Abstract
Of all bacteria, Bartonella quintana has the highest reported in vitro hemin requirement, yet an explanation for this remains elusive. To produce diseases such as trench fever, endocarditis, and bacillary angiomatosis, B. quintana must survive and replicate in the disparate environments of the Pediculus humanus corporis (body louse) gut and the human vasculature. We previously identified a five-member family of hemin binding proteins (Hbps) synthesized by B. quintana that bind hemin on the outer surface but share no similarity to known bacterial heme receptors. In the present study, we examine the transcription, regulation, and synthesis of this virulence factor family by cultivation of the bacterium in environments that simulate natural heme, oxygen, and temperature conditions encountered in the host and insect vector. First, quantitative real-time PCR data show that hbpC expression is regulated by temperature, where a >100-fold increase in transcript quantity was seen at 30°C relative to 37°C, suggesting that HbpC synthesis would be greatest in the cooler temperature of the louse. Second, cultivation at human bloodstream oxygen concentration (5% relative to 21% atmospheric) significantly decreases the transcript quantity of all hbp genes, indicating that expression is influenced by O2 and/or reactive oxygen species. Third, a differential expression pattern within the hbp family is revealed when B. quintana is grown in a range of hemin concentrations: subgroup I (hbpC and hbpB) predominates in a simulated louse environment (high heme), and subgroup II (hbpA, hbpD, and hbpE) is preferentially expressed in a simulated human background (low heme). By using two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotting, and matrix-assisted laser desorption ionization—time of flight mass spectrometry fingerprinting, we demonstrate that synthesis of HbpA correlates with hbpA transcript increases observed at low hemin concentrations. Finally, an hbpA promoter-lacZ reporter construct in B. quintana demonstrates that a transcriptional regulator(s) is controlling the expression of hbpA through a cis-acting regulatory element located in the hbpA promoter region.
Trench fever, the common name for the acute febrile syndrome associated with Bartonella quintana infection, has affected millions of people during war and is presently reemerging in inner cities throughout the world (28, 36) and in AIDS patients. Chronic manifestations of persistent infection by this α-proteobacterium include protracted bacteremia, endocarditis, bacillary angiomatosis, and bacillary peliosis (20, 31). Although B. quintana has been found in small mammals (24), ticks (11), and fleas (41), maintenance in nature is thought to be restricted to humans and body lice (Pediculus humanus corporis). Transmission to humans occurs when louse fecal matter or a crushed louse containing the bacterium is introduced into the bloodstream by breaches in the integument, usually by the itching caused by louse infestation. Living between the clothing and the skin, body lice normally take several meals per day and acquire B. quintana by imbibing the blood of a bacteremic host (9). Unhygienic, overcrowded conditions disseminate infected lice throughout the population and can quickly result in an epidemic.
Of all bacteria, B. quintana has the greatest known requirement for exogenous heme (33, 34, 48). Heme consists of an iron atom contained in the center of a large heterocyclic organic ring called a porphyrin. The Fe3+ oxidation product of heme is called hemin. It is generally accepted that this extraordinary supplement requirement (20 to 40 μg/ml of medium) is similar for all Bartonella species, and erythrocytes, hemoglobin, or hemin is essential for in vitro cultivation (8). Since combinations of iron and porphyrin cannot substitute for heme in Bartonella cultivation, several researchers have hypothesized that high levels of heme are necessary for one or more of the following: a source of iron (10, 42), a precursor for synthesis of porphyrin-containing proteins (34), and a hydrogen peroxide-detoxifying system (33).
To generate disease, B. quintana must survive immune attack, adapt to host and vector environments, and proliferate throughout the human-louse-human cycle. Free heme is quite rare in humans (6), whereas potentially toxic levels are frequently generated following blood meal digestion within the louse gut (9, 19, 37, 47). Considering its extraordinary heme requirement, it is obvious that heme acquisition mechanisms are essential for replication and, ultimately, the pathogenesis of B. quintana.
