Abstract
Campylobacter curvus was isolated from blood cultures of a patient with liver abscesses. Bacterial identification involved Gram staining, biochemical analysis, gas-liquid chromatography, and 16S rRNA sequencing. The difficulty in isolation, identification, and growth of the species confirms previous work that these organisms may be overlooked by conventional detection methods.
CASE REPORT
A52-year-old carpenter with a history of smoking and diverticulitis, but otherwise generally healthy, presented with general malaise and pyrexia. A chest X-ray image revealed minor changes suggestive of pneumonia, and the patient was treated with intravenous cefuroxime. He improved and was discharged.
Three weeks later, he returned with pyrexia (temperature, 39.5°C), night sweats, anorexia, and a 30-lb weight loss. The abdominal exam was unremarkable. Investigations revealed a white blood cell count of 27,000. The chest X-ray image showed some minor changes consistent with chronic obstructive pulmonary disease. Ultrasound of the abdomen and pelvis revealed two lesions on his liver, which may have been cysts or abscesses. Empirically, he was started on ceftriaxone (2 g, given intravenously once daily) and metronidazole (500 mg, given perorally thrice daily). He improved within 48 h and went on to make an uneventful recovery.
At this time the patient was referred to our center, from a rural hospital, because of the diagnostic dilemma, as well as the two lesions on his liver detected by ultrasound. A computed tomography scan could not be performed because of the patient's allergy to the contrast media; however, a repeat ultrasound showed two complex masses within the right lobe of the liver consistent with abscesses. Aspirates were obtained and sent for analysis. Pathology showed “necro-inflammatory” material. Aerobic and anaerobic cultures of the aspirated material were negative. One anaerobic blood culture demonstrated growth of curved gram-negative rods. The organism grew on both tryptic soy and brain heart infusion agars but failed to grow on chocolate agar.
The organism was subjected to Gram straining and oil immersion phase-contrast microscopy, which indicated it to be a gram-negative curved rod. The unknown bacterial strain was grown at 37°C on brain heart infusion (Becton Dickinson, Mississauga, Ontario, Canada) agar (Invitrogen, Burlington, Ontario, Canada) plates with 5% defibrinated sheep blood (Dalynn Biologicals, Calgary, Alberta, Canada) only under strict anaerobic conditions. Three days of uninterrupted growth was required to produce a lawn of confluent growth.
Routine biochemical tests (Table 1) were used to compare the organism with Helicobacter pylori strain J99 and Campylobacter jejuni strain NCTC 11168. Indoxyl acetate hydrolysis was done by using the method described by Popovic-Uroic et al. (19) and commercially prepared disks (American Type Culture Collection [ATCC], Rockville, MD). Nitrate reduction, catalase, and urease tests were all done by using previously described methods (2, 10, 14). Oxidase tests were performed with 1% tetramethyl-p-phenylenediamine (TMPD; Sigma, Oakville, Ontario, Canada) (10). The motility of the organism was assessed by using the agar method (Dalynn Biologicals) (14).
TABLE 1.
Results of biochemical tests of patient isolate
| Test | Actual result
|
Expected resulta
|
||||
|---|---|---|---|---|---|---|
| Unknown | NCTC 11168 | J99 | C. curvusd | C. jejuni | H. pylori | |
| Nitrate reduction | + | + | NRc | + | + | Variable |
| Indoxyl acetateb | + (weak) | + | − | + | + | − |
| Catalase | − | + | + | − | + | + |
| Urease | − | − | + | − | − | + |
| Oxidase | + | + | + | + | + | + |
| Motility | + | + | + | + | + | + |
Expected results, except that indoxyl acetate and C. curvus results were taken from Biochemical Tests for Identification of Medical Bacteria (14).
The indoxyl acetate expected results were obtained from Popovic-Uroic et al. (19).
NR, no result was recorded after several attempts since bacteria would not grow in media.
The expected results for this species were taken from Bergey's Manual of Determinative Bacteriology (8). Wolinella succinogenes results were identical in all tests except for the indoxyl acetate test, in which the result was negative.
