Abstract
Virulence factors of pathogenic Escherichia coli belonging to a recently emerged and disseminated clonal group associated with urinary tract infection (UTI), provisionally designated clonal group A (CGA), have not been experimentally investigated. We used a mouse model of ascending UTI with CGA member strain UCB34 in order to identify genes of CGA that contribute to UTI. iha was identified to be expressed by strain UCB34 in the mouse kidney using selective capture of transcribed sequences. iha from strain UCB34 demonstrated a siderophore receptor phenotype when cloned in a catecholate siderophore receptor-negative E. coli K-12 strain, as shown by growth promotion experiments and uptake of 55Fe complexed to enterobactin or its linear 2, 3-dihydroxybenzoylserine (DHBS) siderophore derivatives. Siderophore-mediated growth promotion by Iha was TonB dependent. Growth and iron uptake were more marked with linear DHBS derivatives than with purified enterobactin. The reported phenotype of adherence to epithelial cells conferred by expressing iha from a multicopy cloning vector in a poorly adherent E. coli K-12 host strain was confirmed to be specific to iha, in comparison with other siderophore receptor genes. iha expression was regulated by the ferric uptake regulator Fur and by iron availability, as shown by real-time reverse transcriptase PCR. In a competitive infection experiment using the mouse UTI model, wild-type strain UCB34 significantly outcompeted an isogenic iha null mutant. Iha thus represents a Fur-regulated catecholate siderophore receptor that, uniquely, exhibits an adherence-enhancing phenotype and is the first described urovirulence factor identified in a CGA strain.
Urinary tract infections (UTIs) are one of the most frequent bacterial infections in industrialized countries, and Escherichia coli is the major causal agent (26, 61). Many virulence factors associated with extraintestinal pathogenic E. coli (ExPEC) strains, the distinctive strains that cause most UTIs, are important for establishing infection. These include adhesins, toxins, iron acquisition systems, and capsular antigens (11, 23, 25). Extraintestinal infections, including UTIs, are caused predominantly by E. coli isolates belonging to phylogenetic group B2 (60 to 70%), whereas the remaining cases are caused mostly by strains belonging to phylogenetic group D (8, 42, 66). Most research into the pathogenic mechanisms of ExPEC has focused on archetype strains, such as CFT073, J96, CP9, and 536, which all belong to group B2. Much less attention has been given to the virulence mechanisms of group D ExPEC strains, which represent the second most important cause of UTI after group B2 strains (8, 41, 66).
Recently, a multidrug-resistant clonal group, termed clonal group A (CGA), was identified as a cause of UTI outbreaks in California, Michigan, and Minnesota (46). It is now known that this clonal group is widespread and quite prevalent throughout the United States and is also widely prevalent, although to a lesser extent, in many other countries (40, 41, 46). This newly emerged clonal group was responsible for up to 50% of the trimethoprim-sulfamethoxazole-resistant isolates identified in some areas (27, 40, 41, 46). The prevalence of resistance to trimethoprim-sulfamethoxazole, which is a commonly used first-line antibiotic therapy for UTIs (61), is increasing (32), which emphasizes the importance of elucidating the virulence mechanisms of CGA. CGA strains derive from E. coli phylogenetic group D (41) and demonstrate a fairly conserved virulence gene profile. Specifically, CGA strains commonly contain the F16 papA allele and papG allele II encoding a major subunit and adhesin of P fimbriae, respectively, iutA encoding the aerobactin siderophore receptor, kpsMTII encoding group II capsule synthesis, and traT encoding a plasmid-associated exclusion protein, whereas they typically lack sfa/foc (S and F1C fimbriae), afa/dra (Dr family adhesins), hly (hemolysin), cnf (cytotoxic necrotizing factor), iroN (siderophore receptor), iss (serum resistance associated), and malX (pathogenicity island marker) (40, 46). Hence, although CGA strains can cause UTIs in healthy women, they lack many of the virulence-associated genes common to group B2 ExPEC strains.
In order to investigate potential genes that may contribute to the capacity of CGA strains to cause UTIs, we used CGA strain UCB34 in a mouse model of ascending UTI for the identification of genes that are expressed in vivo. We used the cDNA capture method selective capture of transcribed sequences (SCOTS) to recover bacterial transcripts from infected tissues (15, 18). This strategy resulted in the capture of iha transcripts during infection in the mouse kidney.
Iha was first described as an adhesin in an enterohemorrhagic E. coli O157:H7 strain and was named “IrgA homologue adhesin,” based on its homology to the IrgA enterobactin siderophore receptor of Vibrio cholerae (49) and its ability to confer epithelial cell adherence capability to a nonadherent K-12 strain when expressed from a multicopy plasmid (60). More recently, Iha was determined to be a urovirulence factor for ExPEC strain CFT073 and its double pap mutant UPEC76, in a mouse UTI model (39). Despite its high homology to siderophore receptors, Iha has thus far been characterized functionally only as a putative adhesin. The prevalence of iha has been reported in a number of studies; overall, 37% to 55% of UTI isolates contained iha or closely related sequences (4, 39, 41, 43, 44). Certain studies have reported an epidemiological association of iha with E. coli isolates causing UTI and other extraintestinal diseases, compared to that with commensal fecal E. coli isolates (4, 30, 39, 41, 43, 44), whereas other reports did not establish such an association (4, 30). However, specifically among CGA isolates from UTIs, iha was present in 92% to 100% of the isolates tested (40, 41), suggesting that Iha might be partly responsible for the virulence of CGA.
The purpose of this study was to investigate the role of the in vivo-expressed iha gene from CGA strain UCB34 as a virulence factor in a mouse UTI coinfection model and to further assess its function and regulation in vitro.
MATERIALS AND METHODS
Bacterial and cell culture.
The bacterial strains and plasmids used are presented in Table 1. UCB34 is an O17/77 CGA strain isolated from a 19-year-old woman with cystitis at the University of California, Berkeley (46). Bacteria were routinely grown in Luria-Bertani (LB) broth, tryptic soy agar, or nutrient broth (NB) (Gibco, Carlsbad, CA). M63 glucose minimal medium [13.6 g/liter KH2PO4, 2 g/liter (NH4)2SO4, 1 mM MgSO4, 0.1 mM CaCl2, and 0.4% glucose, pH 7.4, adjusted with KOH] was used for growth promotion and uptake experiments. Antibiotics were used at 25 μg/ml for chloramphenicol and kanamycin, 10 μg/ml for tetracycline, and 40 μg/ml for carbenicillin. Siderophore production in M63 medium by strains H5058, MG1655, UCB34, QT686, and χ7122 was determined by liquid chromatography/mass spectrometry (LC/MS) (see below) (Table 1). T24 bladder (ATCC HTB-4) epithelial cells were grown in McCoy's 5a medium (modified) with 1.5 mM l-glutamine adjusted to contain 2.2 g/liter sodium bicarbonate and 25 mM HEPES and supplemented with 10% fetal bovine serum. 293 kidney (ATCC CRL-1573) epithelial cells were grown in Eagle's minimum essential medium with 2 mM l-glutamine and Earle's balanced salt solution, adjusted to contain 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and 25 mM HEPES, and supplemented with 10% fetal bovine serum (Gibco).
TABLE 1.