Previously, we discovered a family of hemin binding proteins (HbpA, HbpB, HbpC, HbpD, and HbpE) synthesized by B. quintana that serves as hemin receptors yet shares no similarity to known bacterial heme binding proteins (10, 32). In the present study, we examine the expression, regulation, and synthesis of this virulence factor family in conditions that reflect body louse and human environments. We report that hemin, oxygen, and temperature influence the hbp transcript profile in a differential and coordinated manner.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Table 1 describes the bacterial strains used in this study. B. quintana strain JK31 is a recent low-passage virulent human isolate and was a generous gift from Jane Koehler (University of California at Davis). The Brucella agar-hemin (BA-H) cultivation media for Bartonella consisted of Brucella broth (Becton Dickinson, Sparks, MD) base containing 1.5% Difco agar (Becton Dickinson) and supplemented with hemin chloride (Calbiochem, San Diego, CA). Hemin chloride (hereafter called hemin) was dissolved in 0.02 N NaOH to a final concentration of 5 mg/ml and was added to the media in a range of 0 mM to 5 mM, where 0.15 mM was the typical (control) hemin concentration. Bartonella organisms were routinely grown in a humidified 5% CO2 incubator at 37°C and approximately 21% O2. The growth temperature was modified by growing at 30°C in a humidified 5% CO2 incubator. The O2 concentration of the environment was lowered by cultivation in a Pyrex Vacuum Desiccator jar (Corning, St. Louis, MO) where atmospheric air was replaced with a blood-gas mixture (5% CO2, 5% O2, 90% N2; NorLab, Boise, ID). Atmospheric gasses were evacuated by vacuum (to 50 cm Hg), and blood gas was allowed to fill the desiccator jar to 10 lb/in2. After repeating the evacuation-replacement procedure seven times, the internal pressure was equalized to atmospheric pressure, and the jar was placed at 37°C. Approximately 96 h was required for Bartonella to reach mid-log phase. For electroporation-mediated transformation, strain JK31 was plated on heart infusion broth base (Becton Dickinson) containing 1.5% Difco agar and supplemented with 2% (vol/vol) sheep serum and 4% (vol/vol) defibrinated sheep blood (Quad Five, Ryegate, MT). Filter-sterilized kanamycin sulfate was added to a final concentration of 25 μg/ml for selection of transformants.
TABLE 1.
Bacterial strains, plasmids, and primers
| Strains, plasmids, and primers | Relevant characteristic(s) | Source or reference |
|---|---|---|
| B. quintana | ||
| JK31 | Low-passage virulent human isolate | J. Koehler |
| JK31 pHPRO+ LACZ+ | JK31 with pHPRO+ LACZ+ | This study |
| JK31 pHPRO− LACZ+ | JK31 with pHPRO− LACZ+ | This study |
| JK31 pHPRO+ LACZ− | JK31 with pHPRO+ LACZ− | This study |
| E. coli | ||
| DH5α | Host strain for cloning | Gibco-BRL |
| TOP10 F′ | Host strain for cloning | Invitrogen |
| Plasmids | ||
| pHBP-CMV | pBK-CMV with 3.5-kbp insert containing hbpA and flanking sequence | 32 |
| pCR2.1-TOPO | TA cloning plasmid for PCR products | Invitrogen |
| pCR2.1-HPRO | pCR2.1-TOPO with 240-bp hbpA promoter region | This study |
| pUJ9 | Promoterless ′lacZ fusion plasmid | 12 |
| pUJ-HPRO | 302-bp EcoRV/BamHI fragment from pCR2.1-HPRO into SmaI/BamHI site of pUJ9 | This study |
| pBBR1MCS-2 | Shuttle vector for Bartonella | 22 |
| pHPRO+ LACZ+ | Reporter construct: pBBR1MCS-2 with hbpA-promoted ′lacZ | This study |
| pHPRO- LACZ+ | Control plasmid 1: pHPRO+ LACZ+ minus hbpA-promoter | This study |
| pHPRO+ LACZ- | Control plasmid 2: pHPRO+ LACZ+ minus ′lacZ | This study |
| Primers | ||
| HPRO FOR | 5′-CAGGCAGAATATCGTTACAGC | This study |
| HPRO REV | 5′-AAACTTTGCTCCTTTATTTATGAAG | This study |
| PBBR FOR+AvrII | 5′-ATCCTAGGGCATAAAGTGTAAAGCCTGGGGT | This study |
| PBBR REV+AscI | 5′-ATGGCGCGCCCTGGTGCTACGCCTGAATAAGTG | This study |
| HPROLAC REV+AvrII | 5′-ATCCTAGGACATCCAGAGGCACTTCACCG | This study |
| HPROLAC FOR+AscI | 5′-ATGGCGCGCCCAGGCAGAATATCGTTACACG | This study |
| LINK FOR+AscI | 5′-ATGGCGCGCCTAGGCAATCGATGAATTCAT | This study |
| LINK REV+EcoRI | 5′-ATGAATTCATCGATTGCCTAGGCGCGCCAT | This study |
| HSEQ | 5′-ACCTCGCTAACGGATTCAC | This study |
| LACZ FOR | 5′-AGTTCTGTATGAACGGTCTGGTC | This study |
| LACZ REV | 5′-AGGTATTCGCTGGTCACTTCG | This study |
| LACZ PROBE | 5′-CCGACCGCACGCCGCATCCAG | This study |
Escherichia coli strains TOP10 F′ and DH5α, employed in cloning experiments, were cultivated with Luria-Bertani medium using standard concentrations of antibiotic supplements (5).
