Abstract
In only a few instances has the clonal composition of Staphylococcus aureus collections that include methicillin-susceptible S. aureus (MSSA) been extensively characterized. In order to investigate the clonal composition of MSSA and methicillin-resistant S. aureus (MRSA) and examine whether the infections diagnosed at our hospital were related to internationally distributed S. aureus lineages, we collected 89 clinical S. aureus isolates from patients at a public university hospital in Rio de Janeiro, Brazil, from September 1999 to June 2000. All S. aureus isolates were genotyped by pulsed-field gel electrophoresis and multilocus restriction fragment typing (MLRFT), and a subset (n = 17) was further characterized by multilocus sequence typing (MLST). The 34 MRSA isolates were additionally characterized by SCCmec typing. The MSSA population (n = 55) was grouped into 18 restriction fragment types (RFTs); of these, five RFTs accounted for 67% (37) of the MSSA isolates. MRSA isolates were clustered into only three RFTs (P = 0.02). The majority of MSSA RFTs were related to sequence type 30 (ST30) (12 isolates, 22%), ST1, ST188, and ST432 (6 isolates, 11% each). The predominant MRSA RFT comprised 31 (91%) of 34 isolates; four randomly selected isolates of this RFT were ST239, the previously described widely disseminated Brazilian clone. However, a fifth isolate belonging to this RFT was the ST644, a new single locus variant of ST239. By applying MLRFT and MLST, we found evidence for a clonal structure in MSSA isolates and detected the dissemination of MSSA clonal complexes 1, 5, 8, 30, and 45.
Staphylococcus aureus is an important human pathogen that causes both healthcare-associated infections (HAI) and community-acquired infections (CAI). Shortly after the introduction of methicillin into clinical practice in 1960, S. aureus developed broad resistance to methicillin and related β-lactams through exogenous acquisition of a mobile genetic element called staphylococcal chromosomal cassette mec (SCCmec) (15). Since then, the spread of methicillin-resistant S. aureus (MRSA) has been tracked in hospitals worldwide. In Brazilian hospitals, about 37% of the S. aureus strains causing infections are resistant to methicillin (11). A single MRSA clone accounts for the vast majority of infections by MRSA in Brazilian hospitals (25, 26); the so-called Brazilian clone was characterized by multilocus sequence typing (MLST) as sequence type 239 (ST239) (20). Recently, the first five cases of community-acquired MRSA (CA-MRSA) in Brazil have been identified and were determined to be caused by isolates belonging to ST30 (22). ST30 strains have been associated with CA-MRSA infections worldwide and are noted for their carriage of a virulence determinant, the Panton-Valentine leucocidin (PVL), and a particular allotype of the type IV SCCmec (13, 24, 29).
Few studies have focused on methicillin-susceptible S. aureus because of its susceptibility to effective first-line antibiotics. Renewed interest in the population genetics of MSSA has been fueled by recent studies that reported at least 20 independent acquisitions of a novel type IV SCCmec element by unrelated MSSA lineages (23). It appears that the success of type IV SCCmec-bearing MRSA clones in community settings worldwide is due to the fitness of the community-adapted MSSA precursors. In Brazil, MSSA is still the major cause of nosocomial and community-acquired staphylococcal infections; however, little is known about the molecular epidemiology of MSSA infections.
Due to the complex composition of bacterial populations, an approach consisting of multiple molecular strain typing methods is desirable to better understand S. aureus epidemiology. In the present study, we investigated clinical and genotypic characteristics associated with S. aureus infections in patients admitted to a large university hospital located in the city of Rio de Janeiro, Brazil, by using pulsed-field gel electrophoresis (PFGE), multilocus restriction fragment typing (MLRFT), MLST, and PCR-based genotyping for the presence of the four allotypes of SCCmec and PVL. Our objective was to investigate the clonal composition of MSSA and examine whether any of the infections diagnosed at our hospital were related to major internationally distributed S. aureus lineages.
MATERIALS AND METHODS
Setting.
