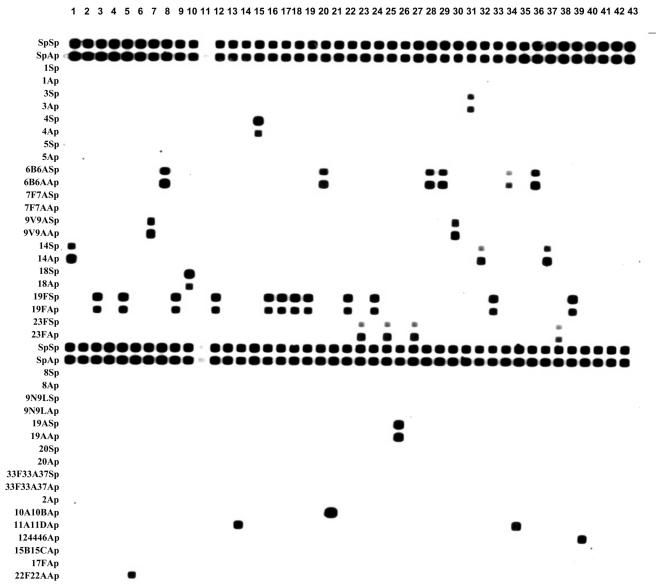
RLB hybridization was based on previously published methods (http://www.nioo.knaw.nl/cl/me/) (8, 11) with the following modifications: the hybridization temperature was 60°C, and 1.25 pM of each probe, in 150 μl of 500 mM NaHCO3 (pH 8.4), was used in each slot to label the membrane (Fig. 1).
FIG. 1.
mPCR/RLB results for a representative sample of 43 clinical isolates. See Table 1 for descriptions and specificities of probes listed at left. The choice of probes for this membrane, in addition to two pairs of S. pneumoniae-specific probes, was based on two for each of the serotypes represented in the 11-valent conjugate vaccine (serotypes 1, 3, 4, 5, 6B, 7F, 9V, 14, 18C, 19F, and 23F [shown in the top half of the membrane]) and others that are identified relatively commonly (serotypes 8, 9N, 19A, 20, and 33F) and single probes for each of the less common serotypes among those represented in the 23-valent vaccine (serotypes 2, 10A, 11A, 12, 15B, 17F, and 22F). The conventional serotypes of the 43 isolates shown on this membrane are (from left to right): 14, NT, 19F, NT, 19F, 22F, 9V, 6A, 19F, 18C, NSP, 19F, 16F, 11A, 4, 19F, 19F, 19F, 19F, 6B, 10A, 19F, 23F, 19F, 23F, 19A, 23F, 6A, 6B, 9V, 3, 14, 19F, 6B, 11A, 6A, 14, 23F, 19F, 46, 47F, 47A, and 48. NT, nontypeable. NSP, not S. pneumoniae.
A collection of 334 reference strains and well-characterized clinical isolates were used to develop the assay, of which 244 had been used in our previous studies (3, 6) and 90 were serotype reference strains, newly purchased from Statens Serum Institut of Denmark (1, 2). Except for serotypes 10C, 11F, 12B, 25A, 33D, and 44, all serotypes were represented by two or more strains.
All of the putative serotype-specific primers or probes yielded mPCR products and RLB signals from isolates with the corresponding serotypes. However, as demonstrated previously, serotype discrimination based solely on the wzy gene (or even the whole cps gene cluster) is not straightforward (3, 6). Primers and probes designed to identify serotypes 6B, 7F, 9N, 9V, 10A, 11A, 12F, 15B, 18C, 22F, and 33F could not distinguish between the target and one or more closely related serotypes, usually but not always in the corresponding serogroup (Table 1 and Fig. 1) because they share virtually identical wzy sequences. Thus, 17 serotypes, in addition to those in the 23-valent polysaccharide vaccine, were amplified and hybridized by the mPCR/RLB system. These cross-reactions are predictable, and individual serotypes can be identified using a limited number of factor antisera. The remaining 12 primer pairs and probe pairs were truly serotype specific, and there was excellent agreement between paired probes for the same serotypes, indicating that single probes would be adequate for most serotypes.
The method was further evaluated using 268 clinical isolates, the serotypes of which were unknown at the time of mPCR/RLB testing. These isolates included 135 consecutive invasive isolates referred to the NSW Pneumococcal Reference Laboratory for serotyping and 133 colonizing isolates from patients with respiratory infections at the Children's Hospital, Westmead, New South Wales, Australia. Conventional serotyping was performed using the Quellung reaction, as previously described (3). Two isolates were not amplified by mPCR/RLB, and phenotypic retesting showed that they were not S. pneumoniae. Of 266 pneumococcus strains, 12 (4.5%) were nontypeable by mPCR/RLB (amplified by mPCR but hybridized only with the pneumococcal control probe); of these, 10 belonged to serotypes not represented in the current mPCR/RLB assay, namely, 16F (one isolate), 23A (three isolates), 35F (four isolates), 35B (one isolate), and 38 (one isolate), and 2 were not serotypeable. Another 4 of the 266 pneumococcus isolates were not serotypeable but were identified by mPCR/RLB as serotype(s) 4, 11A/11D, 14, and 33F/33A/37. The predicted serotype(s) of the other 250 isolates were as follows: 3 (5 isolates), 4 (13 isolates), 6B/6A (45 isolates), 7F/7A (1 isolate), 9V/9A (15 isolates), 10A/10B (2 isolates), 11A/11D (2 isolates), 14 (51 isolates), 15B/15C (4 isolates), 18 (14 isolates), 19F (63 isolates), 19A (12 isolates), 22F/22A (2 isolates), and 23F (21 isolates). Thus, 181 of 254 (71%) isolates that were typeable by mPCR/RLB were identified exactly, and another 73 (29%) were identified to within one to five serotypes, for which a limited number of antisera were needed to distinguish individual serotypes.
