Abstract
Liver and plasma hepatitis C virus (HCV) variability was compared by E2 cloning and sequencing in three patients coinfected with HCV and human immunodeficiency virus (HIV) before and after interferon treatment and in three patients solely infected with HCV. The plasma and liver samples contained unique sequences. In the patients coinfected with HIV, accumulated random mutations produced mostly nonsynonymous substitutions in contrast to the reduced HCV genetic variability seen after treatment.
The course of hepatitis C virus (HCV) infection is modified during coinfection with human immunodeficiency virus (HIV), undergoing enhanced viral replication and accelerated progression to cirrhosis (4, 15, 20, 25). In addition, a sustained virological response occurs in only 25% of the patients coinfected with HCV and HIV that are treated with interferon-ribavarin. HCV has a quasispecies distribution (9, 21, 23) that is best studied in the E2 envelope hypervariable region 1 (HVR1) (14) and is occasionally described as predictive of a favorable outcome (5, 7, 8, 14, 17). However, little is yet known about HCV quasispecies in patients coinfected with HIV (2, 15).
Our study focused on liver samples in three nonresponding patients included in a clinical trial (22) in order (i) to describe liver and plasma HCV variability in patients coinfected with HCV and HIV and in patients solely infected with HCV at baseline and (ii) to compare hepatic and plasmatic quasispecies in patients coinfected with HCV and HIV before and 6 months after completion of anti-HCV treatment.
The three patients coinfected with HIV (P1, P2, and P3) had TCD4 lymphocytes at >250/mm3, undetectable HIV RNA, and chronic HCV hepatitis. Patients P1 and P2 were infected with HCV genotype 3, and patient P3 was infected with HCV genotype 1b. The three HIV-negative genotype 1b-infected patients (P4, P5, and P6) had not received any anti-HCV treatment for several months.
After RNA extraction from plasma and liver (22), a 325-bp fragment encompassing HVR1 region was amplified (11) and cloned (pGEM-T Easy Vector System I; Promega). For the three patients coinfected with HIV 222 clones were evaluated (mean, 18.5 per sample), and for the three HIV-negative patients 95 clones were evaluated. The sequences (CEQ2000; Beckman Coulter) were aligned (CLUSTAL W 1.74), and phylogenetic trees were constructed by the neighbor-joining method. GenBank accession numbers for the original nucleotide sequences presented here are recorded as AY793020 to AY793336.
The quasispecies complexity was calculated by using normalized Shannon entropy (Sn) (24). Diversity was analyzed for (i) the mean genetic distance (d, i.e., the number of nucleotide differences divided by total number of nucleotides) and (ii) synonymous substitutions (dS) and nonsynonymous substitutions (dN). The data were by using t test results (paired t tests or Mann-Whitney nonparametric tests), and correlations were investigated by using Prism 2.01 software.
Table 1 shows detailed complexity and diversity results before treatment for the six patients (baseline) and after interferon treatment for the three patients coinfected with HIV (posttreatment). Before treatment, HCV quasispecies displayed no specific complexity or diversity pattern related to sample types (plasma or liver) or patients characteristics. Table 2 presents the statistical parameters used in this study. Complexity and diversity were significantly correlated. In the three patients coinfected with HIV, synonymous substitutions were the most frequent at baseline. Other researchers previously described a higher diversity in severely immunocompromised patients coinfected with HIV (19) or in patients with end-stage liver disease (1), which was not the case in our patients.
TABLE 1.
Complexity and diversity of the entire sequence encompassing HVR1 for patients P1 to P6
| Patient | Value | Complexity
|
Diversity
|
||
|---|---|---|---|---|---|
| Sn nta | Sn aab | dc | dN/dSd | ||
| P1 | Plasma baseline | 0.975 | 0.514 | 0.026 | 0.309 |
| Liver baseline | 0.889 | 0.337 | 0.011 | 0.053 | |
| Plasma posttreatment | 0.207 | 0.139 | 0.002 | 0.333 | |
| Liver posttreatment | 0.539 | 0.442 | 0.004 | 1.25 | |
| P2 | Plasma baseline | 0.657 | 0.453 | 0.008 | 0.375 |
| Liver baseline | 0.739 | 0.53 | 0.01 | 0.273 | |
| Plasma posttreatment | 0.783 | 0.457 | 0.01 | 0.392 | |
| Liver posttreatment | 0.38 | 0.233 | 0.003 | 0.286 | |
| P3 | Plasma baseline | 0.959 | 0.821 | 0.033 | 0.448 |
| Liver baseline | 0.714 | 0.679 | 0.031 | 0.444 | |
| Plasma posttreatment | 1 | 0.758 | 0.015 | 0.229 | |
| Liver posttreatment | 0.256 | 0.168 | 0.002 | 1 | |
| P4 | Plasma baseline | 0.54 | 0.127 | 0.003 | >1 |
| Liver baseline | 0.722 | 0.616 | 0.007 | 0.555 | |
| P5 | Plasma baseline | 0.958 | 0.792 | 0.022 | 0.576 |
| Liver baseline | 1 | 0.787 | 0.021 | 0.6 | |
| P6 | Plasma baseline | 0.937 | 0.769 | 0.022 | 2.25 |
| Liver baseline | 0.622 | 0.564 | 0.015 | 1.42 | |
Shannon entropy (Sn) at the nucleotide level (nt).
