Abstract
Aspergillus markers are becoming increasingly important for the early diagnosis of invasive aspergillosis. The kinetics of release of these surrogate markers, however, is largely unknown. We investigated the release of β-(1-5)-galactofuranosyl (galf) antigens (Platelia Aspergillus), 1,3-β-d-glucan (BG) (Fungitell), and DNA (PCR) in an in vitro model of Aspergillus fumigatus. The results showed that release is correlated to the growth phase of the fungus, which depends on available nutrients. Whereas galf antigens and BG are released during logarithmic growth, DNA is released only after mycelium breakdown. During early logarithmic growth, galf antigens seem to be released somewhat earlier than BG. Furthermore, galf antigen concentrations of more than 120,000 times the serum cutoff value (0.5 ng/ml) can be measured, while BG concentrations reach a value only 978 times the serum cutoff value (60 pg/ml). During lytical growth, release of galf antigens further increased to a maximum level, which depended on pH. After that, the concentration of galf antigens stayed high (pH 7.4) or decreased to zero within 4 days (pH 5.0). In contrast to galf antigens, BG concentration decreased after 1 day of growth. The decrease of galf components seems to be due to the enzyme β-galactofuranosidase, which is able to destroy galf epitopes and whose activity fluctuates in the culture filtrates in parallel with galf antigen concentration. Fungal DNA seems to be released only due to autolysis caused by nutrient limitation. In conclusion, several factors clearly influence the release of surrogate markers in vitro. These same factors might also play a role at the infection site of Aspergillus disease in humans.
Invasive aspergillosis (IA) has become a leading cause of death among immunocompromised hosts, including transplant patients, those treated for hematological malignancy, and those treated with high-dose corticosteroids (41, 49). In addition, IA is increasingly observed in the nonneutropenic phase after hematopoietic stem cell transplantation and in nonclassic settings, such as intensive care units with critically ill patients (13, 16). The high mortality is due partly to the difficulty in establishing a diagnosis at an early stage of infection, since presenting symptoms are nonspecific and sensitivity of cultures is low. Techniques to improve timely diagnosis have focused on the detection of circulating surrogate markers released by the fungus (43, 45, 50). With the development of nonculture-based methods, such as PCR and antigen detection, circulating markers can be detected at an early stage of infection in patients with invasive disease (12, 27, 28).
The commercially available sandwich enzyme-linked immunosorbent assay (ELISA) (Platelia Aspergillus [PA]; Bio-Rad, Marnes-la-Coquette, France) is based on the rat immunoglobulin M monoclonal antibody EB-A2, which binds the β-(1-5)-galactofuranosyl (galf) side chains of the Aspergillus galactomannan (GM) molecule (24, 50, 51). In addition to GM, fungal glycoproteins also react with the EB-A2 antibody, including phospholipase C and phytase, which were shown to have only one terminal galactofuranose unit that was essential for binding with the EB-A2 antibody (24, 48). These findings implicate that the so-called “GM antigen” is not a single molecule but a family of molecules for which expression could be modulated by the immediate fungal environment (34). However, the actual galf antigens that circulate in vivo in the body fluids of patients have not been characterized. Furthermore, there are several problems related to antigen detection. In some patients with proven IA, circulating antigen is not detected despite repeated sampling (false negative) (55). Also, the performance of antigen detection might be reduced in patients receiving antifungal prophylaxis. These and other factors that influence the release of galf antigens, including their leakage from the site of infection into the blood and their bonding to substances present in the blood, were recently explored (31).
In contrast to the PA ELISA, there is no standardized PCR method for the detection of Aspergillus DNA for early diagnosis of IA. Whereas most methods show good sensitivity and specificity in experimental settings, unresolved issues, such as the optimal specimen (e.g., whole-blood, serum, plasma, and bronchoalveolar lavage specimens) and DNA extraction method, are probably major causes of variability in performance in clinical studies (5, 6, 18, 26). False-negative results are especially often found in patients with proven IA (6), which might be correlated with antifungal drug treatment.
A third promising nonculture-based diagnostic test detects circulating 1,3-β-d-glucan (BG). BG is a cell wall component of most medically important fungi, including Aspergillus fumigatus (35, 36). Furthermore, A. fumigatus releases BG into the culture fluid in parallel with in vitro fungal growth (33) and can be quantified spectrophotometrically by activation of factor G, a coagulation factor of the horseshoe crab (35). A recently FDA-approved commercial method for the determination of BG has become available (Fungitell, formerly Glucatell; Associates of Cape Cod) (38). There are only a limited number of clinical studies published, and a definitive cutoff value for the diagnosis of IA remains to be established (37, 38, 42).
At present, there is insufficient understanding of the in vitro and in vivo kinetics of circulating galf antigens, BG, and fungal DNA. Recently, an in vitro study of the release of galf antigens by A. fumigatus in relation to the fungal biomass (20) showed that the amounts of galf antigens released in the culture supernatant were not different for strains from circulating antigen-positive (CAG+) and circulating antigen-negative (CAG−) IA patients. Other causes for negative antigenemia could be the reduced release of galf antigens due to environmental factors, such as pH, and reduced availability of nutrients at the site of infection. Because the kinetics of release of surrogate markers is largely unknown, we used the same in vitro model to study and compare the release of galf antigens, DNA, and BG by A. fumigatus strains from different patients.
MATERIALS AND METHODS
Fungal strains and culture conditions.
A. fumigatus strains from a CAG+ and a CAG− patient were cultured in flasks containing yeast nitrogen base (YNB) liquid medium with different glucose concentrations (5, 25, and 100 mM) and at different pHs (5.0 to 7.4). The medium was buffered with MOPS (morpholinopropanesulfonic acid) (170 mM) or MES (morpholineethanesulfonic acid) (100 mM) or not buffered. Each growth experiment was performed in duplicate and with one CAG+ (AZN 4684) and one CAG− (AZN 4565) strain. The cultures were incubated for a maximum of 9 days at 37°C and 160 rpm after inoculation with spores to a final concentration of 106 to 107 spores/ml. For each time measurement, one Erlenmeyer flask (500 ml) containing 200 ml of culture was used. The mycelium was harvested by filtration on a filter (0.8 μm; Schleicher & Schuell) and freeze-dried to determine the biomass. The culture filtrate was used for the PA ELISA, pH and glucose measurements, the β-galactofuranosidase assay, and detection of DNA and BG. Furthermore, the release of galf antigens, BG, and DNA was also measured in one Erlenmeyer flask by taking (sterile) samples at serial time points, followed by filtration of the sample (0.2 μm cellulose acetate; Schleicher & Schuell).
PA ELISA.
The culture filtrate samples were serially diluted in water for detection of reactivity by the PA ELISA. Each plate contained a standard calibration curve (0 to 15 ng/ml purified GM) in order to use the ELISA quantitatively. The PA ELISA was performed according to the manufacturer's instructions. Briefly, 50 μl of a reaction mixture containing horseradish peroxidase-conjugated anti-GM monoclonal antibody EB-A2 was added to each well of a microtiter plate coated with the same monoclonal antibody EB-A2, followed by the addition of 50 μl of pretreated sample. After 90 min of incubation at 37°C, the plates were washed five times with washing buffer before 200 μl of buffer containing tetramethylbenzidine solution was added. Then, the plates were incubated for another 30 min in the dark at room temperature, followed by the addition of 100 μl of 1.5 N sulfuric acid to stop the reaction. The optical density (OD) was read at 450 and 620 nm. A test sample was considered positive when the OD at 450 nm was higher than that of the cutoff sample (i.e., 0.5 ng GM). When testing different dilutions, the OD value closest to the OD value of the cutoff sample (i.e., 0.5 ng GM; R4) was interpolated in the calibration curve, and the total PA reactivity was expressed as the amount of GM in ng/ml (in addition to GM, actual PA reactivity in culture filtrates comes from several galf components; however, only purified GM was available for quantification).
