Abstract
The accessory gene regulator (agr) is a crucial regulatory component of Staphylococcus aureus involved in the control of bacterial virulence factor expression. We developed a real-time multiplex quantitative PCR assay for the rapid determination of S. aureus agr type. This assay represents a rapid and affordable alternative to sequence-based strategies for assessing relevant epidemiological information.
Staphylococcus aureus is a major pathogen responsible for both nosocomial and community-acquired infections. The severity of S. aureus-associated infections ranges from benign localized skin abscesses to life-threatening diseases, such as arthritis, osteomyelitis, and endocarditis (10). Population analyses based on molecular characterization have proven useful to establish relatedness between clinical isolates responsible for defined diseases. Recently, the presence of genes encoding either the Panton-Valentine leukocidin or other toxins in strains harboring a type IV staphylococcal cassette element were shown to describe community-onset methicillin-resistant strains (8, 15). Association of a particular agr type in clinical isolates harboring important virulence factors, such as toxic-shock syndrome toxin (TSST-1) (6) or exfoliatin toxin has already been observed (4). More recently, this association was also suggested for specific diseases such as bullous impetigo, involving strains from agr groups II and IV, and TSST-1-mediated diseases, mainly related to agr type III isolates (5). Another important study by von Eiff and colleagues reported the prevalence of agr type II among strains harboring the bicomponent toxin gene, lukD-lukE, isolated from anterior nares or blood (17). Finally, a possible link between specific agr types and vancomycin resistance was suggested (16).
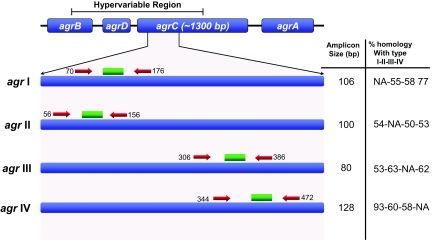
The contribution of agr to S. aureus virulence has been clearly linked to its implication in gene regulation (11, 12) and bacterial interference (6). S. aureus agr is a 3-kb locus showing highly conserved and hypervariable regions (11) among S. aureus strains. The sequence of this hypervariable segment is the target of PCR amplification (5) for defining agr types (11).
The aim of this study was to develop a novel procedure for the rapid typing of agr to be used as a high-throughput epidemiological screening assay. Type-specific oligonucleotides targeting the variable moiety of the agrC gene encoding the receptor of the autoinducing peptide (11) were selected and validated against reference strains. Finally, agr-typed strains were subjected to a recently published genotyping assay (2) that yielded conserved and well-segregated clusters.
Strain collection.
Eighty reference Staphylococcus aureus strains previously characterized by sequencing (1) were used; they consisted of sets of 20 strains of each agr type (types I to IV). In addition, sequenced strains COL, N315, and MW2, belonging to agr types I, II, and III, respectively, were simultaneously analyzed during these experiments.
Sequence analysis and primer and probe selection.
Sequences of agr loci from types I to IV (see Table 1 for accession numbers) were aligned using ClustalW to localize conserved and divergent regions. The designing of type-specific oligonucleotides was performed in variable regions using the software PrimerExpress 2.0 (PE Biosystems, Foster City, CA). Analysis of the agrC and agrD regions (using SIM; http://www.expasy.org/tools/sim-prot.html) showed similarities ranging from 73 to 93% (considering alignment of agr types II and III as well as I and IV, respectively), whereas the conserved moiety showed >93% similarity. Based on these observations, minor groove binder (MGB) probes coupled to dark quenchers were designed to ensure optimal specificity (7) between the different alleles of the agr locus.
TABLE 1.