E. coli strains and plasmids
| Strain or plasmid | Genotype (siderophore[s])a | Source or reference |
|---|---|---|
| E. coli K-12 strains | ||
| DM1187 | lexA51 lexA3 | 50 |
| H5058 | aroB tsx malT cirA fepA fiu (Entb) | 5 |
| MG1655 | F− λ−rph-1 (Ent) | 9 |
| ORN172 | thr-1 leu-6 thi-1 Δ(argF-lac)U169 xyl-7 ara-13 mtl-2 gal-6 rspL tonA2 minA minB Δ(fimEACDFGH)::kan pilG1 | 63 |
| QC2517 | MG1655 recD190::Tn10 Δfur::cat (Ent) | 20 |
| QT1272 | H5058 ΔtonB::kan (Entb) | This study |
| Other E. coli strains | ||
| QT686 | UCB34 Δiha::tetAR(B) (Ent, Aero, Ybtc) | This study |
| QT796 | UCB34 Δfur::cat (Ent, Aero, Ybtc) | This study |
| UCB34 | O17/77 ExPEC CGA isolate (cystitis); possesses F16 papA, papGII, iutA, kpsMTII, fyuA, ybte, iha; lacks sfa/foc, afa/dra, hly, cnf, iroN, iss, malX (Ent, Aero, Ybtc) | Amee Manges, 46 |
| χ7122 | Avian pathogenic E. coli, O78:K80:H9; gyrA Nalr (Ent, Aero, Sal) | 52 |
| Plasmids | ||
| pACYC184 | p15A ori; Tcr Cmr | 14 |
| pAMR18 | Vibrio cholerae CA401 irgA in pACYC184 | 49 |
| pBC SK+ | ColE1 ori, Cmr | Stratagene, La Jolla, CA |
| pBR322 | pMB1 ori; Apr Tcr | 10 |
| pC6 | E. coli rrnB operon in pBR322, Apr | C. Squires |
| pCR2.1-TOPO | pUC ori; Apr Kmr | Invitrogen, Carlsbad, CA |
| pIJ68 | MG1655 fepA in pACYC184, Cmr | This study |
| pIJ82 | UCB34 iha in pCR2.1-TOPO; Apr Kmr | This study |
| pIJ83 | iha::tetAR(B) in pACYC184, Cmr | This study |
| pIJ84 | UCB34 iha in pACYC184, Cmr | This study |
| pIJ111 | UCB34 iha in pBC SK+, Cmr | This study |
| pIJ120 | χ7122 iroN in pBC SK+, Cmr | This study |
| pIJ122 | MG1655 fepA in pBC SK+, Cmr | This study |
| pIJ123 | irgA in pBC SK+, Cmr | This study |
| pIJ159 | H5058 tonB in pBR322; Apr Tcr | This study |
| pKD13 | Rγ ori, FRT-flanked kanamycin resistance; Kmr Apr | 16 |
| pKD46 | pSC101 Ts ori, araBp-gam-bet-exo, Apr | 16 |
| pYA3442 | tetAR(B) in pBSL86, Apr | 19 |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline; Ent, enterobactin; Aero, aerobactin; Sal, salmochelin; Ybt, yersiniabactin. The sidephores listed in parentheses are known siderophores produced.
Only when supplemented with shikimate or dihydroxybenzoic acid.
Possesses yersiniabactin genes, but no product was detected by mass spectrometry.
SCOTS.
Five female CBA/J mice (5 to 6 weeks old) were inoculated via a urethral catheter under nonrefluxing conditions as previously described (37, 38) with a bacterial suspension of UCB34 (approximately 3 × 108 CFU/g of mouse weight). After 24 h, infected tissues were harvested aseptically. Total RNA from the kidneys of the five infected mice was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). RNA samples were treated with DNase using the DNA-free kit (Ambion, Austin, TX) to remove genomic DNA contamination. Five 1-μg samples of each RNA were converted into cDNA by random priming with Superscript II reverse transcriptase (Invitrogen), followed by Klenow treatment (New England Biolabs, Ipswich, MA). Primers with a defined sequence at the 5′ end and random nonamers at the 3′ end (CMD115, 5′-GTGGTACCGCTCTCCGTCCGANNNNNNNNN-3′) were used for first- and second-strand cDNA syntheses. Samples were then pooled, and cDNA from each kidney was then amplified for 25 cycles by PCR using the defined primer. SCOTS was performed as previously described (15, 18). The initial capture round was performed with the five cDNA samples (corresponding to each kidney) in parallel. After the first round, the five cDNA samples were combined and two more rounds of capture were done. This procedure resulted in selection for cDNA corresponding to bacterial transcripts. In the second step of SCOTS, UCB34-specific transcripts (absent from E. coli K-12) were selectively enriched by performing three more rounds of capture while blocking for nonpathogenic transcripts using a 100-fold excess of E. coli MG1655 K-12 genomic DNA. Selected cDNA fragments were then cloned with a TA cloning kit (Invitrogen) and were sequenced.
Cloning the iha, fepA, irgA, iroN, and tonB genes.
For cloning experiments, restriction enzymes and DNA modification enzymes were purchased from and used as recommended by New England Biolabs or Invitrogen. The iha gene from UCB34 was PCR amplified using Elongase (Invitrogen) with the following primers: CMD204, 5′-GGTGGAAATCCGCTTCTACAG-3′; and CMD205, 5′-TAACGAAATCTCATTAGCGGATCG-3′. The amplified product was cloned in pCR2.1-Topo (Invitrogen), resulting in pIJ82. The iha gene was subcloned from pIJ82 following digestion with EcoRV and BamHI. Ligation of the iha-containing fragment into the same sites of cloning vectors pACYC184 and pBC SK+ generated plasmids pIJ84 and pIJ111, respectively. fepA from strain MG1655 was PCR amplified using Elongase with the following primers: CMD127, 5′-GCCAAGCTTCGCGCAATTCGAGGCG-3′; and CMD128, 5′-GAAAAGCTTAACCGCAGTCTGCGAGT-3′. The amplified product was digested with HindIII and was cloned into vectors pACYC184 and pBC SK+, generating plasmids pIJ68 and pIJ122, respectively. irgA from Vibrio cholerae CA401 was subcloned from plasmid pAMR18 (49) into pBC SK+ by using EcoRI, generating plasmid pIJ123. Plasmid pIJ120 was produced by cloning iroN from E. coli strain χ7122 into pBC SK+ at the HincII and XhoI sites. The PCR fragment was amplified with Elongase using primers CMD276 (5′-TACCGCAGTTTAAACAGGCTTTCATAATTCTC-3′) and CMD277 (5′-CGCCCTCGAGACTACGATCAGAATGATGCGGT-3′), which have a PmeI and a XhoI site, respectively, at their 5′ end. tonB from strain H5058 was PCR amplified using Phusion (Finnzymes, Finland) and primers CMD658 (5′-AAGGCCGAATTCAAAGTAAGGGTAATTACGCCAA-3′) and CMD659 (5′-CCTGTTGAATTCTAGTCAAAAGCCTCCGGTCGG-3′). The amplified product was digested with EcoRI and was cloned into vector pBR322, generating plasmid pIJ159.
Construction of iha, fur, and tonB mutants.
A mutant iha allele was created as follows: a 1,154-bp PCR fragment of iha was generated using primers CMD200 (5′-GGGCGGGATCCTGAATATCATTACCAGA-3′) and CMD201 (5′-GTGCCGGATCCTTCCACACCATGCAAC-3′). The amplified product was digested with BamHI and was cloned in pACYC184, resulting in pIJ81. pIJ81 was digested with SacI, which introduced a 96-bp deletion in the middle of iha, and a tetAR(B) cassette derived from Tn10 (obtained from pYA3442 [19]) was ligated into the SacI sites, resulting in pIJ83. The iha::tetAR(B) allele from pIJ83 was PCR amplified using primers CMD200 and CMD201 and was introduced into strain UCB34 by homologous recombination using the λ Red recombinase method (16). An iha mutant strain, designated QT686, was confirmed by PCR using external primers flanking the iha allele. QT686 demonstrated no difference in growth rate, plasmid profile, extended virulence profile (43), or pulsed-field gel electrophoresis restriction digest band pattern of genomic DNA compared with the wild-type parent. A fur mutant of strain UCB34 was created as follows: a Δfur::cat allele derived from E. coli strain QC2517 (20) was amplified with primers CMD18 (5′-ATTCTAGACTGCTGCTGGGCATCCC-3′) and CMD19 (5′-ACTCTAGACACTCCGACATCCCAAGC-3′). The Δfur::cat-containing amplicon was transferred to strain UCB34 by homologous recombination using the λ Red recombinase method (16), as the DNA region encompassing fur is highly conserved among different E. coli strains. The fur mutant of strain UCB34 was designated QT796. A tonB mutant of strain H5058 was created. Primers CMD656 (5′-ATTTAAAATCGAGACCTGGTTTTTCTACTGAAATGATTATGACTTCAATGATTCCGGGGATCCGTCGACC-3′) and CMD657 (5′-CCTCCGGTCGGAGGCTTTTGACTTTCTGCTTACTGAATTTCGGTGGTGCCTGTAGGCTGGAGCTGCTTCG-3′) were used to create a ΔtonB::kan allele by PCR, using the kanamycin resistance cassette from pKD13. The regions underlined show homologies to pKD13, and the remaining 50 bp of each primer are homologous to DNA flanking tonB. Primers were designed from the sequence of E. coli K-12 strain MG1655. The ΔtonB::kan-containing amplicon was transferred to strain H5058 by homologous recombination using the λ Red recombinase method (16). A tonB mutant strain was designated QT1272 and was confirmed by PCR using external primers flanking the tonB allele (CMD658 and CMD659).