Shannon entropy (Sn) at the amino acid level (aa).
Genetic distance.
dN/dS ratio is considered as a measure of immune pressure compared to genetic drifts when >1 (18) and appears in boldface.
TABLE 2.
Statistical results obtained from comparison of the complexity and diversity data in patients solely infected with HCV or coinfected with HIV and HCV
| Parameter | Subset | P and/or r2 |
|---|---|---|
| HVR1 complexity > flanking regions complexity | P = 0.04 | |
| Correlation between nucleotide and amino acid complexity | In patients solely infected with HCV | r2 = 0.6648 |
| In patients coinfected with HIV | r2 = 0.7819 | |
| In both groups of patients | r2 = 0.68 | |
| Diversity d at baseline > d posttreatment in patients coinfected with HIV | In plasma | P < 0.02 |
| In liver | P < 0.02 | |
| In both compartments | P < 0.02 | |
| Correlation between complexity (Sn) and diversity (d) in patients coinfected with HIV | Nucleotide entropy (Sn) | r2 = 0.5021; P = 0.0099 |
| Amino acid entropy (Sn) | r2 = 0.6506; P = 0.0015 |
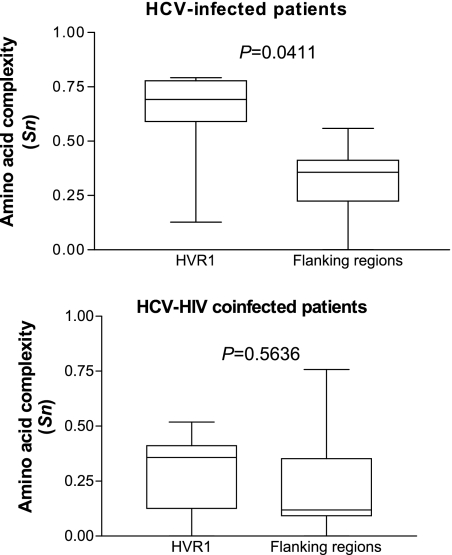
In patients solely infected with HCV, the complexity was significantly higher in HVR1 than in flanking regions (Fig. 1), whereas no difference appeared in patients coinfected with HIV. Since HVR1 is known to harbor both neutralizing and cytotoxic T epitopes, this absence of specific complexity pattern in HVR1 suggests a weak immune pressure, if any, which could result from HIV-related immune deficiency. In these patients, HVR1 quasispecies evolution should therefore more likely be due to a high rate of accumulation of random mutations than to a positive selection pressure.
FIG. 1.
Comparison of amino acid complexity expressed with Shannon entropy (Sn), between HVR1 and in the flanking regions in HCV-solely infected and HCV-HIV coinfected patients. Mann-Whitney test.
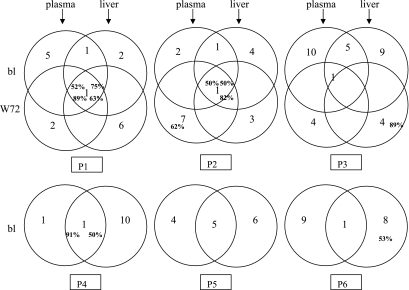
On the phylogenetic trees (data not shown), each patient's sequences clustered independently, thus excluding cross-contamination. The viral variant distribution is presented in Fig. 2. There was not necessarily a dominant variant in each compartment at each time, but each compartment harbored specific variants, as already described (12, 16). For HIV-infected patients P1 and P2, one common dominant variant was present both at baseline and after treatment completion (50 to 89% of the clones). However, in P2, it represented only 5% of the plasma clones after treatment. For patient P3, only one baseline plasma variant was still present after treatment (<50% of the clones), and the liver harbored one predominant but previously undetected variant (89% of the clones). Of the three patients solely infected with HCV, patient P4 displayed a variant distribution similar to that of patients P1 and P2. Patients P5 and P6 had, respectively, 5 and 1 common variants in the plasma and liver, but none predominated. There was one major liver variant in P6 (53% of the clones).