Fungitell BG assay.
The culture filtrate samples were serially diluted in glucan-free reagent grade water (RGW) for detection of BG by the Fungitell test kit (Associates of Cape Cod, Falmouth, Mass.) in a kinetic, chromogenic format as recommended by the manufacturer. All assays were performed in duplicate. Briefly, 25-μl samples of the standards (100 to 6.25 pg/ml pure pachyman, a linear BG) and unknown pretreated samples were added to each well of a microtiter plate. Unknown samples (5 μl) were pretreated for 10 min at 37°C with an alkaline reagent (20 μl; 0.125 M KOH/0.6 M KCl). Fungitell reagent (lyophilized BG-specific Limulus amebocyte lysate) was reconstituted with 2.8 ml of RGW, followed by 2.8 ml of pyrosol reconstitution buffer (2 M Tris HCl, pH 7.4), and 100 μl of this mixture was added to each sample. An OD405 was read for 40 min at 37°C, using an Anthos HT-3 plate reader and MikroWin software (Mikrotek Laborsysteme GmbH), and the concentration in each unknown sample was calculated using the calibration curve after the standards were multiplied by 5, so that the range was from 500 to 31.25 pg/ml. When absorbance was outside the range of the standard curve, the samples were diluted in RGW and tested again.
Glucose assay.
Glucose concentration was measured enzymatically by coupling the glucose oxidase reaction to the peroxidase reaction. Culture filtrate samples (serially diluted in water) and glucose standard samples (0.05 to 1.2 mM glucose) were analyzed by taking 100 μl of sample and by mixing this with 1 ml of reagent containing Na2HPO4 · 2H2O (13.6 g/liter), NaH2PO4 · H2O (7.2 g/liter), glucose oxidase (750 U), peroxidase (150 U), and o-dianisidine dihydrochloride (39.4 mg/liter). After 45 min of incubation at room temperature, the formed brown-colored product (oxidized o-dianisidine) was measured spectrophotometrically at 450 nm. The A450 of the unknown samples was corrected for the blank (diluted) culture medium without glucose, and the amount of glucose was calculated from the standard curve.
β-Galactofuranosidase assay.
β-Galactofuranosidase activity was determined by measuring (in microtiter plates) the hydrolysis of p-nitrophenyl-β-d-galactofuranoside (PNP-galf; Sigma) following incubation at 37°C and pH 4.0 for 1.5 h. The reaction was started by the addition of 60 μl of culture filtrate to 1 mM of substrate in a total volume of 100 μl 100 mM sodium acetate buffer, pH 4.0. The reaction was terminated with 200 μl of stop solution containing 0.25 M Na2CO3 and 0.25 M NaHCO3. The yellow-colored product (p-nitrophenol) was measured spectrophotometrically at 405 nm. One enzyme unit is defined as the amount required to hydrolyze 1 μmol of substrate min−1 under these conditions, and p-nitrophenol (Sigma) was used for the standard curve. Different blanks were used, including water and culture medium.
DNA detection.
DNA extraction of 1-ml culture filtrate samples was performed with and without physical disruption, using MagNA lyser green 1.4-mm ceramic beads (MagNA lyser; Roche), for two cycles of 20 s at 6,500 rpm, followed by isolation with the MagNA Pure LC apparatus (Roche Biochemicals). The total nucleic acid large-volume isolation kit (Roche Biochemicals) was used. We used a starting volume of 1,000 μl eluted in 50 μl, and 10 μl was used for the PCR. A. fumigatus DNA was detected with a previously published PCR protocol which amplifies a 363-bp fragment of the Aspergillus 18S rRNA gene (30).
RESULTS
General growth characteristics.
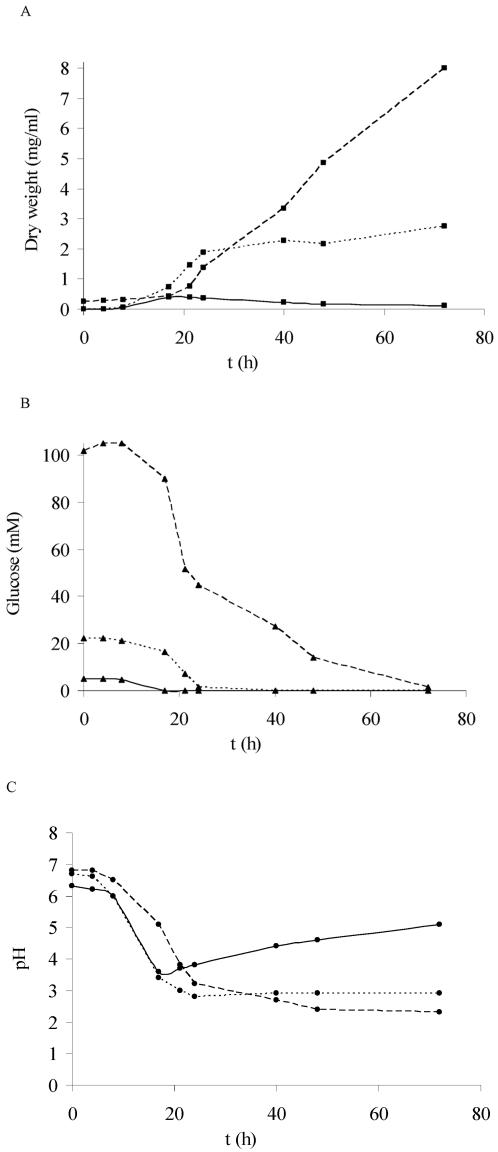
Growth on glucose resulted in an associated drop in pH when the medium was not buffered, followed by a pH increase when glucose became limited (Fig. 1A to C). During the logarithmic growth phase, glucose consumption was correlated to biomass production. Glucose became limited within 17 h (5 mM) or after 1 and 3 days (25 and 100 mM glucose, respectively). In order to study the effect of glucose limitation, the 5 mM concentration (also human blood values) was used for further studies. The effects of different pHs at this glucose concentration were tested (Fig. 2 and 3). In all cases, the maximum dry weight was about 0.4 to 0.5 mg/ml, which was reached within 17 h, the same time needed for glucose consumption. CAG+ and CAG− A. fumigatus strains gave similar results. Glucose limitation initiated the lytical phase, which is shown by the decrease of dry weight (Fig. 2 and 3; glucose concentrations are not shown but were 0 within 1 day of growth). Nine days after glucose became limited, the mycelium seemed to be broken down entirely in most cultures. However, the fungus was still viable after being transferred to a fresh medium.
FIG. 1.
Growth of A. fumigatus (CAG− strain) on YNB supplemented with 5 mM (—), 25 mM (- - -), or 100 mM (- -) of glucose as the carbon source. The medium was not buffered, and the initial pH was 6.3 to 6.8. (A) ▪, dry weight (mg/ml). (B) ▴, glucose concentration in culture filtrate (mM). (C) •, pH of culture filtrate.