Purification of enterobactin and its by-products.
Extraction was done using the method described by Young and Gibson (64) with slight modifications. MG1655 bacterial cells were grown for 18 h in M63 glucose minimal medium with 75 μM 2,2′-dipyridyl. Cells were pelleted, and the supernatant was retained. FeCl3 (5 mM) was added to the supernatant, and samples were filtered. One liter of the clear supernatant was passed through a DE52 DEAE-cellulose column (Whatman, United Kingdom). The column was washed with water and eluted with 2.5 M NH4Cl. Colored fractions were pooled, and a sample was taken for analysis by LC/MS. The siderophores were then acidified with HCl to pH 1.5, and three extractions with ethyl acetate were made. To remove any of the hydrolysis products of enterobactin, an additional extraction step with sodium phosphate buffer (0.1 M, pH 7) was done on the combined ethyl acetate extract. These extractions were then dehydrated with Na2SO4, evaporated, and resuspended in methanol for the sample containing both enterobactin and linear degradation products or in ethyl acetate for the purified enterobactin sample. Analysis by LC/MS revealed the presence of enterobactin and linear 2,3-dihydroxybenzoylserine (DHBS) trimers, dimers, and monomers at a proportion of 3:2:4:1, respectively, in the enterobactin and DHBS product mix. LC/MS analysis also revealed a highly purified enterobactin in the sample extracted with sodium phosphate buffer.
LC/MS.
Analyses by LC/MS were performed by using an Agilent HP 1100 high-pressure liquid chromatograph with a C8 Luna Phenomenex 150-mm by 3-mm column at a flow rate of 400 μl/min, a linear gradient of water-acetonitrile with 1% acetic acid, and a Quattro II (Micromass, Canada) mass spectrometer in electrospray-positive mode. Catecholate siderophores were detected and quantified in full-scan mode between 150 to 1,200 m/z. Enterobactin and linear DHBS trimers, dimers, and monomers were detected at 670, 688, 465, and 242 m/z, respectively (7, 62). The relative quantification of each peak was done by integration of the area under the curve determined by the MassLynx software. The presence of other siderophores, such as aerobactin, salmochelins, and yersiniabactin, was determined from the M63 glucose culture supernatants of strains UCB34, QT686, MG1655, H5058, and χ7122 at 565, 627, and 483 m/z, respectively (28, 33, 35).
Growth in iron-limited medium.
E. coli strain H5058 and derivatives were grown at 37°C with agitation in M63 glucose minimal medium supplemented with aromatic amino acids (20 mg/liter each of l-phenylalanine, l-tryptophan, and l-tyrosine), thiamine (1 mg/liter), appropriate antibiotics, and 50 μM 2,2′-dipyridyl. Enterobactin extracts with or without DHBS products (50 μM) were added to cultures, and bacterial growth was measured by spectrophotometry (an optical density at 600 nm [OD600]) at each hour for 12 h. A final time point was taken at 24 h. Tests were performed in triplicate.
Siderophore-55Fe uptake experiments.
55Fe-radiolabeled siderophore uptake experiments were based on a protocol modified from that described by Sabri et al. (57) and Eisenhauer et al. (21). E. coli strain H5058 and derivatives were grown overnight at 37°C with agitation in supplemented M63 minimal medium (without dipyridyl). Cultures were adjusted to an OD600 of 0.5, corresponding to 108 CFU/ml, and were washed three times with equal volumes of nonsupplemented M63 medium. After the third wash, 50 μM 2,2′-dipyridyl was added to the cells and they were subsequently incubated at 37°C for 60 min without agitation. 55Fe3+ isotope (PerkinElmer, Wellesley, MA) was complexed with enterobactin extracts with or without DHBS products in a 1:3 ratio and incubated for 10 min at room temperature with agitation. The 55Fe-siderophore complex was added to bacterial cells at a 1-nmol/ml concentration, and samples were left to stand for 5 min at room temperature. Samples were centrifuged at 10,000 × g for 2 min, and pellets were washed three times with M63 medium and resuspended in 100 μl of M63 medium. A 2-ml volume of Optiphase scintillation cocktail was added to the resuspended cells, and scintillation was measured in a Beckman LS 1701 (Beckman, Fullerton, CA) scintillation counter on the 0 to 350 channels as suggested for 55Fe by the supplier. All experiments were done in triplicate and controlled by using nonwashed cells with isotope-siderophore mix as a positive control and cells without isotope as a negative control. Statistical significance was calculated by an unpaired t test (two tailed).
Adherence assays.
Quantitative adherence assays were performed essentially as described previously (22, 47). T24 or 293 epithelial cells were grown to confluence in 24-well plates. E. coli K-12 derivative ORN172 or ExPEC strain UCB34 and derivatives were grown on LB plates containing appropriate antibiotics and were resuspended in LB broth to an OD600 of approximately 0.5. Cells were washed twice with serum-free medium and were then incubated for 1 hour with 880 μl of fresh medium (with serum). Next, 20 μl of bacteria (approximately 3 × 107 CFU) was added to six wells per strain (multiplicity of infection, approximately 1/60 to 1/100), and bacteria-host cell contact was enhanced by a 5-min centrifugation at 600 × g. After 2 hours of incubation, three wells per strain were lysed by adding 100 μl of phosphate-buffered saline, pH 7.4, containing 1% (wt/vol) sodium deoxycholate. Lysates were then plated on LB agar with appropriate antibiotics. The bacteria present in these lysates represented total bacteria. Cells in the three other wells per strain were washed three times using serum-free medium and were then similarly lysed. Bacterial adherence was calculated as the number of bacteria after the washes divided by total bacteria. Statistical significance was calculated by an unpaired t test (two tailed).
Quantitative real-time reverse transcriptase (RT) PCR.