Oligonucleotides used in the multiplex quantitative PCR assay
| Primer or probe name | Sequence (5′→3′) | Length (bp) | 5′ Dye | GenBank accession no. | Concn (nM) |
|---|---|---|---|---|---|
| agr type I | AF210055 | ||||
| F_agr | CCAGCTATAATTAGTGGTATTAAGTACAGTAAACT | 35 | 200 | ||
| R_agr | AGGACGCGCTATCAAACATTTT | 22 | 200 | ||
| P_agra | ATAGGAATTTCGACATTATC | 20 | FAM | 100 | |
| agr type II | AF001782 | ||||
| F_agr | CAATAGTAACAATTTTAGTGACCATGATCA | 30 | 100 | ||
| R_agr | GCAGGATCAGTAGTGTATTTTCTTAAAGTT | 30 | 100 | ||
| P_agra | TTGCAACAGTAGGTTTGTT | 19 | TET | 50 | |
| agr type III | AF001783 | ||||
| F_agr | CATTATAACAATTTCACACAGCGTGTT | 27 | 200 | ||
| R_agr | GCAAGTGCATAAGAAATTGATACATACA | 28 | 200 | ||
| P_agra | ATAGTTCTACCAATCTTTTTGG | 22 | VICb | 100 | |
| agr type IV | AF288215 | ||||
| F_agr | GAGTTCTCAAAAAGATTAGCTCATCATATC | 30 | 50 | ||
| R_agr | TAGCTTCATCCGAGTTTATTTGAGAAT | 27 | 50 | ||
| P_agra | TTCTACTGCTTACTTTTTCATTG | 23 | NEDb | 50 |
Minor groove binder probes with a nonfluorescent quencher bound to the 3′ end (Applied Biosystems).
Applied Biosystems.
Bacterial lysis.
Genomic DNA was extracted from one colony suspended in 200 μl Tris-EDTA buffer (10 mM Tris, 1 mM EDTA). A total of 100 mg of glass beads (diameter, 100 μm; Schieritz and Hauenstein, Switzerland) was added to the suspension, and bacteria were lysed by vortexing at maximum power for 45 s. The liquid phase was cleared from beads and bacterial debris by centrifugation and diluted 50-fold, and a 5-μl aliquot was used for real-time multiplex PCR assays.
Nucleic acid detection by real-time multiplex PCR and analysis.
Each analysis was performed in triplicate; the nucleic acids from the reference strains were simultaneously assayed in each run. Conditions for the amplification on the SDS 7700 (Applied Biosystems) were the following: time 1 (t1), 2 min at 50°C; t2, 10 min at 95°C; t3, 15 s at 95°C; and t4, 1 min at 60°C (t3 and t4 were repeated 30 times). The volume of the PCR mixture (Eurogentec, Seraing, Belgium) was 20 μl and contained all primers and probes (Table 1) at the indicated concentrations. Fluorescent values recorded from cycles 3 to 15 were used to define fluorescent background levels (SDS 1.9 software; Applied Biosystems) using spectral compensation. Cycle thresholds were manually adjusted to 0.1, 0.14, 0.1, and 0.1 for 6-carboxyfluorescein (FAM), 6-carboxy-4,7,2′,7′-tetrachlorofluorescein (TET), NED, and VIC, respectively, to avoid possible cross-talk, mainly between FAM and TET. A reaction was considered positive when fluorescence levels exceeded the detection threshold between cycles 15 and 30.
Rapid genotyping.
Rapid genotyping was performed using a recently published variable number of tandem repeats-based (VNTR) method (2), using eight primer pairs and a microcapillary electrophoresis system for the rapid evaluation of the VNTR profile.