FIG. 2.
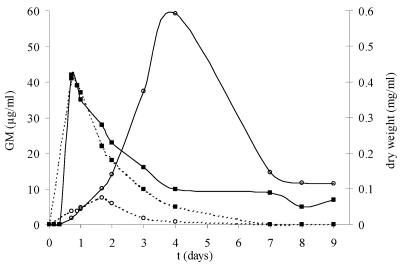
Growth of A. fumigatus on YNB medium supplemented with 5 mM of glucose as the carbon source. Each line is the mean value of a CAG+ and a CAG− strain. Media were not buffered, and the initial pH was 7.4 (—) or pH 6.3 (- - -). (▪) Dry weight (mg/ml); (○) GM (μg/ml), i.e., PA ELISA reactivity due to galf antigens.
FIG. 3.
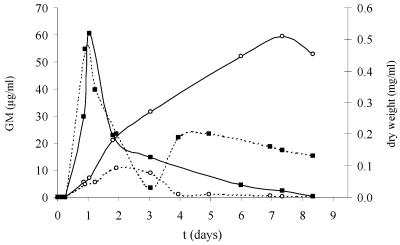
Growth of A. fumigatus on YNB medium supplemented with 5 mM of glucose as the carbon source. Each line is the mean value of a CAG+ and a CAG− strain. Media were buffered with MES at pH 7.4 (—) or pH 5.0 (- - -). (▪) Dry weight (mg/ml); (○) GM (μg/ml), i.e., PA ELISA reactivity of the supernatant due to galf antigens.
Release of galf antigens.
During the logarithmic growth phase, when glucose was consumed (values not shown), galf antigens were released in the culture supernatant (Fig. 2 and 3). After glucose became limited (after 1 day of growth), PA ELISA reactivity further increased, followed by a decrease. The maximum PA ELISA reactivities varied between the different culture conditions. At these maximum levels, the pH was 7.1 for the culture that started at pH 7.4 and pH 4.4 for the culture that started at pH 6.3 (values not shown). PA ELISA reactivity was the highest when pH was 7.1 to 7.4 (about 60 μg/ml) (Fig. 2 and 3) and lower when pHs were 4.4 (7.5 μg/ml) (Fig. 2) and 5.0 (10.9 μg/ml) (Fig. 3). During this lytical phase, PA ELISA reactivity reached the maximum value after 40 h (pH 5 and lower) or after 4 days (pH 7.1) and more than 7 days (pH 7.4), followed by a decrease. Galf antigens became undetectable within 4 days at low pH. In contrast, at pH 7.4, PA ELISA reactivity (expressed as GM concentration) decreased only slightly (Fig. 3).
Release of Aspergillus DNA.
Aspergillus DNA was not detected in the culture supernatant during logarithmic growth (Tables 1 and 2 and Fig. 2 and 3), indicating that DNA was not released by the fungus. DNA was detected only during the lytical phase, which corresponded with autolysis of the fungus due to lack of nutrients (i.e., carbon source). These positive samples were negative after mechanical disruption with beads (MagNA lyser; Roche), which indicates that pretreatment results in loss of “free” DNA. Furthermore, DNA was detected in pretreated mycelial samples (positive control) but not in any untreated mycelial samples, indicating that pretreatment was required to release the fungal DNA from the mycelium.
TABLE 1.
Release of Aspergillus markers during in vitro growth under different conditionsa
| Mediumb | t (h/days) | GM (μg/ml) (n = 2) | BG (pg/ml) (n = 2) | GM (ng/ml)c | BG (pg/ml)d | DNA (PCR) (CAG+/ CAG−)e |
|---|---|---|---|---|---|---|
| I | 0 | 0.01 | 539 | 0 | 0 | Neg/neg |
| 16/0.7 | 3.70 | 1,351 | 7,380 | 14 | Neg/neg | |
| 24/1.0 | 4.85 | 1,351 | 9,680 | 14 | Neg/neg | |
| 40/1.7 | 7.47 | 664 | 14,920 | 2 | Neg/pos | |
| 48/2.0 | 5.89 | 539 | 11,760 | 0 | Neg/pos | |
| 72/3.0 | 1.80 | 1,039 | 3,580 | 8 | Neg/pos | |
| 96/4.0 | 0.73 | 664 | 1,440 | 2 | Pos/pos | |
| 168/7.0 | 0.01 | 40 | 0 | 0 | Pos/pos | |
| 192/8.0 | 0.02 | 165 | 20 | 0 | Pos/pos | |
| 216/9.0 | 0 | 102 | 0 | 0 | Pos/pos | |
| II | 0 | 0.015 | 40 | 0 | 0 | Neg/neg |
| 4/0.2 | 0.025 | 290 | 20 | 4 | Neg/neg | |
| 8/0.3 | 0.068 | 352 | 106 | 5 | Neg/neg | |
| 16/0.7 | 1.71 | 914 | 3,390 | 15 | Neg/neg | |
| 24/1.0 | 4.40 | 477 | 8,770 | 7 | Neg/pos | |
| 40/1.7 | 10.10 | 477 | 20,170 | 7 | Neg/pos | |
| 48/2.0 | 14.10 | 914 | 28,170 | 15 | Neg/pos | |
| 72/3.0 | 37.50 | 914 | 74,970 | 15 | Neg/pos | |
| 96/4.0 | 59.30 | 102 | 18,570 | 1 | Neg/pos | |
| 168/7.0 | 14.70 | 0 | 29,370 | 0 | Pos/pos | |
| 192/8.0 | 11.60 | 165 | 23,170 | 2 | Pos/pos | |
| 216/9.0 | 11.40 | 40 | 22,770 | 0 | Pos/pos |
Results are the mean values from two individual growth experiments (CAG+ and CAG− strains).
Medium I, YNB medium with 5 mM of glucose. The medium was not buffered and started at pH 6.3. Medium II, YNB medium with 5 mM of glucose. The medium was not buffered and started at pH 7.4. See also Fig. 2.
GM amount divided by the Platelia Aspergillus cutoff value (0.5 ng/ml) after correction for value at time zero.
BG amount divided by the Fungitell cutoff value (60 pg/ml) after correction for value at time zero.
Neg, negative; pos, positive.
TABLE 2.