A RapID ANA II Code Compendium identified two possibilities for the unknown organism, Wolinella succinogenes (48.5% probability) or “Bacillus gracilis” (51% probability). Gas and liquid chromatographic analysis (data not shown) revealed a succinic acid peak. This peak is characteristic of Wolinella spp., as well as certain other ɛ-proteobacteria that are capable of reducing fumarate (as a terminal electron acceptor) to succinate in the presence of formate or hydrogen (11, 18). Wolinella sp., a commensal organism related to Campylobacter spp. (25), is generally found in the gastrointestinal tract of cattle and had never been identified as a human pathogen. The positive motility test definitively ruled out “B. gracilis,” since it is nonmotile. In contrast, W. succinogenes is motile, further corroborating the identification and placing the probability of this being the correct species at 98%. Subsequently, 16S rRNA sequencing was performed to confirm the identity of the strain.
The first 600 bp of the unknown gene sequence were compared to the GenBank data (National Center for Biotechnology Information, National Institutes of Health). The sequence was a 100% match to C. curvus strain LMG 11034 (GenBank accession AF550650) (5). The previously described biochemical tests were all consistent with C. curvus.
The MICs for several antibiotics were determined by using the Etest method (Epsilometer gradient agar diffusion test strips; AB Biodisk, Solna, Sweden) according to manufacturer's directions. Antibiotic susceptibility testing was done as follows (MICs in parentheses): clarithromycin (3 μg/ml), erythromycin (2 μg/ml), cefuroxime (>256 μg/ml), ceftriaxone (>32 μg/ml), metronidazole (0.38 μg/ml), amoxicillin-clavulanic acid (>256 μg/ml), imipenem (>32 μg/ml), quinupristin (>32 μg/ml), nalidixic acid (>256 μg/ml), and ciprofloxacin (0.047 μg/ml). Nalidixic acid resistance is characteristic of C. curvus (8). To confirm the differences in MIC for ciprofloxacin and nalidixic acid, these were repeated by using agar dilution testing, which gave similar results (a ciproflaxicin MIC of 0.125 μg/ml and a nalidixic acid MIC of 128 μg/ml). These MICs are similar to those obtained for the Campylobacter fetus subsp. fetus type strain ATCC 27374 (23), suggesting that the resistance profiles for quinolone antibiotics in wild-type C. curvus and C. fetus subsp. fetus may be similar.
Agarose-embedded chromosomal DNA preparation, restriction endonuclease digestion, and pulsed-field gel electrophoresis (PFGE) were performed essentially according to the protocol described by Ribot et al. (20). The unknown isolate was digested with 20 U each of SmaI (10 U/μl; Roche Applied Science, Laval, Quebec, Canada), SnaBI (5 U/μl; New England Biolabs, Inc. [NEB], Mississauga, Ontario, Canada), RsrII (4 U/μl; NEB), or PmeI (10 U/μl; NEB). The size reference strain, Salmonella choleraesuis subsp. choleraesuis serotype Braenderup H9812, was digested with XbaI (20 U/μl; NEB). A 50- to 1,000-kb Lambda Ladder PFG Marker (N0340S; NEB) was included on the gel as standard. DNA fragments were separated by using a CHEF-DR II PFGE system (Bio-Rad Laboratories, Hercules, Calif.). Images were captured by using a Gel Logic 200 Imaging System (Kodak, New Haven, Conn.) and were saved as 8-bit no scaling tagged image file format (tif) images for transfer to a BioNumerics database (Bionumerics Version 3.5; Applied Maths, Austin, Tex.). All lanes were normalized to the lambda ladder PFG marker prior to base pair analysis. The PFGE profile after PmeI digestion was useful for the estimation of genome size of the C. curvus isolate (Fig. 1). The size of C. curvus genome was calculated to be approximately 1.6 Mb (1,555 kb).
FIG. 1.