This study was conducted at Hospital Clementino Fraga Filho (HUCFF), a large (490 bed), tertiary-care teaching hospital of the Universidade Federal do Rio de Janeiro. This hospital handles about 1,200 admissions per month, including 50 at a 10-bed general intensive care unit. It does not provide obstetric or pediatric care. All isolates analyzed in this study were consecutively collected from September 1999 to June 2000 from patients with S. aureus infections as defined below.
Definitions.
We included in this study all patients admitted to HUCFF who had an S. aureus infection detected at admission or during hospitalization. S. aureus infection was defined as the diagnosis by the attending physician, isolation of S. aureus from a normally sterile site, and prescription of antistaphylococcal antibiotic therapy. Hospital-associated infections were classified according to published criteria (12) and included infections diagnosed in patients undergoing hemodialysis. Community-acquired infections were those diagnosed within 48 h of admission and not related to any hospital procedure.
Clinical data.
A retrospective review of each patient's medical records was conducted with a standardized data collection form. Data obtained included age, sex, type of infection, dates of admission, specimen collection and discharge, length of hospitalization, presence of underlying illness, location in the hospital, presence of HAI or CAI, and outcome.
Bacterial isolates.
A total of 89 S. aureus isolates were consecutively obtained from clinical specimens from 87 patients admitted to HUCFF. Only one isolate per patient was included except in two cases where the staphylococcal infection was considered to be polyclonal; two isolates were included in each of those two cases. All isolates were characterized as S. aureus by analysis of cell morphology on Gram stain, free coagulase production, and by the Vitek system GPI card (BioMérieux, Marcy l'Etoile, France). A representative isolate of the MRSA Brazilian clone (HU25) was kindly provided by Agnes M. S. Figueiredo, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil, and used as a reference strain for molecular typing.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility of S. aureus isolates was evaluated by disk diffusion according to Clinical and Laboratory Standards Institute (CLSI; formerly NCCLS) recommendations (4). The following antimicrobial agents were tested: chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamicin, oxacillin, penicillin, rifampin, tetracycline, trimethoprim/sulfametoxazol, and vancomycin. To calculate antimicrobial resistance rates, we used the CLSI/NCCLS guidelines. Oxacillin MICs were determined by an agar dilution technique (18). The presence of the mecA gene was evaluated by PCR with primers MRS1 and MRS2 (5).
PFGE typing.
S. aureus isolates were typed by PFGE after SmaI digestion as previously described (30). Banding patterns were analyzed by visual inspection and by computer-assisted analysis with GelCompar II (version 1.5) software (Applied Maths, Kortrijk, Belgium) using 0.40% optimization and 1% position tolerance. The similarity of banding patterns was assessed using the Dice index and the unweighted pair group method with arithmetic average. Isolates were considered to belong to different genotypes when the banding patterns revealed more than six band differences (27).
MLRFT and MLST.
MLRFT was performed as described by Diep et al. (7) for all S. aureus isolates. Briefly, seven housekeeping genes were amplified in PCRs using primers previously described for MLST (8). Amplicons were subjected to digestion with restriction endonucleases and electrophoresed on 4% Metaphor agarose gels (7). For each locus, unique band patterns were assigned letter codes and restriction fragment types (RFTs) were defined by the combination of alleles at the seven loci (7). A subset of 17 isolates belonging to RFTs containing two or more isolates was further characterized by MLST (8). eBURST (version 2) software (www.mlst.net/) was used to assign sequenced isolates to clonal complexes (CCs), as proposed by Feil et al. (10). Accordingly, isolates included in the same clonal complex had five to seven loci that were identical to at least one other isolate. Isolates that did not belong to any CC were defined as singletons.
SCCmec typing.
Typing of the SCCmec was performed by PCR for all MRSA isolates as described by Oliveira et al. (21). For isolates that were nontypeable by the Oliveira procedure, the protocol described by Okuma et al. (19) was used.
Detection of PVL encoding genes.
PVL encoding genes lukS-PV and lukF-PV were coamplified by PCR reactions as described by Lina et al. (16) with primers luk-PV-1 and luk-PV-2. We used PVL-positive USA300 strain FPR3757 as a positive control for the PVL PCR assay (6).