Release of Aspergillus markers during in vitro growth under different conditionsa
| Mediumb | t (h/days) | GM (μg/ml) (n = 2) | BG (pg/ml) (n = 2) | GM (ng/ml)c | BG (pg/ml)d | DNA (PCR) (CAG+/ CAG−)e |
|---|---|---|---|---|---|---|
| III | 0 | 0.015 | 44 | 0 | 0 | Neg/neg |
| 6/0.3 | 0.080 | 180 | 130 | 2.3 | Neg/neg | |
| 22/0.9 | 4.67 | 588 | 9,307 | 9.1 | Neg/neg | |
| 30/1.3 | 5.61 | 180 | 11,190 | 2.3 | Neg/neg | |
| 47/1.9 | 10.90 | 1,064 | 21,770 | 17.0 | Neg/pos | |
| 73/3.0 | 9.01 | 1,268 | 17,990 | 20.4 | Neg/pos | |
| 95/4.0 | 1.08 | 112 | 2,130 | 1.1 | Pos/pos | |
| 119/5.0 | 1.08 | 44 | 2,130 | 0 | Neg/pos | |
| 167/7.0 | 0.570 | 248 | 1,110 | 3.4 | Pos/pos | |
| 177/7.4 | 0.250 | 248 | 470 | 3.4 | Neg/pos | |
| 200/8.3 | 0.150 | 44 | 270 | 0 | Neg/pos | |
| IV | 0 | 0.010 | 215 | 0 | 0 | Neg/neg |
| 4/0.2 | 0.020 | 44 | 20 | 0 | Neg/neg | |
| 6/0.3 | 0.052 | 84 | Neg/neg | |||
| 21/0.9 | 5.40 | 594 | 10,779 | 6 | Neg/neg | |
| 25/1.0 | 7.08 | 1,177 | 14,140 | 16 | Neg/neg | |
| 44/1.8 | 21.09 | 581 | 42,160 | 6 | Pos/neg | |
| 73/3.0 | 31.50 | 492 | 62,980 | 5 | Neg/neg | |
| 144/6.0 | 52.03 | 1,177 | 104,040 | 16 | Neg/neg | |
| 177/7.4 | 59.43 | 819 | 118,840 | 10 | Neg/neg | |
| 200/8.3 | 52.80 | 492 | 105,580 | 5 | Neg/neg |
Results are the mean values from two individual growth experiments (CAG+ and CAG− strains).
Medium III, YNB medium with 5 mM of glucose. The medium was buffered at pH 5.0. Medium IV, YNB medium with 5 mM of glucose. The medium was buffered at pH 7.4. See also Fig. 3.
GM amount divided by the Platelia Aspergillus cutoff value (0.5 ng/ml) after correction for value at time zero.
BG amount divided by the Fungitell cutoff value (60 pg/ml) after correction for value at time zero.
Neg, negative; pos, positive.
Release of BG.
Like PA ELISA-reactive components, BG was released during logarithmic growth in the culture medium, as shown in Table 1. BG increased to a maximum, ranging from 914 to 1,351 pg/ml, after about 1 day of growth with 5 mM of glucose and at different pHs (mediums I to IV) and was followed by a decrease. After the first decrease, the BG level showed a second fluctuation at all pHs. However, these absolute values are not much higher than background values at time zero. Furthermore, BG levels reached approximately 16 times the serum cutoff value of 60 pg/ml while galf antigen levels reached up to 118,840 times the serum cutoff value of 0.5 ng/ml GM (Table 1).
Release of BG compared to galf antigens and DNA.
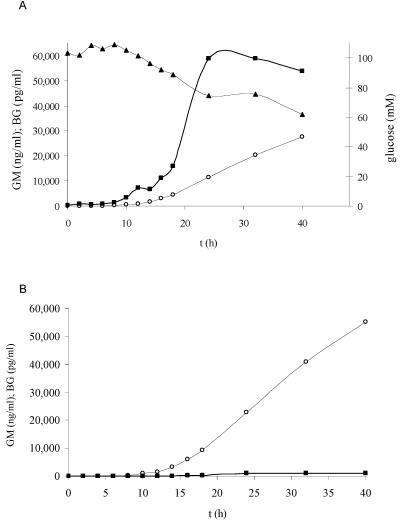
In order to study the release of surrogate markers during early logarithmic growth under nutrient-sufficient conditions (i.e., 100 mM glucose), BG, galf antigens, DNA, and the consumption of glucose were measured during the first 40 h of growth of the fungus in one Erlenmeyer flask. Compared to 5 mM of glucose, the BG levels in the supernatant were much higher when the fungus was grown at 100 mM of glucose, as shown in Fig. 4. Whereas PA ELISA reactivity continued to increase to more than 60,000 ng/ml GM or 120,000 times the serum cutoff value of 0.5 ng/ml after 3 days of growth, when 100 mM glucose is consumed (results shown only for first 40 h), BG reached a maximum of 58,915 pg/ml or 978 times the serum cutoff value after 1 day of growth, when about 30 mM of glucose had been consumed, and decreased after that (Fig. 4A and B). Furthermore, the results of the PA ELISA, Fungitell assay, and PCR are also shown in Table 3. The first indication of PA ELISA reactivity was shown by an increase of the ELISA index (EI) of 0.5 after 2 h of growth. After 4 h of growth, the EI was three times the EI at time zero. After 6 h of growth, the EI was 42 times higher than the value at time zero. The first indication of BG release was more difficult to determine because the BG background at time zero was 213.4 pg/ml (i.e., the mean of 0, 0, 76.1, and 777.5 pg/ml). After 2 h of growth, the BG value was just above the highest background value, and the BG values after 4 and 6 h of growth were below the highest background value. Definitive release was measured after 8 h of growth (Table 3). Aspergillus DNA was not detected in the culture supernatant within the first 40 h of growth (Table 3).
FIG. 4.
(A) Release of galf antigens (expressed as the amount of GM) and BG during growth of A. fumigatus on YNB medium supplemented with 100 mM of glucose as the carbon source in an Erlenmeyer flask. Each line is the mean value of a CAG+ and a CAG− (GM) strain. The medium was buffered with MOPS at pH 7.4. (▴) Glucose concentration in culture filtrate (mM); (○) GM (ng/ml); (▪) BG (pg/ml). (B) Same as panel A, but the amounts of GM and BG are divided by serum cutoff values (CV) 0.5 ng/ml and 60 pg/ml, respectively.
TABLE 3.
Release of Aspergillus markers during in vitro growth in Erlenmeyer flask on YNB with 100 mM of glucose buffered at pH 7.4a
| Time of growth (h) | PA ELISA result (EI)b | Fungitell result (BG in pg/ml)c | PCR (Aspergillus DNA)d |
|---|---|---|---|
| 0 | 0.470 | 213.4 | Neg |
| 2 | 0.970 | 827.6 | Neg |
| 4 | 1.570 | 527.0 | Neg |
| 6 | 21.0 | 727.4 | Neg |
| 8 | 157.5 | 1,278.5 | Neg |
| 10 | 506.0 | 3,274.2 | Neg |
| 12 | 791.5 | 7,278.1 | Neg |
| 14 | 1,663.5 | 6,773.0 | Neg |
| 16 | 3,031.0 | 11,282.0 | Neg |
| 18 | 4,594.0 | 15,791.0 | Neg |
| 24 | 11,440.0 | 58,914.9 | Neg |
| 32 | 20,430.0 | 58,914.9 | Neg |
| 40 | 27,669.5 | 53,904.9 | Neg |
Results are the mean values from two individual growth experiments (CAG+ and CAG− strains).
Cutoff value, 0.5 ng/ml GM.
Cutoff value, 60 pg/ml BG. The values at time zero ranged from 0 to 777 pg/ml. Values at 2, 4, and 6 h were within the same range as those at time zero.
Neg, negative.
In order to confirm that the decrease of BG was caused by an enzyme that was released into the culture medium, the following experiment was performed. A filtered supernatant containing 51,814 pg/ml BG was boiled to reduce the enzyme activity and then incubated for 24 h at 37°C. The remaining BG concentration was measured at 4, 8, and 24 h. There was a 60% (24 h) decrease in BG concentration in nonboiled supernatant but no decrease of BG in the boiled supernatant.
β-Galactofuranosidase activity.