For expression analysis in NB medium, UCB34 and the Δfur::cat derivative strain QT796 were grown for 3 h at 37°C with agitation. Approximately 3 × 107 CFU were pelleted by centrifugation for 5 min at room temperature, and RNA was extracted from whole bacterial cells by using TRIzol reagent according to the manufacturer's instructions (Invitrogen). For cell culture conditions, bacterial cells were suspended in culture medium in the presence or absence of 293 epithelial cells as described above for the adherence assays. For bacteria-eukaryote cell interaction, no washes were done, medium was removed, and TRIzol was added to the wells. Bacteria without eukaryotic cells were harvested by centrifugation. A 10-fold dilution was performed in TRIzol, followed by RNA extraction. RNA was submitted to a second precipitation using lithium chloride (Ambion) and was treated twice with DNase using the DNA-free kit (Ambion) to eliminate any genomic DNA. RNA concentrations were determined using a NanoDrop 1000 apparatus (NanoDrop Technologies, Wilmington, DE), and 15 ng of total RNA was reverse transcribed in triplicate using random hexamers and Superscript II (Invitrogen). Real-time PCR was done using the Dynamo SYBR green qPCR kit (Finnzymes) according to the manufacturer's guidelines. Reactions (25 μl) were performed using 1 μl of cDNA template per reaction. A Rotor-Gene 3000 real-time PCR apparatus (Corbett Research, Sydney, Australia) was used. PCR conditions consisted of an initial incubation step for 15 min at 95°C, followed by 45 cycles for 20 s at 95°C, 20 s at 60°C, and 20 s at 72°C. Melting curve analyses were performed after each reaction to ensure amplification specificity. Differences (n-fold) in transcript were calculated using the relative comparison method, and amplification efficacies of each primer set were verified as previously described (2, 58). RNA levels were normalized by using rpoD as the control gene (3, 49, 67). Statistical significance was calculated on ΔΔCt values (58) with a paired t test, and P values were all <0.0001. Primers used for real-time PCR analysis were CMD306 (5′-GGCTGAATCTGCAGGAAAGCAACA-3′) and CMD307 (5′-TGCAGGCTGACAGAATCATCCACA-3′) for iha, CMD308 (5′-AGCTGACTGACAGCACCATCGTAA-3′) and CMD309 (5′-AAACCTTGCGATATGTTCAGCGCC-3′) for fepA, and CMD310 (5′-TCATGAAGCTCTGCGTTGAGCAGT-3′) and CMD328 (5′-CAATTGCCGCGTTCAACCAGGTAT-3′) for rpoD.
Mouse infection experiment.
Fifteen female CBA/J mice were coinfected with a mixed inoculum, consisting of the strain UCB34 and its isogenic iha mutant QT686 in equal proportions, using the same ascending UTI model used for SCOTS (37, 38). After 48 h, mice were euthanatized. Bacterial colonies recovered from tissues, urine, and the initial inoculum suspension were replica plated to LB agar plates with or without tetracycline to determine the relative proportion of wild-type UCB34 versus iha mutant QT686 in each sample. Data from postinfection cultures (output ratios) were normalized to the input ratios. Statistical differences were calculated with the Wilcoxon matched-pairs test (signed rank).
Nucleotide sequence accession number.
The accession number of the complete nucleotide sequence of iha from strain UCB34 is DQ211582.
RESULTS
iha is expressed by CGA strain UCB34 during UTI.
In order to identify genes expressed in vivo by CGA strain UCB34, mice were infected in a UTI model. RNA was extracted from infected kidneys and was used for SCOTS. This technique of bacterial transcript capture allowed us to determine that iha from UCB34 was expressed in the mouse kidneys. The 470-bp SCOTS fragment clone that was identified by sequencing was identical to a portion of the iha gene from strain CFT073. By using primers derived from the CFT073 and EDL933 iha sequences, the iha gene and promoter region were amplified from genomic DNA of strain UCB34 and were cloned. iha from strain UCB34 is nearly identical to iha from strain CFT073, demonstrating only a single nonsynonymous nucleotide difference that results in a Gly565 to Ser565 substitution in the predicted peptide sequence. We identified a putative Fur box regulatory region located just 3′ of the predicted −35 region. In addition, a putative TonB box has already been identified at the beginning of Iha from strain EDL933 (60). The TonB box is also conserved in the predicted Iha proteins from strains UCB34 and CFT073.
Iha has previously been reported to share 53% similarity to the siderophore receptor IrgA from Vibrio cholerae (60). Additionally, new protein sequence homology searches showed that Iha demonstrates identity/similarity with a number of putative outer membrane receptors from recently completed genomes, as well as to characterized iron-regulated outer membrane proteins, such as Cir2A from Yersinia pestis and Yersinia pseudotuberculosis (73.2% similarity) (13, 17, 59), CfrA from Campylobacter coli and Campylobacter jejuni (54.5%) (31, 51), and BfrA from Bordetella bronchiseptica (50.7%) (6). Known siderophore receptors from E. coli that share identity/similarity with Iha include FepA (44.1%) (45) and IroN (44.2%) (55, 56). The percent similarities reported are Emboss pairwise alignments (full-length sequence alignments) obtained using Needle (European Bioinformatics Institute; http://www.ebi.ac.uk).
Iha from UCB34 functions as a siderophore receptor in E. coli K-12 strain H5058.
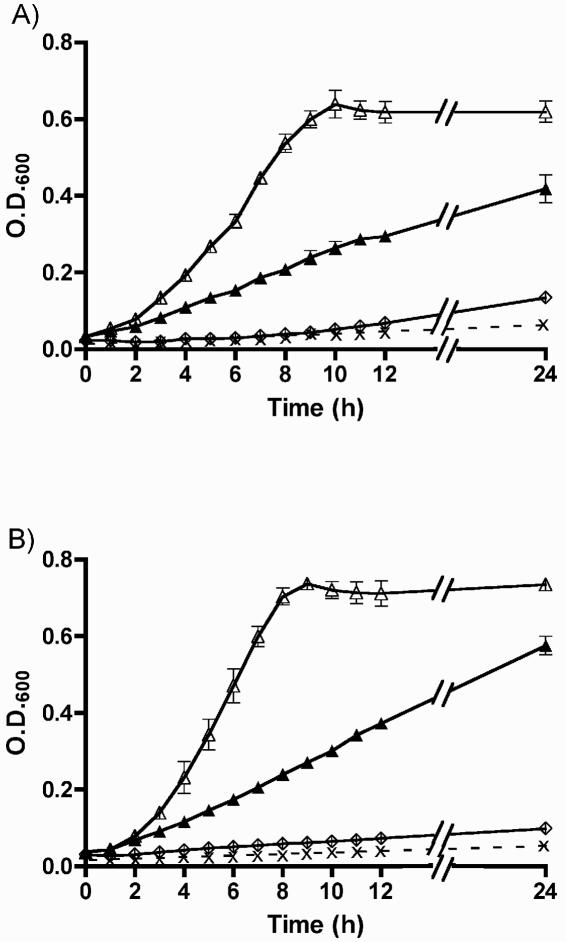
Due to its similarity to other siderophore receptors, we wished to assess whether Iha from UCB34 was able to function as a siderophore receptor. First, growth promotion experiments in iron-limited minimal medium were done using the E. coli K-12 strain H5058. H5058 is an aroB mutant and is therefore not able to synthesize aromatic amino acids or enterobactin. Production of enterobactin by H5058 can be achieved only when the strain is supplemented with shikimate or dihydroxybenzoic acid, which are precursors of enterobactin (29). Strain H5058 is also defective in the uptake of enterobactin and enterobactin's DHBS derivatives, as it lacks the catecholate siderophore receptors FepA, Fiu, and Cir. It therefore grows poorly in iron-limited medium even when supplemented with shikimate or dihydroxybenzoic acid. Introduction of plasmid pIJ84 (iha) increased the growth of strain H5058 in iron-limited minimal medium supplemented with purified enterobactin compared to the negative control (vector only), although growth promotion was less marked than when complemented with the positive control plasmid pIJ68 (fepA) (Fig. 1). The addition of purified enterobactin to iron-limited medium promoted the growth of the iha clone up to 68% of the fepA clone (Fig. 1A), whereas the addition of extract that contained both enterobactin and DHBS products (trimers, dimers, and monomers) promoted the growth of the same clone to 78% of the fepA-complemented positive control (Fig. 1B). Further, from 6 h onward, growth of the iha clone, when supplemented with the enterobactin-DHBS mixture, was significantly greater (P < 0.05) than when supplemented with purified enterobactin alone. These results indicate that iha can promote growth of a catecholate siderophore receptor-negative E. coli K-12 strain in iron-limited medium in the presence, but not the absence, of DHBS products, derivatives of enterobactin, and to a lesser extent, cyclic enterobactin, presumably by facilitating the uptake of iron coupled to these substances.
FIG. 1.