Statistical analysis.
Bivariate analysis was performed by χ2 test or Fisher's exact test for categorical variables and the Student t test or Mann-Whitney test for continuous variables with Epi Info (version 6.0).
RESULTS
Demographic and clinical data.
From September 1999 to June 2000, 87 patients who met the case definition for S. aureus infection were consecutively enrolled into the study. Of these, 74 (85%) patients had HAIs and 13 (15%) had CAIs (Table 1). Fifty-three patients (61%) were males. Patients' ages ranged from 13 to 90 years (mean, 49.5 ± 19.7 years). Thirty-two patients (37%) had MRSA infections, all of which were HAIs. MRSA was associated with coresistance to non-β-lactam drugs (P < 0.001) and a longer time of hospitalization (P = 0.005). Variables, such as age, presence and type of underlying illness, type of infection, outcome, and sex, with MRSA and MSSA infections were not significantly different among patients.
TABLE 1.
Key characteristics of S. aureus-infected patients admitted to HUCFF from September 1999 to June 2000
| Variable | Value of variable for patients with:
|
Pa | ||
|---|---|---|---|---|
| MRSA | MSSA
|
|||
| CAIb | HAIc | |||
| Categorical variables, n (%) | ||||
| Sex (male) | 21 (66) | 7 (54) | 26 (62) | 0.6 |
| Infection | ||||
| Bloodstream | 9 (28) | 2 (15) | 20 (48) | 0.3 |
| Respiratory | 10 (31) | 1 (8) | 7 (33) | 0.06 |
| Surgical site | 3 (9) | 0 (0) | 5 (21) | 0.8 |
| Skin and soft tissue | 2 (6) | 4 (31) | 3 (7) | 0.6 |
| Otherd | 8 (29) | 6 (46) | 7 (17) | 0.6 |
| Outcome | ||||
| Discharge | 20 (62) | 8 (61) | 32 (76) | 0.3 |
| Death | 12 (37) | 3 (23) | 12 (28) | 0.45 |
| Related death | 5 (15) | 0 | 0 | |
| Numerical variables, median (SDe) | ||||
| Age (yr) | 54.2 (20.5) | 38.9 (22.8) | 49.3 (17.5) | 0.09 |
| Duration of hospitalization (days) | 43.8 (42.0) | 22.5 (19.2) | 22.8 (17.5) | 0.01 |
| Time to S. aureus isolationf (days) | 16.6 (22.4) | NAh | 6.8 (12.9) | 0.02 |
| Disease timeg (days) | 35.9 (37.3) | 24.1 (18.3) | 13.6 (49.3) | 0.15 |
Comparison between MRSA-infected (n = 32) and MSSA-infected (n = 55) patients.
Community-acquired infections (n = 13) (infections diagnosed within 48 h of admission and not related to any hospital procedure).
Hospital-associated infections (n = 42) (defined according to published criteria [12]), including infections diagnosed in patients undergoing hemodialysis.
Includes catheter-associated, bone and joint, deep-seated, and urinary tract infections.
SD, standard deviation.
Values calculated for hospital-associated infections only.
Time from S. aureus isolation to discharge (excluding death).
NA, not applicable.
Typing of S. aureus isolates and identification of epidemic clones and clonal complexes.
Table 2 summarizes the genetic relationship between MSSA (n = 55) and MRSA (n = 34) isolates. The 55 MSSA isolates were clustered into 18 RFTs by MLRFT. Of these, 67% (37/55) belonged to only five RFTs. Most (9/13) CA-MSSA isolates belonged to just two RFTs. The 34 MRSA isolates were clustered into just three RFTs (P = 0.02 compared to MSSA population).
TABLE 2.