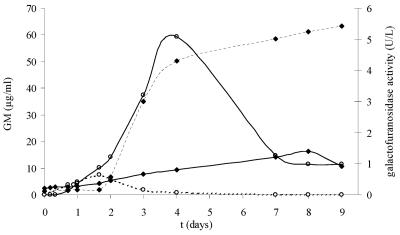
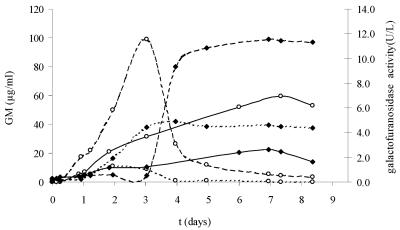
The galactofuranosidase activity of the same culture filtrates as those shown in Fig. 2 and 3 was determined (Fig. 5 and 6). The enzyme activity was low during logarithmic growth (about 0.2 to 0.4 U/liter) and increased during the lytical phase, when the carbon source was limited. Maximal activity was the lowest at higher pHs, being 1.4 U/liter at pH 7.1 to 7.4 (Fig. 5) and 2.6 U/liter at pH 7.4 (Fig. 6), and higher at low pHs, being 5.4 U/liter at pH 3.6 to 4.4 (Fig. 5) and 4.9 U/liter at pH 5.0 (Fig. 6). Activity was even higher when grown on 25 mM of glucose at pH 5.0, being 11.6 U/liter. Furthermore, a strong increase in enzyme activity seems to be correlated with a decrease in PA ELISA reactivity. This phenomenon is most clearly seen with 25 mM of glucose (Fig. 6). In order to confirm that β-galactofuranosidase was released into the culture medium and reduced the amount of PA ELISA-reactive material, a filtered supernatant with a galactofuranosidase activity of 10 U/liter and a PA ELISA reactivity of 2.1 μg/ml GM was boiled to reduce the enzyme activity and then incubated for 24 h at 37°C. The remaining enzyme activity and PA ELISA reactivity were measured at 4, 8, and 24 h. The enzyme activity showed a 100% reduction due to the boiling of the supernatant. There was a 69.8% (24 h) decrease in PA ELISA reactivity in the nonboiled supernatant but no reduction in PA ELISA reactivity in the boiled supernatant.
FIG. 5.
Release of GM and β-galactofuranosidase during growth of A. fumigatus on YNB medium supplemented with 5 mM of glucose as the carbon source. Each line is the mean value of a CAG+ and a CAG− strain. Media were not buffered and started at pH 7.4 (—) or pH 6.3 (- - -). (♦) β-Galactofuranosidase activity (U/liter); (○) GM (μg/ml).
FIG. 6.
Release of GM and β-galactofuranosidase during growth of A. fumigatus on YNB medium supplemented with 5 mM (— and - - -) or 25 mM (- -) of glucose as the carbon source. Each line is the mean value of a CAG+ and a CAG− strain. Media were buffered with MOPS (170 mM) at pH 7.4 (—) or with MES (100 mM) at pH 5.0 (- - - and - - -). (♦) β-Galactofuranosidase (U/liter); (○) GM (μg/ml).
DISCUSSION
In this study, the glucose concentration and buffering capacity of the medium determined the pH during fungal growth. In Aspergillus species, growth in the presence of glucose is normally associated with organic acid production and an associated drop in pH (21). In addition, growth in the presence of ammonium, the main nitrogen source of YNB, also decreased medium pH because uptake of ammonia is accompanied by the release of a proton from the cell, as shown for other fungi (1, 17). This study shows that the release of GM and other galf antigens by A. fumigatus during the logarithmic growth phase corresponds with the increase in fungal biomass and the glucose concentration used. The same is true for BG release during the first 24 h of logarithmic growth. However, BG was detected somewhat later than galf antigens. Furthermore, BG showed a decrease after 24 h, which is not due to nutrient limitation (i.e., glucose). A decrease of BG after 24 h of in vitro growth has already been shown by Miyazaki et al. (33) for Candida albicans and was suggested to be caused by the enzyme β-1,3-glucanase. Those authors also measured a maximum BG release by A. fumigatus of about 10,000 pg/ml after 48 h of growth at 30°C. A. fumigatus has been shown to produce cell wall-associated exo-1,3-β-glucanases and an endo-1,3-β-glucanase, which might have a role in cell wall morphogenesis (14, 15). In other filamentous fungi, exocellular β-glucanases have also been found and seem to have a role in hydrolyzing exocellular BGs for fungal catabolism. However, these enzymes are expressed only under glucose limitation (15). The BG decrease in this study is clearly enzyme related, but it is not clear which of the glucanases is responsible.
When glucose becomes limited, mycelium is broken down. This results in a further release of galf antigens (expressed as GM concentration with equivalent PA ELISA reactivities). A part of these antigens probably originate from the cell wall, including GM and peptidoGM (25); however, other galf antigens might be induced due to nutrient limitation, e.g., glucose and phosphate. Phospholipase C, a possible virulence factor, is repressed by phosphate (34) and might be induced by phosphate limitation. Together with phytase and alkaline phosphatase, this enzyme might be part of a phosphate-scavenging system as suggested by Morelle et al. (34). The exact cause of increase of PA ELISA reactivity in the culture supernatants remains unknown because not all in vitro galf antigens and their contributions to the total PA ELISA reactivity are characterized. In addition to glucose, pH also has an influence on the release of galf antigens. More PA ELISA reactivity is found in the medium when it is cultured at a higher pH than at a low pH. Galactomannan is a cell wall component that is also released in the environment, probably as a carbon overflow mechanism (31, 40). The role of some extracellular polysaccharides of fungi may be to reserve nutrient sources of carbon. During C limitation, GM and other galf epitopes might be broken down by the enzyme exo-β-d-galactofuranosidase to produce galactose, which can then be used as a secondary C source, as suggested for Aspergillus niger (57) and Penicillium fellutanum (40). The production of this enzyme seemed to be medium dependent, with glucose as a repressor (10). Furthermore, a galactofuranosidase was purified from A. niger with an optimum pH of 3 to 4 (56). The results of this study suggest that β-galactofuranosidase activity is involved in the decrease of galf epitopes in the culture supernatant. The activity rises after glucose becomes limited and seems to be influenced by the pH of the culture medium. As a consequence, the maximum galf antigen concentration is lower at a low pH than at a higher pH. Furthermore, lowering the pH gives a fast decrease of galf antigen concentration compared to buffering the medium at pH 7.4 (Fig. 2 and 3). The β-galactofuranosidase activity in the culture filtrates is probably even higher because the assay uses synthetic PNP-galf as a substrate. The purified galactofuranosidase from Trichoderma harzianum showed a low activity on synthetic PNP-galf, indicating that the enzyme needs more than one galf residue for binding (53). Furthermore, the presence of galf antigens as a concurrent substrate in the culture supernatant might also decrease the measured enzyme activity.
A specific β-d-galactofuranosidase has been detected in only a few species of fungi: P. fellutanum (44), Penicillum and Aspergillus species (10), T. harzianum (53), Helminthosporium sacchari (11), and A. niger (56). A comparison between enzyme levels shows that A. fumigatus (this study) produced about the same level as the Penicillium species (5 to 50 U/liter) but not as much as A. niger (250 U/liter) (56). However, growth conditions and substrates differed between different studies. The β-d-galactofuranosidase in the culture supernatant probably has an autocatalytic role because activity increased under starvation conditions. A polyclonal antibody against the enzyme of P. fellutanum showed that the enzyme was present throughout the hyphae, including the walls (32). The production of extracellular β-d-galactofuranosidase by Aspergillus species could interfere with their ability to be detected in immunoassays because the enzyme degrades the epitopes. Similar activity could also be used in vivo against galf components from competing microorganisms, as suggested by Wallis et al. (56). This study shows no difference in galactofuranosidase activity between CAG+ and CAG− A. fumigatus strains, which argues against a role of this enzyme in the cause of false-negative reactivity.