Three isolates identified by mPCR/RLB as serotypes 19A, 9V/9A, and 23F had been serotyped, initially, as 19F, 19A, and 6B, respectively. The discrepancies were resolved by repeating the serotyping, which confirmed that the mPCR/RLB results were correct.
Our molecular serotype prediction mPCR/RLB assay for 23 pneumococcal “vaccine” serotypes is clearly less discriminatory than conventional serotyping (1-3, 6; Kong et al., Australian Patent Office) because individual serotype-specific targets are not available within cps gene clusters of some serotypes. However, there are obvious advantages of the mPCR/RLB. First, it uses reagents and techniques that are available in many microbiology laboratories; an uncomplicated, rapid DNA preparation method (4); and a single mPCR/RLB reaction (5). Second, it provides more consistent and objective results than immunological methods such as the Quellung reaction; cross-reactions are predictable and can be resolved with a small number of antisera. Serotypes that are nontypeable by the mPCR/RLB are isolated uncommonly from clinical specimens (∼5% of invasive isolates in NSW in the past 3 years [data not shown]). Third, the method, potentially, could to be used directly to test clinical specimens and so rapidly identify possible vaccine failure.
Currently, there is no single, ideal technique for pneumococcal serotyping, but this mPCR/RLB format is convenient, rapid, objective, reproducible, and discriminatory when used in conjunction with a limited number of antisera.
Acknowledgments
We thank Denise Murphy, Queensland Health Scientific Services, Brisbane, Australia; Diana Martin, Institute of Environmental Science Research, Porirua, New Zealand for providing isolates; and Ping Zhu for technical assistance.
This study was funded, in part, by the National Centre for Immunization Research and Surveillance of Vaccine Preventable Disease, Children's Hospital at Westmead, New South Wales, Australia.
REFERENCES
- 1.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrichsen, J. 1999. Typing of Streptococcus pneumoniae: past, present, and future. Am. J. Med. 107:50S-54S. [DOI] [PubMed] [Google Scholar]
- 3.Kong, F., and G. L. Gilbert. 2003. Using cpsA-cpsB sequence polymorphisms and serotype-/group-specific PCR to predict 51 Streptococcus pneumoniae capsular serotypes. J. Med. Microbiol. 52:1047-1058. [DOI] [PubMed] [Google Scholar]
- 4.Kong, F., S. Gowan, D. Martin, G. James, and G. L. Gilbert. 2002. Serotype identification of group B streptococci by PCR and sequencing. J. Clin. Microbiol. 40:216-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong, F., L. Ma, and G. L. Gilbert. 2005. Simultaneous detection and serotype identification of Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization. J. Med. Microbiol. 54:1133-1138. [DOI] [PubMed] [Google Scholar]
- 6.Kong, F., W. Wang, J. Tao, L. Wang, Q. Wang, A. Sabananthan, and G. L. Gilbert. 2005. A molecular-capsular-type prediction system for 90 Streptococcus pneumoniae serotypes using partial cpsA-cpsB sequencing and wzy- or wzx-specific PCR. J. Med. Microbiol. 54:351-356. [DOI] [PubMed] [Google Scholar]
- 7.Salo, P., A. Ortqvist, and M. Leinonen. 1995. Diagnosis of bacteremic pneumococcal pneumonia by amplification of pneumolysin gene fragment in serum. J. Infect. Dis. 171:479-482. [DOI] [PubMed] [Google Scholar]
- 8.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeng, X., F. Kong, H. Wang, A. Darbar, and G. L. Gilbert. 2006. Simultaneous detection of nine antibiotic resistance-related genes in Streptococcus agalactiae using multiplex PCR and reverse line blot hybridization assay. Antimicrob. Agents Chemother. 50:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao, Z., F. Kong, and G. L. Gilbert. 2006. Reverse line blot assay for direct identification of seven Streptococcus agalactiae major surface protein antigen genes. Clin. Vaccine Immunol. 13:145-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zwart, G., E. J. van Hannen, M. P. Kamst-van Agterveld, G. K. Van der, E. S. Lindstrom, J. Van Wichelen, T. Lauridsen, B. C. Crump, S. K. Han, and S. Declerck. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]