Growth of E. coli K-12 catecholate siderophore receptor-negative strain H5058 containing a cloned copy of iha (pIJ84, ▴), fepA (pIJ68, ▵), or the control vector (pACYC184, ⋄) in iron-limited minimal medium (M63 glucose) supplemented with 50 μM 2,2′-dipyridyl and 50 μM of purified enterobactin (A) or 50 μM of enterobactin and its DHBS breakdown products, trimers, dimers, and monomers (B). Growth of H5058 (pIJ84) expressing iha in medium without siderophore supplements is indicated (x) on dashed lines.
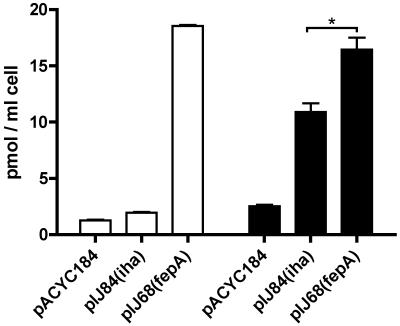
Siderophore-55Fe complex uptake assays using strain H5058 containing plasmids carrying iha or fepA, or the control vector alone, were done to further investigate the role of Iha as a siderophore receptor. The iha clone mediated uptake of the cyclic enterobactin-55Fe complex slightly, 1.96 ± 0.04 pmol/ml (mean ± standard deviation) compared to 1.28 ± 0.04 pmol/ml for the negative control. This uptake was minimal compared to that of the fepA-complemented positive control, i.e., 18.55 ± 0.09 pmol/ml (Fig. 2). By contrast, when a siderophore extract comprised of a mixture of enterobactin and its DHBS products was used for the uptake experiments, the iha clone demonstrated an increased uptake level of 10.9 ± 0.77 pmol/ml, compared to 16.4 ± 1.06 pmol/ml for the fepA-complemented positive control. These results correlate with the growth experiments and indicate that Iha functions as a catecholate siderophore receptor that demonstrates a greater specificity for linear enterobactin DHBS products than for tricyclic enterobactin. As Iha demonstrated a catecholate siderophore receptor function, we also compared the ability of ExPEC strain UCB34 or its isogenic Δiha derivative QT686 to mediate the uptake of catecholate siderophores from the enterobactin-DHBS mixture. Interestingly, QT686 demonstrated a decreased transport capacity of 25.6% ± 2.6% compared to the wild-type parent (P < 0.0001). Taken together, these results support a role for Iha in the uptake of catecholate siderophores in both E. coli K-12 and ExPEC strain UCB34.
FIG. 2.
Enterobactin-55Fe transport of E. coli K-12 catecholate siderophore receptor-negative strain H5058 containing a cloned copy of iha (pIJ84), fepA (pIJ68), or the control vector (pACYC184) in iron-limited minimal medium (M63 glucose) supplemented with 50 μM 2,2′-dipyridyl using an enterobactin-55Fe complex (white bars) or enterobactin and its DHBS products and 55Fe (black bars). Compared to those for vector controls, values are significantly different (P < 0.0001). *, P = 0.002.
The iha siderophore receptor phenotype is tonB dependent.
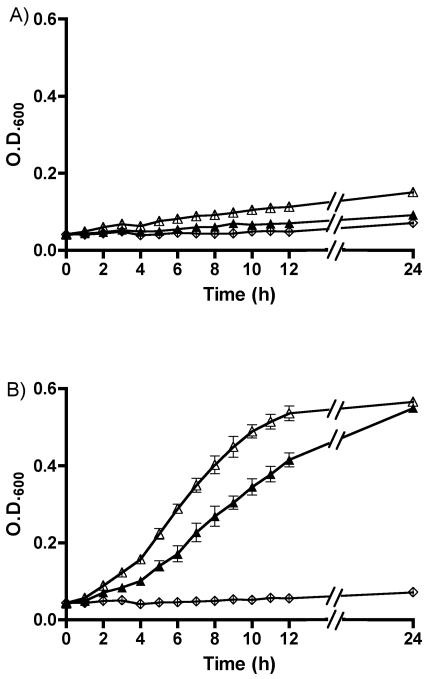
As gram-negative bacteria typically require the TonB-ExbB-ExbD energy complex to import siderophores into the cytoplasm (1, 12), we assessed whether the capacity of Iha to transport siderophores was dependent on the TonB complex. pIJ84 (iha), pIJ68 (fepA), and control vector pACYC184 were introduced into the H5058 tonB mutant strain QT1272. As expected, strains exhibited poor growth in iron-depleted minimal medium supplemented with enterobactin and its DHBS products, even when complemented with fepA or iha (Fig. 3A). However, introduction of a cloned copy of the tonB gene (pIJ159) into these strains restored growth of the iha- and fepA-complemented strain in the same medium (Fig. 3B), suggesting that siderophore transport by Iha is TonB dependent.
FIG. 3.
Growth of E. coli K-12 catecholate siderophore receptor-negative ΔtonB H5058 strain QT1272 (A) or QT1272 complemented with tonB (pIJ159) (B) containing a cloned copy of iha (pIJ84, ▴), fepA (pIJ68, ▵), or the control vector (pACYC184, ⋄) in minimal medium (M63 glucose) supplemented with 50 μM 2,2′-dipyridyl and 50 μM of enterobactin and its DHBS products.
Iha from UCB34 promotes epithelial cell adherence in E. coli K-12 strain ORN172.
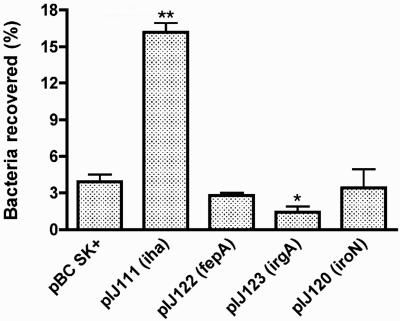
In previous reports, introduction of Iha from strains EDL933 and CFT073, when cloned into a high-copy vector, conferred increased adherence of the poorly adhering E. coli K-12 fim mutant strain ORN172 to epithelial cells (39, 60). These results suggested that Iha may act directly (or indirectly) as an adhesin. As Iha shares sequence similarities with other siderophore receptors, this suggested (by extension) that other siderophore receptors also might promote adherence. Accordingly, we compared the adherence-enhancing phenotype of Iha with other siderophore receptors. Adhesion assays were performed on T24 human bladder epithelial cells using high-copy clones of iha, fepA, iroN, and irgA. Except for the iha-containing clone (pIJ111), no increase in adherence was conferred by the presence of any of the other siderophore receptors on high-copy plasmids (Fig. 4). Similar results were obtained using the 293 human kidney epithelial cell lines. To determine whether plasmid copy number had an effect on adherence to epithelial cells, iha and fepA (as a control) were cloned into the medium-copy vector pBR322. Quantitative adherence assays were performed, and similar adherence results were obtained using the mid-copy clones or the high-copy clones (data not shown). However, no difference in adherence to T24 or 293 cells was observed between (iha-positive) wild-type strain UCB34 and its Δiha::tetAR(B) derivative strain QT686, with or without the addition of 2.5% mannose to the medium (data not shown), consistent with the known presence of additional mannose-resistant adhesins (e.g., P fimbriae) in strain UCB34.
FIG. 4.
Quantitative adherence of E. coli K-12 fim ORN172 containing iha (pIJ111), fepA (pIJ122), irgA (pIJ123), or iroN (pIJ120) cloned on high-copy vector pBC SK+ or the control vector on T24 bladder epithelial cells. Statistical differences compared to the vector control pBC SK+ were as follows: **, P < 0.0002; *, P < 0.03.