Since galactofuranosyl residues and galactofuranosidases are absent in mammalian species, it was suggested that galf residues may have a role in the survival of fungi by preventing the action of hosts' glycosidases on fungal galf components, including GM and the majority of extracellular proteins (52). Marino et al. showed that the P. fellutanum peptidophosphogalactomannan could not be detected in vitro after the addition of galactofuranosidase inhibitors (29). Furthermore, the content of galactofuranose in the cell wall was significantly decreased, and cell structure was strongly disturbed. This suggests that the metabolism of galf components might be a target for development of therapeutic agents, for instance, UDP-galactopyranose mutase, which converts UDP-galactopyranose to UDP-galf. This mutase enzyme is essential for the viability of mycobacteria (46), and activity of the enzyme was shown some time ago in penicillin fungus (39).
As opposed to galf components and BG, Aspergillus DNA was not detected during the logarithmic growth phase of this fungus in vitro. Fungal DNA seemed to be released only after mycelium breakdown, due to autolysis caused by nutrient limitation. However, not all PCR samples were positive during the lytical phase. DNA might be below the detection limit of the PCR test, and DNases secreted by the fungus could lower the DNA level in the culture filtrate. The results suggest that during invasive infection, damage of hyphae is required in order to release fungal DNA, for instance, by human host defense or autolysis. The actual form in which DNA circulates in the blood of an IA patient is not known. Blood cultures are rarely positive and could result only from viable hyphal fragments because conidia are not present in the blood. So the most likely circulating form seems to be free DNA (i.e., naked DNA or DNA bound to fungal cell walls), also suggested by Costa et al. (9). Furthermore, Bougnoux and coworkers showed that intravenously injected Candida DNA is detectable in rabbit serum for hours, which argues against a rapid degradation of foreign DNA in the blood (3).
The form that circulates has a consequence for the DNA isolation method used for PCR. Our results showed that the method normally used for pretreatment of fungal mycelium in order to release DNA resulted in the loss of free DNA after pretreatment of PCR-positive culture filtrate samples. Furthermore, many DNA extraction methods result in decanting of free DNA. This could explain the fact that especially antifungal therapy, which probably releases much free DNA, is often combined with negative PCR results (7, 22, 23). In contrast, Kami et al. used an extraction method that includes free DNA and found no negative PCRs in sera from patients receiving antifungal therapy (18). In conclusion, two extraction methods should be used on EDTA whole-blood samples, one setup to isolate free DNA and another to isolate DNA from fungal cells, in order to include all possible forms of circulating DNA.
During an early stage of fungal infection, detection of circulating galf antigens seems to be superior to that of PCR and BG because of much higher amounts that are released. However, other factors play a role, for instance, the level of host defense, the ease by which the antigen reaches the circulation, and its circulating half-lives. Many studies have been performed to compare the different diagnostic methods for detection of circulating markers. While some studies show that the PA ELISA was more frequently positive (2, 8, 47, 54), other studies seem to be in favor of DNA detection (6, 18). The combination of PCR with the PA ELISA should improve the diagnosis of IA, as also suggested by other authors (4, 7, 8, 47). Only a limited number of comparative studies have been performed with the BG test (18, 19, 42). In a prospective comparison between the diagnostic potentials of real-time PCR, the PA ELISA, and the BG test for IA in hematological patients, the PA ELISA was the most sensitive, using a reduced cutoff of 0.6 ng/ml GM (19). Another study retrospectively compared the BG test with the PA ELISA and showed improved specificity when the two tests were combined (42). However, the usefulness of the BG test for early diagnosis of IA remains to be determined.
This study suggests that during invasive infection, the conditions at the infection site may determine the actual amounts of galf antigens, DNA, and BG that are released. Further studies will be focused on in vitro experiments with nutrients that mimic the clinical situation, for instance, lung proteins, and in vivo (animal) experiments in order to determine the conditions at the infection site and their impact on the release of surrogate markers.
Acknowledgments
We thank Marc Tabouret (Bio-Rad, Steenvoorde, France) for kindly providing the purified GM.
REFERENCES
- 1.Baars, J. J. P., H. J. M. Op den Camp, J. M. H. Hermans, V. Mikês, C. van der Drift, L. J. L. D. Van Griensven, and G. D. Vogels. 1994. Nitrogen assimilating enzymes in the white button mushroom Agaricus bisporus. Microbiology 140:1161-1168. [Google Scholar]
- 2.Becker, M. J., S. de Marie, D. Willemse, H. A. Verbrugh, and I. A. Bakker-Woudenberg. 2000. Quantitative galactomannan detection is superior to PCR in diagnosing and monitoring invasive pulmonary aspergillosis in an experimental rat model. J. Clin. Microbiol. 38:1434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bougnoux, M., C. Dupont, J. Mateo, P. Saulnier, V. Faivre, D. Payen, and M. Nicolas-Chanoine. 1999. Serum is more suitable than whole blood for diagnosis of systemic candidiasis by nested PCR. J. Clin. Microbiol. 37:925-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretagne, S., J. M. Costa, E. Bart-Delabesse, N. Dhedin, C. Rieux, and C. Cordonnier. 1998. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407-1412. [DOI] [PubMed] [Google Scholar]
- 5.Buchheidt, D., M. Hummel, D. Schleiermacher, B. Spiess, and R. Hehlmann. 2004. Current molecular diagnostic approaches to systemic infections with aspergillus species in patients with hematological malignancies. Leuk. Lymphoma 45:463-468. [DOI] [PubMed] [Google Scholar]
- 6.Buchheidt, D., M. Hummel, D. Schleiermacher, B. Spiess, R. Schwerdtfeger, O. A. Cornely, S. Wilhelm, S. Reuter, W. Kern, T. Sudhoff, H. Morz, and R. Hehlmann. 2004. Prospective clinical evaluation of a LightCycler-mediated polymerase chain reaction assay, a nested-PCR assay and a galactomannan enzyme-linked immunosorbent assay for detection of invasive aspergillosis in neutropenic cancer patients and haematological stem cell transplant recipients. Br. J. Haematol. 125:196-202. [DOI] [PubMed] [Google Scholar]
