Coagulase-negative staphylococci (CoNS), and especially Staphylococcus epidermidis, are now considered to be important nosocomial pathogens (7, 10). They usually belong to the human commensal flora and are resistant to multiple antimicrobial agents, including methicillin and other drugs commonly used for the treatment of staphylococcal infections. Given the high frequency of methicillin-resistant isolates, glycopeptide antibiotics have been largely recommended for the empirical treatment of CoNS infections (4).
From January 2000 to December 2004, all nonrepetitive staphylococci isolated and identified in our laboratory were collected and screened for teicoplanin resistance as recommended by the National Breakpoint Committee of the French Society for Microbiology (3). The test consisted of spotting 10 μl of 6 × 108 CFU/ml bacterial suspension (McFarland 2) on Mueller-Hinton agar plates containing 5 μg/ml teicoplanin. Readings were made after 24 h at 37°C. Staphylococcus aureus ATCC 25923 and Staphylococcus haemolyticus CIP 107204 were used as negative and positive controls, respectively. MICs of vancomycin and teicoplanin were determined by the agar dilution method for all strains detected positive with the screen test (8). Classification of strains as susceptible or resistant was performed according to the European consensus (see the EUCAST website [http://www.escmid.org]). A total of 2,476 staphylococci were included in the study, 1,437 S. aureus bacteria and 1,039 CoNS. By using the ID32 Staph gallery (bioMérieux, Marcy l'Etoile, France), the 1,039 CoNS were categorized as 632 (60.8%) S. epidermidis strains, 103 (9.9%) S. haemolyticus, 137 (13.2%) S. hominis, and 167 (16.1%) other CoNS. Within S. epidermidis, 142 (22.5%) strains were isolated from urinary tract infections and 17 (2.7%) were from other nosocomial infections, while 473 (74.8%) were either contaminants of blood cultures or noninvasive colonizers of bladder and peripherically inserted catheters. The vast majority of S. haemolyticus and S. hominis strains (91%) had no clinical significance.
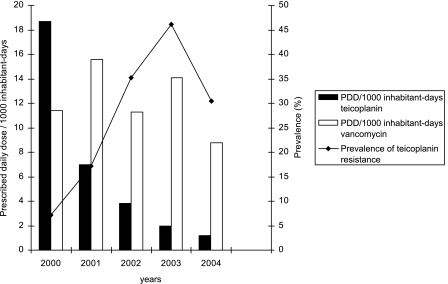
Among 300 S. epidermidis strains resistant to methicillin, 165 (55%) were also resistant to teicoplanin (MIC > 8 μg/ml) (Table 1). Among these, only one strain isolated in 2002 showed a decreased susceptibility to vancomycin (MIC = 8 μg/ml). As observed in other French hospitals (1), there was a very low prevalence of teicoplanin resistance in S. aureus during the time of the study (Table 1). By contrast, the prevalence of teicoplanin resistance among S. epidermidis strains increased from 7.2% in 2000 to 46.1% in 2003 and then subsequently decreased (30.4%) in 2004. Susceptibilities to erythromycin, ciprofloxacin, and gentamicin for S. epidermidis did not vary in the same time period. The pulsed-field gel electrophoresis analysis of 20 S. epidermidis teicoplanin-resistant strains yielded 20 distinct SmaI pulsotypes. The rise of teicoplanin resistance in S. epidermidis was therefore not related to the spread of predominant clones, and this emergence cannot be explained by extensive use of glycopeptides in our hospital. Teicoplanin consumption decreased from 2000 to 2004, whereas vancomycin consumption remained constant (Fig. 1). The trait of teicoplanin resistance mainly occurs among S. epidermidis methicillin-resistant strains (Table 1), and coresistance to ciprofloxacin occurs in 60% of these strains. It is likely that such strains are disseminated to the hospital environment. The extensive use of fluoroquinolones in our hospital could bring local antibiotic pressure, since their consumption was significant during this period (100 prescribed daily doses/1,000 inhabitant days in 2001 and 97 prescribed daily doses/1,000 inhabitant days in 2004). Other recent reports have also pointed out reduced susceptibility to glycopeptides within S. haemolyticus and S. epidermidis in various European countries (2, 5, 6, 9). These alarming data suggest that such strains might be highly disseminated in the community and in hospitals. Large prospective studies are needed to generate a list of risk factors for the emergence of teicoplanin resistance in CoNS isolates.
TABLE 1.