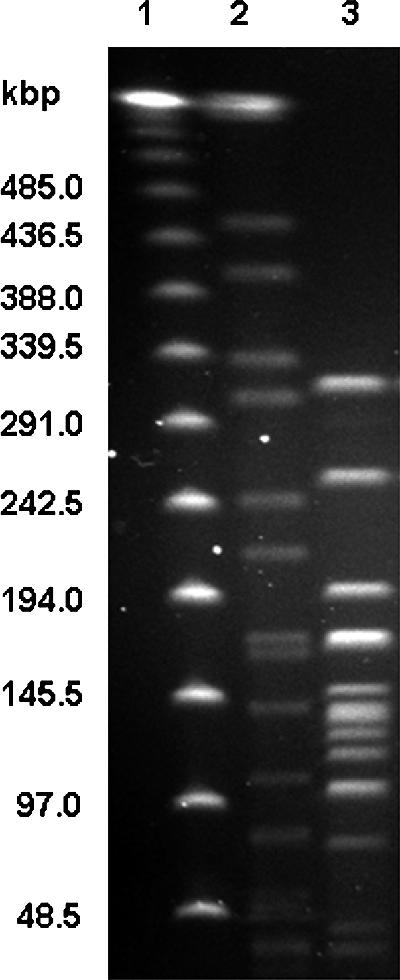
PFGE of the C. curvus sample DNA. Lanes: 1, lambda DNA ladder run as size marker; 2, S. choleraesuis subsp. choleraesuis serotype Braenderup H9812 DNA digested with XbaI (size marker); 3, unknown C. curvus DNA digested with PmeI.
Several attempts were made to isolate plasmid DNA using both midi- and maxikits from QIAGEN (Mississauga, Ontario, Canada). The isolation protocol for plasmid preparation by alkaline lysis with sodium dodecyl sulfate as outlined previously (21) was also attempted. No plasmids were found to be present.
C. curvus was discovered in 1984 and originally classified as Wolinella curva (22) The organism was reclassified as Campylobacter curvus in 1991 based on DNA-DNA hybridization and immunotyping analysis (24). Recently, Han et al. found isolates of C. curvus causing liver abscesses, in conjunction with Streptococcus species, in a lung cancer patient with a postobstructive bronchial abscess, as well as in an ovarian cancer patient with a liver abscess (6).
C. curvus has been found in a wide variety of clinical presentations (1-3), but culture and identification difficulties may be the cause of its under-recognition (16, 17). The species has been isolated from the blood (6) and the oral cavity, although it is noninvasive orally and is not believed to be a causative agent of periodontal disease (7, 15). Although not well documented, the gastrointestinal tract has also been shown previously to be a reservoir for this species (1, 9) and is a far more likely source in this case than the well-documented oral cavity (7, 15, 18).
The genome size for C. curvus has not been published; however, PFGE revealed the genome size for the patient's isolate to be approximately 1.6 Mb. C. jejuni is the only completely sequenced strain of this genus; however, C. coli, C. lari, and C. upsaliensis have all been partially sequenced. Their genome sizes were estimated to be approximately 1.68, 1.5, and 1.66 Mb, respectively (4). Thus, the C. curvus genome is in a size range similar to that of other Campylobacter species.
These findings support other evidence that C. curvus may be more pathogenic than originally believed (1, 6). Selective media for Campylobacter usually contains antibiotics to which species other than C. jejuni, C. coli, and C. lari are susceptible (13). This may explain the difficulties found in culturing and maintaining C. curvus in a laboratory setting. Several species, including C. curvus, do not respond well to conventional isolation methods and, therefore, may remain undetected, particularly if present in conjunction with another Campylobacter species that may be cultured under microaerobic conditions unsuitable for C. curvus (1, 9, 16, 17). Based on our results, this organism is likely being under-reported due to both selective media and unfavorable growth conditions (13). In a few selected cases, higher than usual numbers of C. curvus isolates have been isolated by using filtration methods (1, 12), lending support to the addition of filtration to standard isolation methods. To increase the isolation rate of C. curvus, we recommend a longer incubation period, filtration, and the use of brain heart infusion-5% sheep blood plates (without selection) with incubation under anaerobic conditions. This should be carried out in conjunction with the selective plates that are currently used.
Acknowledgments
We thank Robert White, the family practice physician who collected blood cultures, as well as Kathy Pon and the laboratory staff of the Westlock Healthcare Centre for the isolation of the bacterium. Joanne Simala-Grant and Kelly Baptista are acknowledged for critical reading of the manuscript.
D.E.T. is a scientist with Alberta Heritage Foundation for Medical Research.
REFERENCES
- 1.Abbott, S. L., M. Waddington, D. Lindquist, J. Ware, W. Cheung, J. Ely, and J. M. Janda. 2005. Description of Campylobacter curvus and C. curvus-like strains associated with sporadic episodes of bloody gastroenteritis and Brainerd's diarrhea. J. Clin. Microbiol. 43:585-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altlas, R. M. 2004. Handbook of microbiological media. CRC Press, Boca Raton, Fla.