This report describes a novel multiplex PCR assay that permits rapid determination of agr types based on the discriminatory capacities of short oligonucleotides selected within the hypervariable region of the agr locus. To date, the only procedure allowing agr typing relies on PCR amplification and separation of synthesized products (9) or gene sequencing (5, 14), a time-consuming and labor-intensive strategy. Particular attention was paid to the variable moiety of the agrC gene, providing sufficient divergence between each type to allow the selection of type-specific oligonucleotide sequences (Fig. 1). This rapid method was used on a collection of 80 strains originating from the collection of the French National Reference Centre for Staphylococci (this center receives around 1,000 S. aureus isolates per year for toxin gene determination) and constituted 20 strains of each type (I to IV). The procedure was reproducible, specific, and fast, providing results similar to those of conventional sequence-based analysis. All type II to IV strains from the collection were correctly identified by the multiplex PCR assay. However, 18 out of 20 type I strains were confirmed by the multiplex assay, whereas 2 isolates were defined as type IV. Sequencing of the hypervariable region of the agr locus revealed that both strains were similar to agr type Ic, showing sequence segments of agr types I and IV (3) (G. Lina, personal communication). Finally, this new amplification reaction allowed us to rapidly classify agr types in approximately 3 h for a reasonable cost of $4.00 to $5.00 per strain, including the four probes and primers and the multiplex PCR enzymatic mixture. This unitary cost represents a drastic reduction compared to that of sequencing using fluorescently labeled nucleotide mixtures.
FIG. 1.
Schematic representation of the S. aureus agr locus that contains variable and conserved regions. Numbers indicate oligonucleotide positions based on the 5′ to 3′ sequence. All primers and probes were selected in the first half of the agrC gene, in a region containing enough sequence divergence between the four agr groups to allow specificity of the PCR. The exact length of amplicon obtained for each agr type and the percentage of homology observed in the PCR-targeted region are also indicated. NA, not applicable.
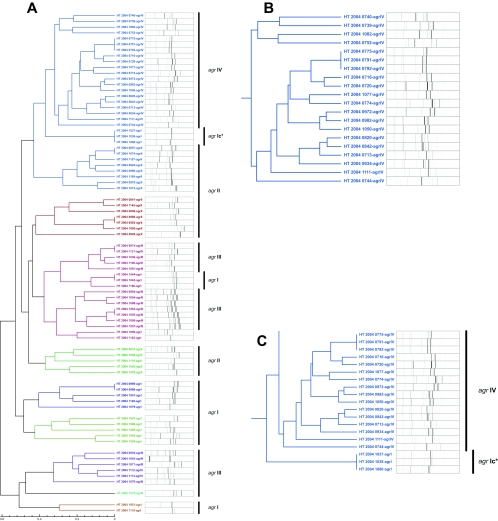
Rapid genotyping was performed to evaluate a possible link between the agr type and the genetic background of the four groups of strains. Figure 2 shows a clear cluster of agr group IV strains (Fig. 2B). Other strains are obviously not distributed randomly but constitute large or smaller clusters identified as related groups of strains segregating based on their agr type. Figure 2C shows a small group of two strains formerly identified as agr type I. Multiplex quantitative PCR reactions classified these strains between isolates from group I and group IV, as previously reported (3, 13). These rare strains harbored elements of variable regions from agr I and agr IV, and our primers and probe system hybridized with the agr IV moiety, thus explaining apparent discrepancies. These discrepancies are, however, rapidly identifiable by molecular assays. Rapid typing results are in concordance with the work of Goerke and colleagues showing that the diversity of type IV isolates is limited and that variation in types I and II is more frequent (3). As a consequence, Fig. 2 clearly shows small clusters of type I and II strains within type III isolates. Strains harboring the agr I locus showed the most dispersed pattern, a finding in accordance with the study of Robinson et al., suggesting that this type is probably the ancestor of other agr loci (13).
FIG. 2.
Clustering tree obtained using high-throughput VNTR assay. (A) General view showing that strains segregated based on their agr type in groups of various sizes. (B) Magnification showing a single cluster of agr type IV. (C) Small cluster of misclassified agr type I coclustered with type IV strains. An asterisk indicates that precise typing was obtained from sequence data (GenBank accession number DQ435772) performed with primers F_agr type I and R_agr type IV (see Table 1).