- 7.Challier, S., S. Boyer, E. Abachin, and P. Berche. 2004. Development of a serum-based Taqman real-time PCR assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 42:844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa, C., J. M. Costa, C. Desterke, F. Botterel, C. Cordonnier, and S. Bretagne. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa, C., D. Vidaud, M. Olivi, E. Bart-Delabesse, M. Vidaud, and S. Bretagne. 2001. Development of two real-time quantitative TaqMan PCR assays to detect circulating Aspergillus fumigatus DNA in serum. J. Microbiol. Methods 44:263-269. [DOI] [PubMed] [Google Scholar]
- 10.Cousin, M. A., S. Notermans, P. Hoogerhout, and J. H. Van Boom. 1989. Detection of beta-galactofuranosidase production by Penicillium and Aspergillus species using 4-nitrophenyl beta-d-galactofuranoside. J. Appl. Bacteriol. 66:311-317. [DOI] [PubMed] [Google Scholar]
- 11.Daley, L. S., and G. A. Strobel. 1983. β-Galactofuranosidase activity in Helminthosporium sacchari and its relationship to the production of helminthosporoside. Plant Sci. Lett. 30:145-154. [Google Scholar]
- 12.Denning, D. W. 2000. Early diagnosis of invasive aspergillosis. Lancet 355:423-424. [DOI] [PubMed] [Google Scholar]
- 13.Denning, D. W. 2004. Aspergillosis in “nonimmunocompromised” critically ill patients. Am. J. Respir. Crit. Care Med. 170:580-581. [DOI] [PubMed] [Google Scholar]
- 14.Fontaine, T., R. P. Hartland, A. Beauvais, M. Diaquin, and J. P. Latge. 1997. Purification and characterization of an endo-1,3-beta-glucanase from Aspergillus fumigatus. Eur. J. Biochem. 243:315-321. [DOI] [PubMed] [Google Scholar]
- 15.Fontaine, T., R. P. Hartland, M. Diaquin, C. Simenel, and J. P. Latge. 1997. Differential patterns of activity displayed by two exo-beta-1,3-glucanases associated with the Aspergillus fumigatus cell wall. J. Bacteriol. 179:3154-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hope, W., T. Walsh, and D. Denning. 2005. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 5:609-622. [DOI] [PubMed] [Google Scholar]
- 17.Huth, J., S. Werner, and H. G. Muller. 1994. The proton extrusion of growing yeast cultures as an on-line parameter in fermentation processes. J. Basic Microbiol. 30:561-567. [Google Scholar]
- 18.Kami, M., T. Fukui, S. Ogawa, Y. Kazuyama, U. Machida, Y. Tanaka, Y. Kanda, T. Kashima, Y. Yamazaki, T. Hamaki, S. Mori, H. Akiyama, Y. Mutou, H. Sakamaki, K. Osumi, S. Kimura, and H. Hirai. 2001. Use of real-time PCR on blood samples for diagnosis of invasive aspergillosis. Clin. Infect. Dis. 33:1504-1512. [DOI] [PubMed] [Google Scholar]
- 19.Kawazu, M., Y. Kanda, Y. Nannya, K. Aoki, M. Kurokawa, S. Chiba, T. Motokura, H. Hirai, and S. Ogawa. 2004. Prospective comparison of the diagnostic potential of real-time PCR, double-sandwich enzyme-linked immunosorbent assay for galactomannan, and a (1→3)-β-d-glucan test in weekly screening for invasive aspergillosis in patients with hematological disorders. J. Clin. Microbiol. 42:2733-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klont, R. R., M. A. Mennink-Kersten, and P. E. Verweij. 2003. In vitro model for studying the release of Aspergillus galactomannan. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-1016. American Society for Microbiology, Washington, D.C.
- 21.Kubicek, C. P., C. F. B. Witteveen, and J. Visser. 1994. Regulation of organic acid production by aspergilli, p. 135-145. In K. A. Powell, A. Renwick, and J. F. Peberdy (ed.), The genus Aspergillus. Plenum, New York, N.Y.
- 22.Lass-Florl, C., J. Aigner, E. Gunsilius, A. Petzer, D. Nachbaur, G. Gastl, H. Einsele, J. Loffler, M. P. Dierich, and R. Wurzner. 2001. Screening for Aspergillus spp. using polymerase chain reaction of whole blood samples from patients with haematological malignancies. Br. J. Haematol. 113:180-184. [DOI] [PubMed] [Google Scholar]
- 23.Lass-Florl, C., E. Gunsilius, G. Gastl, H. Bonatti, M. C. Freund, A. Gschwendtner, G. Kropshofer, M. P. Dierich, and A. Petzer. 2004. Diagnosing invasive aspergillosis during antifungal therapy by PCR analysis of blood samples. J. Clin. Microbiol. 42:4154-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latgé J. P., H. Kobayashi, J. P. Debeaupuis, M. Diaquin, J. Sarfati, J. M. Wieruszeski, E. Parra, J. P. Bouchara, and B. Fournet. 1994. Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 62:5424-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leitao, E. A., V. C. Bittencourt, R. M. Haido, A. P. Valente, J. Peter-Katalinic, M. Letzel, L. M. de Souza, and E. Barreto-Bergter. 2003. Beta-galactofuranose-containing O-linked oligosaccharides present in the cell wall peptidogalactomannan of Aspergillus fumigatus contain immunodominant epitopes. Glycobiology 13:681-692. [DOI] [PubMed] [Google Scholar]
- 26.Loeffler, J., H. Hebart, U. Brauchle, U. Schumacher, and H. Einsele. 2000. Comparison between plasma and whole blood specimens for detection of Aspergillus DNA by PCR. J. Clin. Microbiol. 38:3830-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maertens, J., J. Van Eldere, J. Verhaegen, E. Verbeken, J. Verschakelen, and M. Boogaerts. 2002. Use of circulating galactomannan screening for early diagnosis of invasive aspergillosis in allogeneic stem cell transplant recipients. J. Infect. Dis. 186:1297-1306. [DOI] [PubMed] [Google Scholar]
- 28.Maertens, J., J. Verhaegen, K. Lagrou, J. Van Eldere, and M. Boogaerts. 2001. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: a prospective validation. Blood 97:1604-1610. [DOI] [PubMed] [Google Scholar]
- 29.Marino, K., C. Lima, S. Maldonado, C. Marino, and R. M. de Lederkremer. 2002. Influence of exo beta-d-galactofuranosidase inhibitors in cultures of Penicillium fellutanum and modifications in hyphal cell structure. Carbohydr. Res. 337:891-897. [DOI] [PubMed] [Google Scholar]
- 30.Melchers, W. J., P. E. Verweij, H. P. van den, A. van Belkum, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1994. General primer-mediated PCR for detection of Aspergillus species. J. Clin. Microbiol. 32:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mennink-Kersten, M. A., J. P. Donnelly, and P. E. Verweij. 2004. Detection of circulating galactomannan for the diagnosis and management of invasive aspergillosis. Lancet Infect. Dis. 4:349-357. [DOI] [PubMed] [Google Scholar]
- 32.Miletti, L. C., C. Marino, K. Marino, R. M. de Lederkremer, W. Colli, and M. J. Alves. 1999. Immobilized 4-aminophenyl 1-thio-beta-d-galactofuranoside as a matrix for affinity purification of an exo-beta-d-galactofuranosidase. Carbohydr. Res. 320:176-182. [DOI] [PubMed] [Google Scholar]
- 33.Miyazaki, T., S. Kohno, K. Mitsutake, S. Maesaki, K. Tanaka, and K. Hara. 1995. (1→3)-beta-d-glucan in culture fluid of fungi activates factor G, a limulus coagulation factor. J. Clin. Lab. Anal. 9:334-339.8531015 [Google Scholar]