Frequency (%) of teicoplanin resistance (MIC > 8 μg/ml) according to staphylococcal species and to methicillin resistance from January 2000 to December 2004
| Speciesa | Frequency (%) of strains with teicoplanin MICs > 8 μg/ml by year
|
||||
|---|---|---|---|---|---|
| 2000 | 2001 | 2002 | 2003 | 2004 | |
| S. epidermidis | 7.2 | 17.2 | 35.2 | 46.1 | 30.4 |
| TR/MRI | 20 | 33.9 | 71.7 | 84.4 | 60.9 |
| S. haemolyticus | 17.9 | 25 | 55.5 | 31.2 | 35.7 |
| TR/MRI | 31.8 | 40 | 76.9 | 55.5 | 55.5 |
| S. hominis | 9.5 | 7.4 | 21 | 20.9 | 14.8 |
| TR/MRI | 25 | 16.7 | 80 | 33.3 | 44.4 |
| Other CoNS | 4 | 10.3 | 14.6 | 5.9 | 8.7 |
| TR/MRI | 15.4 | 37.5 | 62.5 | 25 | 100 |
| S. aureus | 2.8 | 1.3 | 0.9 | 0.9 | 1.2 |
| TR/MRI | 2.8 | 3.6 | 6.7 | 2.7 | 4.2 |
TR/MRI, teicoplanin resistance among methicillin-resistant isolates.
FIG. 1.
Relationship between the incidence of teicoplanin resistance in S. epidermidis and glycopeptide use in prescribed daily doses (PDD)/1,000 inhabitant days.
REFERENCES
- 1.Cartolano, G. L., M. Cheron, D. Benabid, M. Leneveu, A. Boisivon, and Association of Hospital Bacteriologists, Virologists, and Hygiene Professionals. 2004. Methicillin-resistant Staphylococcus aureus (MRSA) with reduced susceptibility to glycopeptides (GISA) in 63 French general hospitals. Clin. Microbiol. Infect. 10:448-451. [DOI] [PubMed] [Google Scholar]
- 2.Del'Alamo, L., R. F. Cereda, I. Tosin, E. A. Miranda, and H. S. Sader. 1999. Antimicrobial susceptibility of coagulase-negative staphylococci and characterization of isolates with reduced susceptibility to glycopeptides. Diagn. Microbiol. Infect. Dis. 34:185-191. [DOI] [PubMed] [Google Scholar]
- 3.El Solh, N., M. Davi, A. Morvan, H. Aubry-Damon, N. Marty, and GISA Group, RAISIN Subgroup. 2003. Characteristics of French methicillin-resistant Staphylococcus aureus isolates with decreased susceptibility or resistance to glycopeptides. J. Antimicrob. Chemother. 52:691-694. [DOI] [PubMed] [Google Scholar]
- 4.John, M. A., C. Pletch, and Z. Hussain. 2002. In vitro activity of quinupristin/dalfopristin, linezolid, telithromycin and comparator antimicrobial agents against 13 species of coagulase-negative staphylococci. J. Antimicrob. Chemother. 50:933-938. [DOI] [PubMed] [Google Scholar]
- 5.Lallemand, S., M. Thouverez, K. Boisson, D. Talon, and X. Bertrand. 2002. Bacteraemia caused by coagulase-negative staphylococci exhibiting decreased susceptibility to teicoplanin. J. Hosp. Infect. 51:207-214. [DOI] [PubMed] [Google Scholar]
- 6.Maniati, M., E. Petinaki, I. Spiliopoulou, F. Kontos, D. Petropoulou-Mylona, L. Spaliara, H. Malamou-Lada, C. Koutsia-Carouzou, and A. N. Maniatis. 2005. Rapid increase in numbers of Staphylococcus epidermidis strains with reduced susceptibility to teicoplanin in Greece. Int. J. Antimicrob. Agents 25:346-348. [DOI] [PubMed] [Google Scholar]
- 7.Mohanty, S. S., and P. R. Kay. 2004. Infection in total joint replacements. Why we screen MRSA when MRSE is the problem. J. Bone Joint Surg. Br. 6:266-268. [DOI] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing, 6th informational supplement. M100S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Tacconelli, E., M. Tumbarello, K. De Gaetano Donati, M. Bettio, T. Spanu, F. Leone, L. A. Sechi, S. Zanetti, G. Fadda, and R. Cauda. 2001. Glycopeptide resistance among coagulase-negative staphylococci that cause bacteremia: epidemiological and clinical findings from a case-control study. Clin. Infect. Dis. 33:1628-1635. [DOI] [PubMed] [Google Scholar]
- 10.Von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2:677-685. [DOI] [PubMed] [Google Scholar]