Characteristics of S. aureus clones isolated from 87 patients admitted to HUCFFa
| Clonal complex | Charactaristic of clone (no. of isolates)
|
Pandemic clone | ||||||
|---|---|---|---|---|---|---|---|---|
| RFT | Oxacillin susceptibilityb | SSCmec type | Genotypec | PVL statusd | ST (isolates sequenced) | Resistance profile | ||
| CC8 | BAAACAC (31) | R (31) | IIIA (28) | A (28) | + (1) | 239 (1) | CHL CIP CLI ERY GEN SXT TET (22) | Brazilian |
| + (1) | 644/239 SLV (1) | CHL CIP CLI ERY GEN RIF SXT (2) | ||||||
| None | CHL CIP CLI ERY GEN SXT TET (4) | |||||||
| NT (1) | A (1) | None | 239 (1) | CHL CIP CLI ERY GEN SXT TET (1) | ||||
| IIIB (1) | A (1) | + (1) | 239 (1) | CIP CLI ERY GEN SXT (1) | Hungarian | |||
| IIIA (1) | C (1) | None | 239 (1) | CIP CLI ERY SXT GEN (1) | Brazilian | |||
| CC30 | BBBBBAB (12)e | R (1) | NT (1) | E (1) | + (1) | 30 (1) | CHL CIP CLI ERY GEN SXT TET (1) | |
| S (11) | NA | E (11) | + (1) | 642/30 SLV (1) | CHL CIP CLI ERY PEN SXT TET (1) | |||
| None | CLI ERY PEN (1) | |||||||
| + (1) | ERY PEN (1) | |||||||
| + (1) | GEN PEN (1) | |||||||
| + (3) | PEN (6) | |||||||
| + (1) | Φ (1) | |||||||
| CC5 | AAACCAA (11)e | R (2) | IV (2) | D (2) | None | 5 (1) | CHL ERY (2) | Pediatric |
| S (9) | NA | D (9) | None | CHL CIP CLI ERY GEN PEN SXT TET (1) | ||||
| None | CHL PEN TET (1) | |||||||
| None | CHL CIP ERY (1) | |||||||
| None | ERY PEN TET (1) | |||||||
| None | 641/5 SLV (1) | CLI PEN (1) | ||||||
| + (1) | 641/5 SLV (1) | ERY GEN (1) | ||||||
| None | ERY PEN (1) | |||||||
| None | PEN (2) | |||||||
| CC1 | AAAAAAA (6)e | S (6) | NA | B (6) | + (1) | 1 (2) | CIP CLI ERY GEN PEN SXT (1) | |
| + (1) | ERY PEN TET (2) | |||||||
| None | ERY PEN (1) | |||||||
| + (1) | PEN (1) | |||||||
| None | Φ (1) | |||||||
| CC1 | CAACAAA (6)e | S (6) | NA | J (6) | None | 188/432 SLV (1) | CHL ERY PEN TET (1) | |
| None | CHL ERY PEN (1) | |||||||
| + (1) | ERY PEN (1) | |||||||
| + (1) | 432 (1) | PEN (3) | ||||||
| CC15 | ACAACAA (5) | S (5) | NA | F (5) | None | ERY PEN (2) | ||
| None | ERY GEN PEN (1) | |||||||
| None | 333 (1) | PEN (2) | ||||||
| CC45 | ABDBCAB (3) | S (3) | NA | G (3) | None | 45 (1) | CHL PEN (1) Φ (2) | |
| Singletonf | CADBCAB (3) | S (3) | NA | I (3) | + (2) | 643 (1) | ERY (3) | |
| Othere,g (12) | S (12) | NA | G to R (15) | + (3) | NT | h | ||
R, resistant; S, susceptible; NT, not typeable; NA, not applicable; CHL, chloramphenicol; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; PEN, penicillin; Φ, none; RIF, rifampin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline.
According to presence of mecA gene.
As determined by PFGE.
+, PVL gene-positive isolates (n = 22).
RFTs that include community-acquired isolates.
Singleton indicates an isolate not assigned to any clonal complex.
Includes RFTs CBAACAC and BBBBAAB (2 isolates each), AAAAABC, AAACAAC, AAACCAC, ABABCAB, ABDCCAB, CAABCCA, CAABCCB, and CCACBAA (1 isolate each).