FIG. 2.
Schematic representation of variant repartition in plasma and liver compartments and at baseline (bl) and posttreatment (W72). The numbers of the different variants are in black. The percentage of a variant is indicated in boldface only when it predominates (i.e., >50%).
Previous studies of HCV variability over time also showed little evolution in plasma complexity in most instances, both in patients coinfected with HIV and in patients solely infected with HCV (2, 13). Important quasispecies differences between blood and liver, correlated with hepatic fibrosis, have been described (3). Other researchers found no significant variation (12).
After treatment completion, liver complexity was notably reduced in the HIV-infected patients (Table 2). Although unsuccessful, interferon treatment seemed more efficient for eliminating minor liver variants than plasma ones (6). The predominant variant persisting over time in two patients coinfected with HIV appeared to be the closest to the common node connecting all the clones in each patient. This variant could be more stable on an evolutionary level, as if best adapted to its host environment (10, 13) or more pathogenic (6).
In patients P1 and P3, nonsynonymous mutations markedly increased in the liver after treatment (dN/dS liver ratio of >1): in addition to viral fitness alteration, a possible immunological reaction targeting the liver during interferon treatment and contributing to the selection of major viral variants cannot be excluded.
Finally, diversity decreased significantly after treatment in all patients coinfected with HIV, both in plasma and in liver, as already described (1). Thus, interferon seemed to favor the emergence of more closely related clones, reducing HCV's ability to diversify its genetic repertoire.
In conclusion, the plasma and liver already harbored different HCV quasispecies before treatment, both in patients solely infected with HCV and in patients coinfected with HIV. After unsuccessful interferon treatment, HCV complexity and diversity were both markedly reduced in the HIV-infected patients, but the liver compartment displayed unique evolutionary features.
Acknowledgments
This study was supported by two grants from the Ensemble Contre le SIDA and the Etablissement Français des Greffes and was conducted with the approval of the Bordeaux Ethical Committee (Comite Consultatif Pour la Protection des Personnes en Recherche Biomedicale, Bordeaux A).
We thank J. F. Moreau for providing the TCD4 lymphocyte count results.
REFERENCES
- 1.Alfonso, V., D. M. Flichman, S. Sookoian, V. A. Mbayed, and R. H. Campos. 2004. Evolutionary study of HVR1 of E2 in chronic hepatitis C virus infection. J. Gen. Virol. 85:39-46. [DOI] [PubMed] [Google Scholar]
- 2.Babik, J. M., and M. Holodniy. 2003. Impact of highly active antiretroviral therapy and immunologic status on hepatitis C virus quasispecies diversity in human immunodeficiency virus/hepatitis C virus-coinfected patients. J. Virol. 77:1940-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cabot, B., M. Martell, J. I. Esteban, S. Sauleda, T. Otero, R. Esteban, J. Guardia, and J. Gomez. 2000. Nucleotide and amino acid complexity of hepatitis C virus quasispecies in serum and liver. J. Virol. 74:805-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cribier, B., C. Schmitt, D. Rey, G. Uhl, J. M. Lang, D. Vetter, A. Kirn, and F. Stoll-Keller. 1997. HIV increases hepatitis C viraemia irrespective of the hepatitis C virus genotype. Res. Virol. 148:267-271. [DOI] [PubMed] [Google Scholar]
- 5.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 6.Gretch, D. R., S. J. Polyak, J. J. Wilson, R. L. Carithers, Jr., J. D. Perkins, and L. Corey. 1996. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J. Virol. 70:7622-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hino, K., Y. Yamaguchi, D. Fujiwara, Y. Katoh, M. Korenaga, M. Okazaki, M. Okuda, and K. Okita. 2000. Hepatitis C virus quasispecies and response to interferon therapy in patients with chronic hepatitis C: a prospective study. J. Viral Hepat. 7:36-42. [DOI] [PubMed] [Google Scholar]
- 8.Le Guen, B., G. Squadrito, B. Nalpas, P. Berthelot, S. Pol, and C. Brechot. 1997. Hepatitis C virus genome complexity correlates with response to interferon therapy: a study in French patients with chronic hepatitis C. Hepatology 25:1250-1254. [DOI] [PubMed] [Google Scholar]