- 3.Baar, C., M. Eppinger, G. Raddatz, J. Simon, C. Lanz, O. Klimmek, R. Nandakumar, R. Gross, A. Rosinus, H. Keller, P. Jagtap, B. Linke, F. Meyer, H. Lederer, and S. C. Schuster. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. USA 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.GenBank. 2003. GenBank: BX571656. National Center for Biotechnology Information, Bethesda, Md. [Online.] http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=323.
- 6.Han, X. Y., J. J. Tarrand, and D. C. Rice. 2005. Oral Campylobacter species involved in extraoral abscess: a report of three cases. J. Clin. Microbiol. 43:2513-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han, Y. W., W. Shi, G. T. Huang, S. Kinder Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Bergey's manual of determinative bacteriology, 9th ed. The Williams & Wilkins Co., Baltimore, Md.
- 9.Koga, M., N. Yuki, M. Takahashi, K. Saito, and K. Hirata. 1999. Are Campylobacter curvus and Campylobacter upsaliensis antecedent infectious agents in Guillain-Barre and Fisher syndromes? J. Neurol. Sci. 163:53-57. [DOI] [PubMed] [Google Scholar]
- 10.Koneman, E. W., S. D. Allen, W. M. Janda, P. C. Schreckenberger, and W. C. Winn. 1997. Color atlas and textbook of diagnostic microbiology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 11.Lancaster, C. R., and J. Simon. 2002. Succinate:quinone oxidoreductases from epsilon-proteobacteria. Biochim. Biophys. Acta 1553:84-101. [DOI] [PubMed] [Google Scholar]
- 12.Lastovica, A. J., and E. le Roux. 2000. Efficient isolation of campylobacteria from stools. J. Clin. Microbiol. 38:2798-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loades, C. J., L. E. Reiman, and C. W. Keevil. 2005. Abstr. 105th Gen. Meet. Am. Soc. Microbiol. 2005, abstr. D-198, p. 234-235.
- 14.MacFaddin, J. F. 2000. Biochemical tests for identification of medical bacteria, 3rd ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 15.Macuch, P. J., and A. C. Tanner. 2000. Campylobacter species in health, gingivitis, and periodontitis. J. Dent. Res. 79:785-792. [DOI] [PubMed] [Google Scholar]
- 16.Maher, M., C. Finnegan, E. Collins, B. Ward, C. Carroll, and M. Cormican. 2003. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter species in clinical specimens of feces. J. Clin. Microbiol. 41:2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng, L. K., D. E. Taylor, and M. E. Stiles. 1988. Characterization of freshly isolated Campylobacter coli strains and suitability of selective media for their growth. J. Clin. Microbiol. 26:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paster, B. J., and R. J. Gibbons. 1986. Chemotactic response to formate by Campylobacter concisus and its potential role in gingival colonization. Infect. Immun. 52:378-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popovic-Uroic, T., C. M. Patton, M. A. Nicholson, and J. A. Kiehlbauch. 1990. Evaluation of the indoxyl acetate hydrolysis test for rapid differentiation of Campylobacter, Helicobacter, and Wolinella species. J. Clin. Microbiol. 28:2335-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ribot, E. M., C. Fitzgerald, K. Kubota, B. Swaminathan, and T. J. Barrett. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Tanner, A. C. R., M. A. Listgarten, and J. L. Ebersole. 1984. Wolinella curva sp. nov.: “Vibrio succinogenes” of human origin. Int. J. Syst. Bacteriol. 34:275-282. [Google Scholar]
- 23.Taylor, D. E., and A. S. Chau. 1997. Cloning and nucleotide sequence of the gyrA gene from Campylobacter fetus subsp. fetus ATCC 27374 and characterization of ciprofloxacin-resistant laboratory and clinical isolates. Antimicrob. Agents Chemother. 41:665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandamme, P., E. Falsen, R. Rossau, B. Hoste, P. Segers, R. Tytgat, and J. De Ley. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88-103. [DOI] [PubMed] [Google Scholar]
- 25.Wolin, M. J., Wolin, E. A., and M. J. Jacobs. 1961. Cytochrome-producing anaerobic vibrio, Vibrio succinogenes sp. n. J. Bacteriol. 81:911-917. [DOI] [PMC free article] [PubMed] [Google Scholar]