Includes PEN (4 isolates), ERY GEN PEN, CHL GEN PEN and Φ (2 isolates each), ERY PEN, GEN PEN, CLI GEN PEN, ERY GEN PEN and CHL CLI ERY GEN PEN TET (1 isolate each).
The predominant RFT, BAAACAC, accounted for 91% (31/34) of the MRSA isolates and included HU25, a representative Brazilian clone (Table 2). We further characterized five isolates belonging to this predominant RFT by MLST; of these, four isolates were ST239 and one isolate was newly designated as ST644, a single locus variant (SLV) of ST239 differing in the gmk gene allele. Both ST239 and ST644 belong to CC8 lineage (Fig. 1), confirming that the vast majority of MRSA isolates at our hospital are clonally related to the Brazilian clone. By PFGE, 30 (97%) isolates belonged to the unique genotype A, which also included the Brazilian clone reference strain HU25 (26). This predominant RFT was the only one to include an isolate of a second PFGE genotype, C, which showed 74% similarity to genotype A.
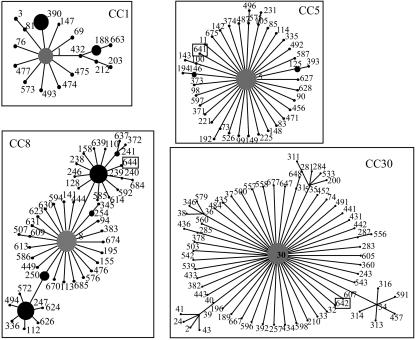
FIG. 1.
Identification of clonal complexes of Staphylococcus aureus-sequenced isolates. The SLVs of the ancestor (gray) are arranged in a circle. Black circles show SLVs of the ancestor ST in the clonal complex. The larger the circle, the larger the number of isolates belonging to that ST registered in the MLST database. Squares represent new STs described in this work.
SCCmec typing revealed that 29 Brazilian clone isolates (including the genotype C isolate) harbored SCCmec IIIA like isolate HU25 (21), while one isolate had SCCmec IIIB associated with the Hungarian clone and one other isolate harbored a nontypeable SCCmec. All isolates harboring SCCmec types IIIA and IIIB had oxacillin MICs of ≥256 μg/ml and were additionally resistant to five or more non-β-lactam antibiotics.
The second most frequent RFT (13% of all isolates), BBBBBAB, included 11 (22%) MSSA isolates and 1 (3%) MRSA. The MRSA isolate was ST30, with a nontypeable SCCmec. The MSSA isolate evaluated by MLST was an SLV (in aroE) of ST30, newly designated as ST642 of the CC30 lineage (Fig. 1).
The third most frequent RFT (12% of all isolates), AAACCAA, included nine (16%) MSSA isolates and two (6%) MRSA isolates. The MRSA isolate evaluated by MLST was ST5. The two MRSA strains had SCCmec IV, low-level oxacillin resistance (MIC, 16 μg/ml) with additional resistance to only erythromycin and chloramphenicol. The association of SCCmec IV with ST5 has previously been associated with the pediatric clone. The sequence-typed MSSA was the newly designated ST641, an SLV (in aroE) of CC5 lineage (Fig. 1). Other less frequent MSSA RFTs were AAAAAAA (ST1 of the CC1 lineage), CAACAAA (ST188 and ST432 of CC1 lineage), and ACAACAA (ST333, a singleton) as described in Table 2.
Genotypic profiles of all S. aureus sequenced isolates are shown in Fig. 2. Apart from RFT BAAACAC, which incorporated two PFGE genotypes, isolates included in the same RFT showed at least 80% similarity by PFGE. With the exception of two isolates belonging to RFTs BBBBBAB and AAACCAA, MSSA isolates showed coresistance to a smaller number of antimicrobial agents than did MRSA isolates. No clusters of RFTs were observed over time.
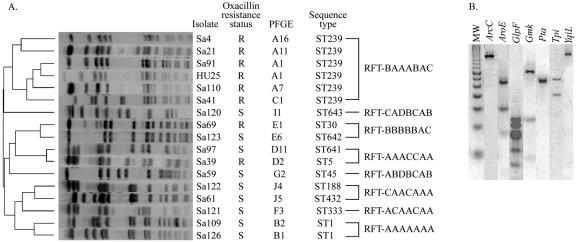
FIG. 2.