As mentioned earlier, the iha promoter region contains a predicted Fur-binding site. The presence of Fur-binding sites on high-copy plasmids has the potential to titrate the Fur regulatory protein and hypothetically could result in the deregulated expression of many proteins in E. coli ORN172, which in turn could potentially be responsible for the increased adherence observed in this and previous studies (39, 60). To assess whether Fur exerts a regulatory effect on adhesion, fur null mutants of strains UCB34 and ORN172 were constructed and used in adherence assays. No differences in adherence to epithelial cells were observed compared to that of the parental strains (data not shown), providing evidence against this hypothesis.
iha expression is modulated by iron availability and Fur but not by contact with host cells.
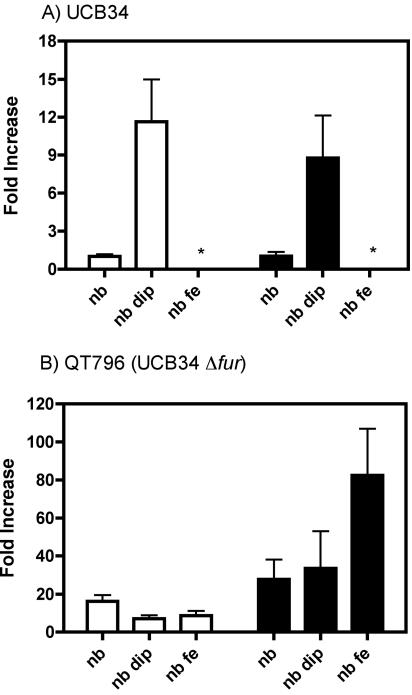
The expression level of iha by CGA strain UCB34 under different culture conditions was determined using real-time RT-PCR experiments and was compared to the level of expression of fepA. The wild-type strain UCB34 and isogenic fur null mutant QT796 were grown in NB medium, iron-limited medium (NB supplemented with 75 μM 2,2′-dipyridyl), or iron-replete medium (NB supplemented with 30 μM FeCl3). Results shown in Fig. 5 represent the increases (n-fold) in expression level compared to the expression level of the wild-type strain grown in NB (onefold increase). In strain UCB34, we observed a 12-fold increase in iha expression following growth in iron-depleted medium, compared to a 9-fold increase for fepA. In contrast, no expression for either iha or fepA was detectable in iron-replete medium. Additionally, no expression of iha by UCB34 was detected in LB medium (data not shown). In the fur mutant QT796, iha and fepA expression were increased 16- and 28-fold, respectively, in NB medium, and regulation by iron of both iha and fepA expression was abrogated, since expression could be detected under all culture conditions.
FIG. 5.
Expression levels of iha (white bars) and fepA (black bars), as determined by real-time PCR under different culture conditions for wild-type strain UCB34 (A) and its fur derivative QT796 (B). Results are presented as increases (n-fold) in expression levels compared to that of the wild-type UCB34 strain grown in NB (onefold increase), NB supplemented with 75 μM dipyridyl (nb dip), and NB supplemented with 30 μM FeCl3 (nb fe). *, No expression could be detected.
Since contact with host cells has been shown to induce expression of iron-regulated proteins by pathogenic E. coli (65), the influence of bacterial interaction with host cells on the expression level of iha was investigated by real-time RT-PCR. In a representative adhesion assay, the level of expression by UCB34 following a 2-h incubation in minimal essential medium was not any different when the incubation was done in the presence, versus the absence, of 293 cells (data not shown).
Iha contributes to virulence of CGA strain UCB34 during UTI.
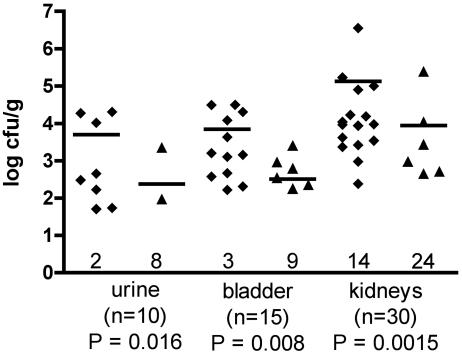
To assess whether Iha contributes to virulence of the CGA strain in the mouse UTI model, dual-strain competition infections were done in female CBA/J mice using wild-type strain UCB34 and its isogenic iha::tetAR(B) derivative QT686. Fifteen mice were challenged via the urethra under nonrefluxing conditions. Figure 6 shows the bacterial numbers for the wild-type or mutant strain in bladders, kidneys, and urine. Wild-type strain UCB34 significantly outcompeted the iha mutant at each site assessed (Fig. 6). For many of the samples, only wild-type strain UCB34 was recovered, indicating a marked deficiency in persistence of the iha mutant in the host compared to that of the wild-type parent strain.
FIG. 6.
Bacterial numbers (log CFU/g) present in the urine, bladder, and kidneys of mice coinfected with wild-type strain UCB34 (♦) and isogenic iha mutant QT686 (▴). Data points represent bacterial adjusted counts (normalized with the input ratio) from tissues and urine isolated from different mice. Horizontal bars represent the means. Numbers near the log 0 limit indicate samples in which no bacteria where detected. n indicates the number of tissue or urine samples harvested from the 15 mice (urine samples were available for only 10 mice). Statistical analysis was done using the Wilcoxon matched-pairs test. P values represent statistical significance between the wild type and the mutant in each sample.
DISCUSSION
In the current report, we have studied Iha from strain UCB34, a representative of the recently emerged ExPEC clonal group designated CGA. Our results demonstrate that Iha functions as a catecholate siderophore receptor, based on growth promotion and uptake experiments using enterobactin and DHBS siderophore by-products. We also investigated the previously reported adhesin phenotype associated with Iha in comparison to other siderophore receptors. Further, coinfection experiments between an iha null mutant and its isogenic parent demonstrated that Iha contributes to colonization and virulence in the mouse urinary tract. Iha thus represents the first characterized virulence factor for a CGA ExPEC strain.
Iha shares high identity/similarity with putative and confirmed siderophore receptors of gram-negative bacteria, including IrgA from Vibrio cholerae. Iha also contains a putative TonB interaction box at the N-terminal region (60), and there is a putative Fur-binding regulatory site in the iha promoter region. Our results demonstrate the previously hypothesized but unconfirmed role of Iha as a siderophore receptor. We investigated this phenotype by determining growth in iron-limited defined minimal medium supplemented with enterobactin and DHBS derivatives (monomers, dimers, and trimers) and by performing uptake experiments using 55Fe-siderophore complexes. Both sets of experiments demonstrated that Iha functions as a siderophore receptor for linear DHBS products and, to a lesser extent, for enterobactin in a catecholate siderophore receptor-negative E. coli K-12 strain (Fig. 1 and 2). However, in these tests, Iha was not as efficient as FepA, the native enterobactin siderophore receptor. Typically, siderophore-mediated iron uptake is TonB dependent. Since Iha possesses a predicted TonB box (60), enterobactin-mediated growth promotion observed in iron-limited medium should be dependent on TonB. Consistent with this, a cloned copy of iha did not restore growth of a catecholate siderophore receptor-negative E. coli K-12 tonB mutant in iron-limited medium, whereas reintroduction of a cloned copy of tonB in this strain, when complemented with either iha or fepA, restored growth (Fig. 3).
Although Iha functions as a catecholate siderophore receptor, an iha mutant of UCB34 grew, as well as its isogenic wild-type parent, in iron-depleted medium (data not shown). This is most likely due to the redundancy of iron acquisition systems present in most ExPEC strains (41). UCB34 is known to possess the enterobactin, aerobactin, and yersiniabactin siderophore systems (Table 1), additional siderophore receptors, and possibly other iron transporters. By contrast, compared to wild-type CGA strain UCB34, the iha mutant demonstrated a significantly reduced capacity to take up iron from an enterobactin-DHBS siderophore mixture, suggesting a direct role in siderophore-mediated iron transport for Iha in the CGA strain. Potential advantages for siderophore receptor redundancy have been previously suggested and include the possibility that multiple acquisition systems may maximize iron uptake, thereby increasing the ability of a pathogenic strain to acquire iron during host infection (54, 56). In addition, some systems might be adapted to specific conditions in different host niches during infection or permit utilization of siderophores produced by other bacterial species within the microbial flora during colonization of host tissues (54, 56).