- 34.Morelle, W., M. Bernard, J. P. Debeaupuis, M. Buitrago, M. Tabouret, and J. P. Latgé. 2005. Galactomannoproteins of Aspergillus fumigatus. Eukaryot. Cell 4:1308-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obayashi, T., M. Yoshida, T. Mori, H. Goto, A. Yasuoka, H. Iwasaki, H. Teshima, S. Kohno, A. Horiuchi, A. Ito, et al. 1995. Plasma (1→3)-beta-d-glucan measurement in diagnosis of invasive deep mycosis and fungal febrile episodes. Lancet 345:17-20. [DOI] [PubMed] [Google Scholar]
- 36.Obayashi, T., M. Yoshida, H. Tamura, J. Aketagawa, S. Tanaka, and T. Kawai. 1992. Determination of plasma (1→3)-beta-d-glucan: a new diagnostic aid to deep mycosis. J. Med. Vet. Mycol. 30:275-280. [DOI] [PubMed] [Google Scholar]
- 37.Odabasi, Z., G. Mattiuzzi, E. Estey, H. Kantarjian, F. Saeki, R. J. Ridge, P. A. Ketchum, M. A. Finkelman, J. H. Rex, and L. Ostrosky-Zeichner. 2004. Beta-d-glucan as a diagnostic adjunct for invasive fungal infections: validation, cutoff development, and performance in patients with acute myelogenous leukemia and myelodysplastic syndrome. Clin. Infect. Dis. 39:199-205. [DOI] [PubMed] [Google Scholar]
- 38.Ostrosky-Zeichner, L., B. D. Alexander, D. H. Kett, J. Vazquez, P. G. Pappas, F. Saeki, P. A. Ketchum, J. Wingard, R. Schiff, H. Tamura, M. A. Finkelman, and J. H. Rex. 2005. Multicenter clinical evaluation of the (1→3) beta-d-glucan assay as an aid to diagnosis of fungal infections in humans. Clin. Infect. Dis. 41:654-659. [DOI] [PubMed] [Google Scholar]
- 39.Pan, F., M. Jackson, Y. Ma, and M. McNeil. 2001. Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183:3991-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, Y. I., M. L. Buszko, and J. E. Gander. 1997. Utilization of phosphocholine from extracellular complex polysaccharide as a source of cytoplasmic choline derivatives in Penicillium fellutanum. J. Bacteriol. 179:1186-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patterson, T. F., W. R. Kirkpatrick, M. White, J. W. Hiemenz, J. R. Wingard, B. Dupont, M. G. Rinaldi, D. A. Stevens, and J. R. Graybill. 2000. Invasive aspergillosis. Disease spectrum, treatment practices, and outcomes. I3 Aspergillus Study Group. Medicine (Baltimore) 79:250-260. [DOI] [PubMed] [Google Scholar]
- 42.Pazos, C., J. Pontón, and A. Del Palacio. 2005. Contribution of (1→3)-β-d-glucan chromogenic assay to diagnosis and therapeutic monitoring of invasive aspergillosis in neutropenic adult patients: a comparison with serial screening for circulating galactomannan. J. Clin. Microbiol. 43:299-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reiss, E., T. Obayashi, K. Orle, M. Yoshida, and R. M. Zancope-Oliveira. 2000. Non-culture based diagnostic tests for mycotic infections. Med. Mycol. 38(Suppl. 1):147-159. [PubMed] [Google Scholar]
- 44.Rietschel-Berst, M., N. H. Jentoft, P. D. Rick, C. Pletcher, F. Fang, and J. E. Gander. 1977. Extracellular exo-beta-galactofuranosidase from Penicillium charlesii: isolation, purification, and properties. J. Biol. Chem. 252:3219-3226. [PubMed] [Google Scholar]
- 45.Ruhnke, M., and G. Maschmeyer. 2002. Management of mycoses in patients with hematologic disease and cancer-review of the literature. Eur. J. Med. Res. 7:227-235. [PubMed] [Google Scholar]
- 46.Sanders, D. A., A. G. Staines, S. A. McMahon, M. R. McNeil, C. Whitfield, and J. H. Naismith. 2001. UDP-galactopyranose mutase has a novel structure and mechanism. Nat. Struct. Biol. 8:858-863. [DOI] [PubMed] [Google Scholar]
- 47.Sanguinetti, M., B. Posteraro, L. Pagano, G. Pagliari, L. Fianchi, L. Mele, M. La Sorda, A. Franco, and G. Fadda. 2003. Comparison of real-time PCR, conventional PCR, and galactomannan antigen detection by enzyme-linked immunosorbent assay using bronchoalveolar lavage fluid samples from hematology patients for diagnosis of invasive pulmonary aspergillosis. J. Clin. Microbiol. 41:3922-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sarfati, J., D. G. Boucias, and J. P. Latgé. 1995. Antigens of Aspergillus fumigatus produced in vivo. J. Med. Vet. Mycol. 33:9-14. [PubMed] [Google Scholar]
- 49.Steinbach, W. J., D. A. Stevens, D. W. Denning, and R. B. Moss. 2003. Advances against aspergillosis. Clin. Infect. Dis. 37(Suppl. 3):S155—S156. [DOI] [PubMed] [Google Scholar]
- 50.Stynen, D., A. Goris, J. Sarfati, and J. P. Latgé. 1995. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J. Clin. Microbiol. 33:497-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stynen, D., J. Sarfati, A. Goris, M. C. Prevost, M. Lesourd, H. Kamphuis, V. Darras, and J. P. Latgé. 1992. Rat monoclonal antibodies against Aspergillus galactomannan. Infect. Immun. 60:2237-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki, E., M. S. Toledo, H. K. Takahashi, and A. H. Straus. 1997. A monoclonal antibody directed to terminal residue of beta-galactofuranose of a glycolipid antigen isolated from Paracoccidioides brasiliensis: cross-reactivity with Leishmania major and Trypanosoma cruzi. Glycobiology 7:463-468. [DOI] [PubMed] [Google Scholar]
- 53.Van Bruggen-Van Der Lugt, A. W., H. J. Kamphuis, G. A. De Ruiter, P. Mischnick, J. H. Van Boom, and F. M. Rombouts. 1992. New structural features of the antigenic extracellular polysaccharides of Penicillium and Aspergillus species revealed with exo-β-d-galactofuranosidase. J. Bacteriol. 174:6096-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verweij, P. E., J. P. Latgé, A. J. Rijs, W. J. Melchers, B. E. De Pauw, J. A. Hoogkamp-Korstanje, and J. F. Meis. 1995. Comparison of antigen detection and PCR assay using bronchoalveolar lavage fluid for diagnosing invasive pulmonary aspergillosis in patients receiving treatment for hematological malignancies. J. Clin. Microbiol. 33:3150-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verweij, P. E., C. M. Weemaes, J. H. A. J. Curfs, S. Bretagne, and J. F. G. M. Meis. 2000. Failure to detect circulating Aspergillus markers in a patient with chronic granulomatous disease and invasive aspergillosis. J. Clin. Microbiol. 38:3900-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wallis, G. L., F. W. Hemming, and J. F. Peberdy. 2001. An extracellular beta-galactofuranosidase from Aspergillus niger and its use as a tool for glycoconjugate analysis. Biochim. Biophys. Acta 1525:19-28.11342249 [Google Scholar]
- 57.Wallis, G. L., R. J. Swift, R. Atterbury, S. Trappe, U. Rinas, F. W. Hemming, M. G. Wiebe, A. P. Trinci, and J. F. Peberdy. 2001. The effect of pH on glucoamylase production, glycosylation and chemostat evolution of Aspergillus niger. Biochim. Biophys. Acta 1527:112-122. [DOI] [PubMed] [Google Scholar]