Genotypic profiles of representative Staphylococcus aureus isolates belonging to major restriction fragment types. (A) Dendrogram from computer-assisted analysis of PFGE profiles obtained for S. aureus isolates. R, resistant; S, susceptible; ST, sequence type. (B) Multilocus restriction fragment type profile of RFT BAAACAC. MW, 50-bp molecular size marker; ArcC, carbamate kinase; AroE, shikimate dehydrogenase; GlpF, glycerol kinase; Gmk, guanylate kinase; Pta, phosphate acetyltransferase; Tpi, triosephosphato isomerase; YqiL, acetyl coenzyme A acetyltransferase.
In the present study, only two MRSA isolates (RFT BBBBBAB isolate Sa69 and RFT BAAACAC isolate Sa88) had a nontypeable SCCmec by use of the multiplex PCR protocol described by Oliveira and coworkers (21). These isolates had only one 162-bp band corresponding to the mecA amplicon in the multiplex reaction. Both isolates remained nontypeable when the alternative protocol consisting of uniplex reactions was applied.
Presence of PVL-encoding genes.
Twenty-two isolates (25%), distributed in 12 different RFTs, had 476-bp amplicons that corresponded to PVL-encoding genes. Five (20%) of the PVL-positive isolates were CAI. Percentages of PVL-positive isolates in RFTs harboring this gene ranged from 1 to 67%. PVL-positive isolates were significantly associated with RFT BBBBBAB: 8 (67%) positive isolates were among 12 in this RFT (P = 0.002), but not with any specific infections or community-acquired isolates.
Statistical analysis of demographic and clinical data of predominant RFTs.
Relative to all other RFTs, BAAACAC was associated with HAI (P = 0.01) and coresistance to non-β-lactam drugs (P ≤ 0.0001) (Table 3). However, most (91%) of the MRSA isolates were included in this RFT, and the same associations were found when MRSA isolates were compared to MSSA isolates, as stated above, indicating that the associations observed for RFT BAAACAC were dependent on the methicillin resistance status.
TABLE 3.
Variables with significant association to methicillin resistance status and being predominant RFT- BAAACAC by univariate analysis
| Variable | No. (%) of isolates
|
|||||
|---|---|---|---|---|---|---|
| Methicillin resistance
|
P | RFT
|
P | |||
| MRSA (n = 34) | MSSA (n = 55) | BAAACAC (n = 31) | Other (n = 58) | |||
| HAI | 34 (100) | 42 (76) | 0.005 | 31 (100) | 45 (77) | 0.01 |
| Co resistance to 5 or more non-β-lactam drugsa | 32 (94) | 4 (7) | <0.0001 | 31 (100) | 5 (9) | <0.0001 |
Including chloramphenicol, ciprofloxacin, clindamycin, erythromycin, gentamicin, rifampin, tetracycline, trimethoprim/sulfametoxazol and vancomycin.
DISCUSSION
In this study, we used MLRFT, MLST, and PFGE to evaluate the clonal composition of MSSA and MRSA isolates from a hospital in Brazil. The majority (64%) of S. aureus infections in our hospital were caused by MSSA.
A diversity of PFGE profiles was observed for MSSA in the present study as described by others (2, 14, 28). In fact, our study isolates were collected in an endemic setting and did not show any clear predominant genotypes. Therefore, Tenover's criteria (27) were not easily applicable since they were formulated for outbreak situations, where PFGE banding profiles are compared to that of an outbreak strain. In addition, the dendrogram built by computer analysis of PFGE profiles did not allow us to delineate clonal groups either. Nevertheless, MLFRT was able to cluster PFGE patterns with fewer than seven band differences into well-delineated clonal types that were supported by MLST: 67% of all MSSA isolates were included in five RFTs. Three of the five RFTs were determined to include multiple STs; these were SLVs, indicating that MLRFT retains clonal cluster relationships. For example, ST239 and its SLV, ST644, are both clustered in RFT BAAACAC.