In addition to transporting specific siderophores, numerous siderophore receptors can also mediate transport of DHBS (34, 53). Interestingly, the ability to take up DHBS by siderophore receptors (FepA, IroN, and Cir) was shown to be required for full virulence of Salmonella enterica serovars in both murine and avian infection models (53). Therefore, DHBS products may be an important means of iron acquisition in vivo, and one role Iha may play in virulence, based on our results, could be the uptake of iron via DHBS products. There might also be some other as-yet-unidentified siderophore(s) produced by certain E. coli strains or other bacteria specific to Iha, and uptake of DHBS products or enterobactin by Iha may be only ancillary. For example, IroN is a urovirulence factor and is the specific siderophore receptor for salmochelins but can also mediate the uptake of enterobactin and DHBS (35, 53, 56).
To further investigate Iha as a potential iron-regulated siderophore receptor, we analyzed iha and fepA expression under different conditions of iron availability. As with fepA, iha expression increased when iron availability was reduced, whereas it was suppressed to below our detection limits when iron was replete in the medium. An analogous increase in expression of fepA in iron-limited medium has been previously reported (36, 48). However, in contrast to the responsiveness of iha expression to iron concentrations, iha expression was not increased in cell adherence assays and is therefore apparently independent of cell-cell contact. Additionally, in accordance with results obtained by Johnson et al. (39), who did not detect Iha from extracts of ExPEC strain CFT073 by Western blotting following growth in LB, no expression of iha by UCB34 was detected in LB medium. Since LB medium is iron rich, it is not surprising that growth in LB represses iha expression.
Fur, the “ferric uptake regulator,” is an established regulator of genes involved in iron homeostasis (1, 24). As we identified a region upstream of the iha start codon that could be a Fur box for iha, we investigated the role of Fur on iha and fepA (as a control) expression by using quantitative RT-PCR. Previous studies have reported an increased expression level for fepA in an E. coli fur mutant compared to that in its parental strain by using microarrays and real-time RT-PCR (67), macroarrays (48), or protein quantification (36). Our results similarly demonstrate a role for fur in the regulation of iha and fepA expression. In the UCB34 fur mutant, iha and fepA expression were constitutive. However, under all growth conditions, the increased expression observed in the fur mutant was considerably higher for fepA than for iha (Fig. 6). Based on these results, Fur regulation of fepA may be more stringent than that of iha, and/or iha may be coregulated by other regulators in addition to Fur. In any case, it is clear that Iha is negatively regulated by iron and that inactivation of the fur gene derepresses iha expression under conditions of iron availability.
Despite the fact that Iha from two archetypal strains (39, 60) has been demonstrated to promote adherence, Iha exhibits no similarity to any characterized adhesins in the available sequence databases (January 2006). Siderophore receptors have not typically been described or investigated as potential adhesins. Based on the considerable identity of Iha with siderophore receptors, we investigated whether three other siderophore receptor-encoding genes (fepA, iroN, and irgA), when cloned in high-copy vectors, could also confer adherence of E. coli ORN172 to uroepithelial cells. Among these siderophore receptors, only iha conferred increased adherence (Fig. 4). These results confirmed those reported previously for Iha obtained from E. coli O157:H7 strain EDL933 and ExPEC strain CFT073 (39, 60) and showed that this phenotype is not simply an artifact broadly characteristic of high-copy siderophore receptor clones in this adhesion model. In addition, when iha from UCB34 was cloned on the medium-copy cloning vector pBR322, the increase in adherence to epithelial cells was similar to what was observed with a high-copy clone. Hence, our results suggest that Iha confers an adhesin function, which is lacking in other siderophore receptors, and is not dependent on high copy number.
The ability of CGA strain UCB34 to colonize the mouse urinary tract was significantly lower for a UCB34 iha null mutant than for the wild-type strain, as demonstrated by competitive infection experiments. This result confirmed the importance of iha in vivo. Additionally, Iha is the first virulence factor to be demonstrated as important for a representative of CGA, or for any phylogenetic group D strain, in the mouse UTI model. In conclusion, Iha may be a dual-function urovirulence factor for E. coli CGA strains and other pathogenic E. coli. Whether the siderophore receptor activity, the adhesin phenotype, or both are important for Iha's demonstrated enhancement of in vivo persistence within the urinary tract remains to be defined.
Acknowledgments
We give kind thanks to A. R. Manges, E. E. Wyckoff, K. Hantke, P. E. Orndorff, and M. Cellier for the gift of strains or plasmids. We give special thanks to F. Lépine for the use of the mass spectrometer.
S.L. was funded by a Fonds de recherches en Santé du Québec (FRSQ) master's level scholarship. M.C. and M.S. were funded by Fondation Armand-Frappier scholarships. Funding for this project was provided by the Canadian Institutes of Health Research (CIHR) and the Canadian Foundation for Innovation (C.M.D.) and by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.).
Editor: A. D. O'Brien
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 1997. Applied Biosystems user bulletin 2. The Perkin-Elmer Corp., Norwalk, Conn.
- 3.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 4.Bauer, R. J., L. Zhang, B. Foxman, A. Siitonen, M. E. Jantunen, H. Saxen, and C. F. Marrs. 2002. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection—usp, iha, and iroNE. coli. J. Infect. Dis. 185:1521-1524. [DOI] [PubMed] [Google Scholar]
- 5.Baumler, A. J., T. L. Norris, T. Lasco, W. Voight, R. Reissbrodt, W. Rabsch, and F. Heffron. 1998. IroN, a novel outer membrane siderophore receptor characteristic of Salmonella enterica. J. Bacteriol. 180:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beall, B., and T. Hoenes. 1997. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology 143:135-145. [DOI] [PubMed] [Google Scholar]
- 7.Berner, I., M. Greiner, J. Metzger, G. Jung, and G. Winkelmann. 1991. Identification of enterobactin and linear dihydroxybenzoylserine compounds by HPLC and ion spray mass spectrometry (LC/MS and MS/MS). Biol. Met. 4:113-118. [DOI] [PubMed] [Google Scholar]
- 8.Bingen-Bidois, M., O. Clermont, S. Bonacorsi, M. Terki, N. Brahimi, C. Loukil, D. Barraud, and E. Bingen. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70:3216-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 10.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 11.Bower, J. M., D. S. Eto, and M. A. Mulvey. 2005. Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 6:18-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun, V., and M. Braun. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529:78-85. [DOI] [PubMed] [Google Scholar]
- 13.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daigle, F., J. Y. Hou, and J. E. Clark-Curtiss. 2002. Microbial gene expression elucidated by selective capture of transcribed sequences (SCOTS). Methods Enzymol. 358:108-122. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N.T.Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dozois, C. M., M. Dho-Moulin, A. Bree, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eisenhauer, H. A., S. Shames, P. D. Pawelek, and J. W. Coulton. 2005. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J. Biol. Chem. 280:30574-30580. [DOI] [PubMed] [Google Scholar]
- 22.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 23.Emody, L., M. Kerenyi, and G. Nagy. 2003. Virulence factors of uropathogenic Escherichia coli. Int. J. Antimicrob. Agents 22(Suppl. 2):29-33. [DOI] [PubMed] [Google Scholar]