- 9.Martell, M., J. I. Esteban, J. Quer, J. Genesca, A. Weiner, R. Esteban, J. Guardia, and J. Gomez. 1992. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J. Virol. 66:3225-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizokami, M., T. Ohno, K. Ohba, G. L. Davis, K. Suzuki, E. Orito, and J. Y. Lau. 1999. Interferon-alpha therapy exerts selective pressure on hepatitis C virus quasispecies equilibrium. Antivir. Ther. 4:15-19. [PubMed] [Google Scholar]
- 11.Neau, D., A. C. Jouvencel, E. Legrand, P. Trimoulet, T. Galperine, I. Chitty, M. Ventura, B. Le Bail, P. Morlat, J. Y. Lacut, J. M. Ragnaud, M. Dupon, H. Fleury, and M. E. Lafon. 2003. Hepatitis C virus genetic variability in 52 human immunodeficiency virus-coinfected patients. J. Med. Virol. 71:41-48. [DOI] [PubMed] [Google Scholar]
- 12.Okuda, M., K. Hino, M. Korenaga, Y. Yamaguchi, Y. Katoh, and K. Okita. 1999. Differences in hypervariable region 1 quasispecies of hepatitis C virus in human serum, peripheral blood mononuclear cells, and liver. Hepatology 29:217-222. [DOI] [PubMed] [Google Scholar]
- 13.Pawlotsky, J. M., G. Germanidis, P. O. Frainais, M. Bouvier, A. Soulier, M. Pellerin, and D. Dhumeaux. 1999. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J. Virol. 73:6490-6499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pawlotsky, J. M., M. Pellerin, M. Bouvier, F. Roudot-Thoraval, G. Germanidis, A. Bastie, F. Darthuy, J. Remire, C. J. Soussy, and D. Dhumeaux. 1998. Genetic complexity of the hypervariable region 1 (HVR1) of hepatitis C virus (HCV): influence on the characteristics of the infection and responses to interferon alfa therapy in patients with chronic hepatitis C. J. Med. Virol. 54:256-264. [PubMed] [Google Scholar]
- 15.Roque-Afonso, A. M., M. Robain, D. Simoneau, P. Rodriguez-Mathieu, M. Gigou, L. Meyer, and E. Dussaix. 2002. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J. Infect. Dis. 185:728-733. [DOI] [PubMed] [Google Scholar]
- 16.Sakai, A., S. Kaneko, M. Honda, E. Matsushita, and K. Kobayashi. 1999. Quasispecies of hepatitis C virus in serum and in three different parts of the liver of patients with chronic hepatitis. Hepatology 30:556-561. [DOI] [PubMed] [Google Scholar]
- 17.Sandres, K., M. Dubois, C. Pasquier, J. L. Payen, L. Alric, M. Duffaut, J. P. Vinel, J. P. Pascal, J. Puel, and J. Izopet. 2000. Genetic heterogeneity of hypervariable region 1 of the hepatitis C virus (HCV) genome and sensitivity of HCV to alpha interferon therapy. J. Virol. 74:661-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 19.Sherman, K. E., C. Andreatta, J. O'Brien, A. Gutierrez, and R. Harris. 1996. Hepatitis C in human immunodeficiency virus-coinfected patients: increased variability in the hypervariable envelope coding domain. Hepatology 23:688-694. [DOI] [PubMed] [Google Scholar]
- 20.Soto, B., A. Sanchez-Quijano, L. Rodrigo, J. A. del Olmo, M. Garcia-Bengoechea, J. Hernandez-Quero, C. Rey, M. A. Abad, M. Rodriguez, M. Sales Gilabert, F. Gonzalez, P. Miron, A. Caruz, F. Relimpio, R. Torronteras, M. Leal, and E. Lissen. 1997. Human immunodeficiency virus infection modifies the natural history of chronic parenterally-acquired hepatitis C with an unusually rapid progression to cirrhosis. J. Hepatol. 26:1-5. [DOI] [PubMed] [Google Scholar]
- 21.Steinhauer, D. A., E. Domingo, and J. J. Holland. 1992. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 122:281-288. [DOI] [PubMed] [Google Scholar]
- 22.Trimoulet, P., D. Neau, B. Le Bail, A. Rullier, M. Winnock, T. Galperine, E. Legrand, E. Schvoerer, M. Dupon, J. M. Ragnaud, P. Bioulac-Sage, G. Chene, H. Fleury, and M. E. Lafon. 2002. Intrahepatic HCV RNA loads in 37 HIV-HCV coinfected patients with controlled HIV infection. J. Med. Virol. 67:143-151. [DOI] [PubMed] [Google Scholar]
- 23.Weiner, A. J., H. M. Geysen, C. Christopherson, J. E. Hall, T. J. Mason, G. Saracco, F. Bonino, K. Crawford, C. D. Marion, K. A. Crawford, et al. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. USA 89:3468-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 25.Zylberberg, H., and S. Pol. 1996. Reciprocal interactions between human immunodeficiency virus and hepatitis C virus infections. Clin. Infect. Dis. 23:1117-1125. [DOI] [PubMed] [Google Scholar]