It is noteworthy that, among the 13 CA-MSSA isolates, 12 were included in the five RFTs that included most isolates. CA-MSSA isolates were not linked by time or place in the hospital with any of the hospital-acquired MSSA isolates. Thus, high rates of hospital transmission or the occurrence of an outbreak are not the likely explanations for the clustering observed.
Based on MLRFT data supported by MLST, a clonal structure is proposed for MSSA. Graphic representation of MLST data by eBURST placed 74 isolates of major RFTs into six clonal complexes with worldwide distribution, showing that these clones have the ability to spread to distant areas. However, the clonality defined by MLRFT for MRSA was significantly greater than that observed for MSSA. We note that two of these RFTs comprised both MRSA and MSSA, further reinforcing the notion that MRSA emerged from MSSA progenitors that acquired SCCmec through horizontal gene transfer (9, 10, 17).
We observed PVL genes in eight MSSA isolates (67%) of RFT BBBBBAB (ST30, CC30). The CA-MRSA isolates reported from several countries, including Brazil, showed PVL genes and SCCmec IV. Such isolates were also characterized as ST30 (CC30) (3, 22, 29). According to Robinson and coworkers (24), the insertion of SSCmec IV into PVL-positive ST30 MSSA has produced CA-MRSA. In the present study, we have observed a high prevalence of RFT BBBBBAB MSSA isolates carrying PLV genes, which leads us to conclude that the model of Robinson and coworkers is supportive of the emergence of CA-MRSA in Brazil. The PVL-positive ST30 MSSA isolates may constitute a reservoir for the horizontal transfer of PVL into other lineages, such as into the three PVL-positive isolates belonging to the Brazilian clone we found at HUCFF.
In the present study, RFT BAAACAC was the only RFT that showed significant association with HAI. However, this association probably reflects the fact that all isolates included in this RFT are MRSA. On the other hand, all RFTs causing CAI also caused HAI. In addition to RFT BAAACAC, RFTs ACAACAA, ABDCCAB, ABDBCAB, and CADBCAB were found among only HAI, but the association of these RFTs with HAI was not significant. Feil et al. (10), studying well-defined populations of S. aureus causing HAI, CAI, or community-acquired colonization, also reported the lack of association between specific clones and any of the three categories of isolates studied.
The analysis of isolates from a geographic region not previously surveyed using such an array of tools resulted in the description of new housekeeping gene alleles and new STs. Most of these new STs are SLVs of already-described STs, such as ST1, ST188, and ST30. Some of the clones of MSSA causing disease at HUCFF are also found causing infection and colonization in other geographic regions of the world (9, 10). For example, the predominant MSSA clones found at our hospital, including CC5 and CC30 clones, also accounted for many invasive cases in Nigerian hospitals (1). It is not surprising that the predominant MSSA clone in Nigerian hospitals and predominant Brazilian MRSA clone in our hospital both belonged to CC8, attesting to the fundamental biological fitness of this clonal lineage.
Since it is possible to systematically link MLRFT results to the MLST database (7), MLRFT provides an important alternative approach to identify MSSA clones and compare data from different geographic regions of the world. In summary, in addition to demonstrating a clonal structure for MSSA, we report additional geographical areas where CCs 1, 5, 8, 15, and 45 have disseminated and describe new STs.
Acknowledgments
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) of Brazil, and the Fogarty International Program in Global Infectious Diseases (TW006563) of the National Institutes of Health.