- 24.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finer, G., and D. Landau. 2004. Pathogenesis of urinary tract infections with normal female anatomy. Lancet Infect. Dis. 4:631-635. [DOI] [PubMed] [Google Scholar]
- 26.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S-13S. [DOI] [PubMed] [Google Scholar]
- 27.France, A. M., K. M. Kugeler, A. Freeman, C. A. Zalewski, M. Blahna, L. Zhang, C. F. Marrs, and B. Foxman. 2005. Clonal groups and the spread of resistance to trimethoprim-sulfamethoxazole in uropathogenic Escherichia coli. Clin. Infect. Dis. 40:1101-1107. [DOI] [PubMed] [Google Scholar]
- 28.Gibson, F., and D. I. Magrath. 1969. The isolation and characterization of a hydroxamic acid (aerobactin) formed by Aerobacter aerogenes 62-I. Biochim. Biophys. Acta 192:175-184. [DOI] [PubMed] [Google Scholar]
- 29.Gibson, F., and J. Pittard. 1968. Pathways of biosynthesis of aromatic amino acids and vitamins and their control in microorganisms. Bacteriol. Rev. 32:465-492. [PMC free article] [PubMed] [Google Scholar]
- 30.Gordon, D. M., S. E. Stern, and P. J. Collignon. 2005. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151:15-23. [DOI] [PubMed] [Google Scholar]
- 31.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 179:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta, K., T. M. Hooton, and W. E. Stamm. 2001. Increasing antimicrobial resistance and the management of uncomplicated community-acquired urinary tract infections. Ann. Intern. Med. 135:41-50. [DOI] [PubMed] [Google Scholar]
- 33.Haag, H., K. Hantke, H. Drechsel, I. Stojiljkovic, G. Jung, and H. Zahner. 1993. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J. Gen. Microbiol. 139:2159-2165. [DOI] [PubMed] [Google Scholar]
- 34.Hantke, K. 1990. Dihydroxybenzoylserine—a siderophore for E. coli. FEMS Microbiol. Lett. 55:5-8. [DOI] [PubMed] [Google Scholar]
- 35.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Higgs, P. I., R. A. Larsen, and K. Postle. 2002. Quantification of known components of the Escherichia coli TonB energy transduction system: TonB, ExbB, ExbD and FepA. Mol. Microbiol. 44:271-281. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, J. R., T. Berggren, and J. C. Manivel. 1992. Histopathologic-microbiologic correlates of invasiveness in a mouse model of ascending unobstructed urinary tract infection. J. Infect. Dis. 165:299-305. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, J. R., and J. J. Brown. 1996. Defining inoculation conditions for the mouse model of ascending urinary tract infection that avoid immediate vesicoureteral reflux yet produce renal and bladder infection. J. Infect. Dis. 173:746-749. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, J. R., S. Jelacic, L. M. Schoening, C. Clabots, N. Shaikh, H. L. Mobley, and P. I. Tarr. 2005. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 73:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson, J. R., A. R. Manges, T. T. O'Bryan, and L. W. Riley. 2002. A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 359:2249-2251. [DOI] [PubMed] [Google Scholar]
- 41.Johnson, J. R., A. C. Murray, M. A. Kuskowski, S. Schubert, M. F. Prere, B. Picard, R. Colodner, and R. Raz. 2005. Distribution and characteristics of Escherichia coli clonal group A. Emerg. Infect. Dis. 11:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson, J. R., T. T. O'Bryan, P. Delavari, M. Kuskowski, A. Stapleton, U. Carlino, and T. A. Russo. 2001. Clonal relationships and extended virulence genotypes among Escherichia coli isolates from women with a first or recurrent episode of cystitis. J. Infect. Dis. 183:1508-1517. [DOI] [PubMed] [Google Scholar]
- 43.Johnson, J. R., T. A. Russo, P. I. Tarr, U. Carlino, S. S. Bilge, J. C. Vary, Jr., and A. L. Stell. 2000. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroNE. coli, among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 68:3040-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanamaru, S., H. Kurazono, S. Ishitoya, A. Terai, T. Habuchi, M. Nakano, O. Ogawa, and S. Yamamoto. 2003. Distribution and genetic association of putative uropathogenic virulence factors iroN, iha, kpsMT, ompT and usp in Escherichia coli isolated from urinary tract infections in Japan. J. Urol. 170:2490-2493. [DOI] [PubMed] [Google Scholar]
- 45.Lundrigan, M. D., and R. J. Kadner. 1986. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J. Biol. Chem. 261:10797-10801. [PubMed] [Google Scholar]
- 46.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 47.Martinez, J. J., M. A. Mulvey, J. D. Schilling, J. S. Pinkner, and S. J. Hultgren. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19:2803-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McHugh, J. P., F. Rodriguez-Quinones, H. Abdul-Tehrani, D. A. Svistunenko, R. K. Poole, C. E. Cooper, and S. C. Andrews. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 49.Mey, A. R., E. E. Wyckoff, A. G. Oglesby, E. Rab, R. K. Taylor, and S. M. Payne. 2002. Identification of the Vibrio cholerae enterobactin receptors VctA and IrgA: IrgA is not required for virulence. Infect. Immun. 70:3419-3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mount, D. W. 1977. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proc. Natl. Acad. Sci. USA 74:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 52.Provence, D. L., and R. Curtiss III. 1992. Role of crl in avian pathogenic Escherichia coli: a knockout mutation of crl does not affect hemagglutination activity, fibronectin binding, or curli production. Infect. Immun. 60:4460-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rabsch, W., U. Methner, W. Voigt, H. Tschape, R. Reissbrodt, and P. H. Williams. 2003. Role of receptor proteins for enterobactin and 2,3-dihydroxybenzoylserine in virulence of Salmonella enterica. Infect. Immun. 71:6953-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo, T. A., U. B. Carlino, and J. R. Johnson. 2001. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69:6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabri, M., S. Léveillé, and C. M. Dozois. A plasmid-encoded SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology 152:745-758. [DOI] [PubMed]
- 58.Schmittgen, T. D., B. A. Zakrajsek, A. G. Mills, V. Gorn, M. J. Singer, and M. W. Reed. 2000. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Anal. Biochem. 285:194-204. [DOI] [PubMed] [Google Scholar]
- 59.Song, Y., Z. Tong, J. Wang, L. Wang, Z. Guo, Y. Han, J. Zhang, D. Pei, D. Zhou, H. Qin, X. Pang, Y. Han, J. Zhai, M. Li, B. Cui, Z. Qi, L. Jin, R. Dai, F. Chen, S. Li, C. Ye, Z. Du, W. Lin, J. Wang, J. Yu, H. Yang, J. Wang, P. Huang, and R. Yang. 2004. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res. 11:179-197. [DOI] [PubMed] [Google Scholar]
- 60.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Warren, J. W., E. Abrutyn, J. R. Hebel, J. R. Johnson, A. J. Schaeffer, and W. E. Stamm. 1999. Guidelines for antimicrobial treatment of uncomplicated acute bacterial cystitis and acute pyelonephritis in women. Infectious Diseases Society of America (IDSA). Clin. Infect. Dis. 29:745-758. [DOI] [PubMed] [Google Scholar]
- 62.Winkelmann, G., A. Cansier, W. Beck, and G. Jung. 1994. HPLC separation of enterobactin and linear 2,3-dihydroxybenzoylserine derivatives: a study on mutants of Escherichia coli defective in regulation (fur), esterase (fes) and transport (fepA). Biometals 7:149-154. [DOI] [PubMed] [Google Scholar]
- 63.Woodall, L. D., P. W. Russell, S. L. Harris, and P. E. Orndorff. 1993. Rapid, synchronous, and stable induction of type 1 piliation in Escherichia coli by using a chromosomal lacUV5 promoter. J. Bacteriol. 175:2770-2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young, I. G., and F. Gibson. 1979. Isolation of enterochelin from Escherichia coli. Methods Enzymol. 56:394-398. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, J. P., and S. Normark. 1996. Induction of gene expression in Escherichia coli after pilus-mediated adherence. Science 273:1234-1238. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, L., B. Foxman, and C. Marrs. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, Z., G. Gosset, R. Barabote, C. S. Gonzalez, W. A. Cuevas, and M. H. Saier, Jr. 2005. Functional interactions between the carbon and iron utilization regulators, Crp and Fur, in Escherichia coli. J. Bacteriol. 187:980-990. [DOI] [PMC free article] [PubMed] [Google Scholar]