Overall, our data suggest that different agr types are distributed in various but limited numbers of genetic backgrounds. This observation is supported by Jarraud and colleagues, who showed a link between toxin patterns and genome contents, as obtained by amplified fragment length polymorphism, suggesting a possible relationship between genomic contents of isolates and specific human infections (5). In this context, our results suggest that a limited number of molecular tests allow the collecting of relevant epidemiological information useful for the characterization of clinical isolates.
In summary, this report describes a rapid, specific, and efficient multiplex PCR allowing the deciphering of important epidemiological information about S. aureus clinical isolates. The moderate turnaround time and reagent cost appear compatible with the utilization of this assay in routine laboratories, allowing rapid molecular determination of the agr loci of S. aureus.
Nucleotide sequence accession number.
The nucleotide sequence referred to in the legend of Fig. 2 has been submitted to GenBank under the number DQ435772.
Acknowledgments
This work was supported by the Swiss National Science Foundation grant nos. PP 00B-103002/1 and 632-057950.99 (J.S.), 32-63710.00 (P.V.), 404940-106296/1 (P.F.), and 31-55344.98 (D.L.), as well as grants from the University of Geneva Hospitals Quality Improvement Research program (CI 70889, CI 70903, and CI 70897).
REFERENCES
- 1.Dufour, P., S. Jarraud, F. Vandenesch, T. Greenland, R. P. Novick, M. Bes, J. Etienne, and G. Lina. 2002. High genetic variability of the agr locus in Staphylococcus species. J. Bacteriol. 184:1180-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francois, P., A. Huyghe, Y. Charbonnier, M. Bento, S. Herzig, I. Topolski, B. Fleury, D. Lew, P. Vaudaux, S. Harbarth, W. Van Leeuwen, A. Van Belkum, D. S. Blanc, D. Pittet, and J. Schrenzel. 2005. Use of an automated multiple-locus, variable-number tandem repeat-based method for rapid and high-throughput genotyping of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goerke, C., S. Esser, M. Kummel, and C. Wolz. 2005. Staphylococcus aureus strain designation by agr and cap polymorphism typing and delineation of agr diversification by sequence analysis. Int. J. Med. Microbiol. 295:67-75. [DOI] [PubMed] [Google Scholar]
- 4.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 7.Kutyavin, I. V., I. A. Afonina, A. Mills, V. V. Gorn, E. A. Lukhtanov, E. S. Belousov, M. J. Singer, D. K. Walburger, S. G. Lokhov, A. A. Gall, R. Dempcy, M. W. Reed, R. B. Meyer, and J. Hedgpeth. 2000. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28:655-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liassine, N., R. Auckenthaler, M. C. Descombes, M. Bes, F. Vandenesch, and J. Etienne. 2004. Community-acquired methicillin-resistant Staphylococcus aureus isolated in Switzerland contains the Panton-Valentine leukocidin or exfoliative toxin genes. J. Clin. Microbiol. 42:825-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 11.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 12.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human diseases. Churchill Livingstone, New York, N.Y.
- 13.Robinson, D. A., A. B. Monk, J. E. Cooper, E. J. Feil, and M. C. Enright. 2005. Evolutionary genetics of the accessory gene regulator (agr) locus in Staphylococcus aureus. J. Bacteriol. 187:8312-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shopsin, B., B. Mathema, P. Alcabes, B. Said-Salim, G. Lina, A. Matsuka, J. Martinez, and B. N. Kreiswirth. 2003. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J. Clin. Microbiol. 41:456-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdier, I., M. E. Reverdy, J. Etienne, G. Lina, M. Bes, and F. Vandenesch. 2004. Staphylococcus aureus isolates with reduced susceptibility to glycopeptides belong to accessory gene regulator group I or II. Antimicrob. Agents Chemother. 48:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Eiff, C., A. W. Friedrich, G. Peters, and K. Becker. 2004. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 49:157-162. [DOI] [PubMed] [Google Scholar]