REFERENCES
- 1.Adesida, S., H. Boelens, B. Babajide, A. Kehinde, S. Snijders, W. van Leeuwen, A. Coker, H. Verbrugh, and A. van Belkum. 2005. Major epidemic clones of Staphylococcus aureus in Nigeria. Microb. Drug Resist. 11:115-121. [DOI] [PubMed] [Google Scholar]
- 2.Blumberg, H. M., D. Rimland, J. A. Kiehlbauch, P. M. Terry, and I. K. Wachsmuth. 1992. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable, ciprofloxacin-resistant, methicillin-susceptible S. aureus isolates. J. Clin. Microbiol. 30:362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charlebois, E. D., F. Perdreau-Remington, B. Kreiswirth, D. R. Bangsberg, D. Ciccarone, B. A. Diep, V. L. Ng, K. Chansky, B. Edlin, and H. F. Chambers. 2004. Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 39:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. M100-S15. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 5.Del Vecchio, V. G., J. M. Petroziello, M. J. Gress, F. K. McCleskey, G. P. Melcher, H. K. Crouch, and J. R. Lupski. 1995. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J. Clin. Microbiol. 33:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, G. F. Sensabaugh, and F. Perdreau-Remington The complete genome sequence of USA300, an epidemic clone of community-acquired methicillin resistant S. aureus. Lancet, in press. [DOI] [PubMed]
- 7.Diep, B. A., F. Perdreau-Remington, and G. F. Sensabaugh. 2003. Clonal characterization of Staphylococcus aureus by multilocus restriction fragment typing, a rapid screening approach for molecular epidemiology. J. Clin. Microbiol. 41:4559-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gales, A. C., S. S. Andrade, H. S. Sader, and R. N. Jones. 2004. Activity of mupirocin and 14 additional antibiotics against staphylococci isolated from Latin American hospitals: report from the SENTRY antimicrobial surveillance program. J. Chemother. 16:323-328. [DOI] [PubMed] [Google Scholar]
- 12.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 13.Gillet, Y., B. Issartel, P. Vanhems, J. C. Fournet, G. Lina, M. Bes, F. Vandenesch, Y. Piemont, N. Brousse, D. Floret, and J. Etienne. 2002. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet 359:753-759. [DOI] [PubMed] [Google Scholar]
- 14.Graham, P. L., III, A. S. Morel, J. Zhou, F. Wu, P. Della-Latta, D. Rubenstein, and L. Saiman. 2002. Epidemiology of methicillin-susceptible Staphylococcus aureus in the neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 23:677-682. [DOI] [PubMed] [Google Scholar]
- 15.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 17.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2001. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus: identification of two ancestral genetic backgrounds and the associated mec elements. Microb. Drug Resist. 7:349-361. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of meticillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 22.Ribeiro, A., C. Dias, M. C. Silva-Carvalho, L. Berquó, F. A. Ferreira, R. N. S. Santos, B. T. Ferreira-Carvalho, and A. M. S. Figueiredo. 2005. First report of infection with community-acquired methicillin-resistant Staphylococcus aureus in South America. J. Clin. Microbiol. 43:1985-1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson, D. A., A. M. Kearns, A. Holmes, D. Morrison, H. Grundmann, G. Edwards, F. G. O'Brien, F. C. Tenover, L. K. McDougal, A. B. Monk, and M. C. Enright. 2005. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet 365:1256-1258. [DOI] [PubMed] [Google Scholar]
- 25.Santos, K. R., L. M. Teixeira, G. S. Leal, L. S. Fonseca, and P. P. Gontijo Filho. 1999. DNA typing of methicillin-resistant Staphylococcus aureus: isolates and factors associated with nosocomial acquisition in two Brazilian university hospitals. J. Med. Microbiol. 48:17-23. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueiredo, H. de Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Dijk, Y., C. L. Wielders, A. C. Fluit, A. Paauw, R. J. Diepersloot, and E. M. Mascini. 2002. Genotyping of clinical methicillin-susceptible Staphylococcus aureus isolates in a Dutch teaching hospital. J. Clin. Microbiol. 40:663-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vivoni, A. M., K. R. Netto-dos-Santos, M. P. de-Oliveira, M. Giambiagi-deMarval, A. L. P. Ferreira, L. W. Riley, and B. M. Moreira. 2005. Mupirocin for controlling methicillin resistant Staphylococcus aureus: lessons from a decade of use at a university hospital. Infect. Control Hosp. Epidemiol. 26:662-667. [DOI] [PubMed] [Google Scholar]