Nucleic acid isolation, purification, and manipulation.
RNA used for quantitative real-time PCR (qRT-PCR) analysis was isolated by using the RiboPure-Bacteria kit with Turbo-DNaseI treatment (Ambion, Austin, TX) and a FastPrep bead homogenizer (Q-Biogene, Carlsbad, CA) per the manufacturers' instructions. Primers and probes used for qRT-PCR analysis of the hbp family were previously described (32). The lacZ primer-probe set was designed with Beacon Designer version 4.0 (Bio-Rad, Hercules, CA). The dual-labeled lacZ probe was synthesized with fluorescent tags as described for the hbp family, where 5-carboxyfluorescein and N,N′,N′-tetramethyl-6-carboxyrhodamine were covalently linked to the 5′ and 3′ ends, respectively (Sigma-Genosys, Woodlands, TX). The lacZ primer pair (Table 1) was synthesized by Applied Biosystems (ABI, Foster City, CA).
Plasmids and primers used in this study are described in Table 1. Standard PCR and cloning procedures were employed for the construction of plasmids (5) with the exception of the Expand Long Template PCR system (Roche Diagnostics, Indianapolis, IN), utilized for high-fidelity amplicon production in inverse PCR-mediated cloning per the manufacturer's instructions. For routine cloning, the Perfectprep Plasmid Mini kit (Eppendorf, Hamburg, Germany) and the QIAquick Spin kit (QIAGEN, Valencia, CA) were used for plasmid isolation and DNA purification, respectively. The Wizard Midiprep kit (Promega, Madison, WI) was used to purify plasmids employed in electroporation-mediated transformation of B. quintana. Bacterial genomic DNA was prepared with a DNeasy Tissue kit (QIAGEN) per the manufacturer's instructions. Quantification of nucleic acids was accomplished by spectrophotometric analysis using a Spectronic Genesys 2 (Milton Roy, Rochester, NY).
Construction of HbpA reporter construct.
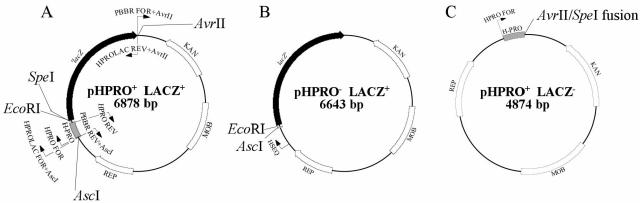
Diagrams of the reporter plasmid pHPRO+ LACZ+ and two control plasmids (pHPRO- LACZ+ and pHPRO+ LACZ−) are shown in Fig. 6. Primers HPRO FOR and HPRO REV were designed to generate a PCR fragment containing the promoter region of hbpA (HPRO). Using pHBP-CMV (32) as the template, the resulting amplicon was cloned into pCR2.1-TOPO per the manufacturer's instructions (Invitrogen, Carlsbad, CA), resulting in pCR2.1-HPRO. Sequence analysis of both strands with M13 universal primers verified that pCR2.1-HPRO contained the HPRO, 240 bp immediately 5′ to the hbpA start site, and it was identical to published sequences (1, 32).
FIG. 6.
A. Reporter construct (pHPRO+ LACZ+) containing the 240-bp hbpA promoter region, HPRO, fused to ′lacZ. Primers used for inverse PCR cloning and sequence analysis are indicated with small arrows and are described in Table 1. Restriction endonuclease sites and plasmid ORFs (KAN, kanamycin resistance cassette; MOB, mobilization gene; REP, origin of replication) are also illustrated. B. Control plasmid 1 derived from the reporter construct by removing the EcoRI-AscI fragment containing the HPRO. C. Control plasmid 2 derived from the reporter construct by removal of the SpeI-AvrII fragment containing ′lacZ.
The HPRO was then recloned into the promoterless ′lacZ fusion plasmid, pUJ9 (12). Specifically, the 302-bp EcoRV/BamHI fragment of pCR2.1-HPRO was ligated to SmaI/BamHI-restricted pUJ9, resulting in pUJ-HPRO. Orientation of HPRO relative to ′lacZ was confirmed by sequence analysis of pUJ-HPRO with the HPRO FOR primer. Finally, inverse PCR-mediated cloning was utilized to move the HPRO-′lacZ fragment into the Bartonella shuttle vector, pBBR1MCS-2 (7, 22). Specifically, primers (PBBR FOR+AvrII and PBBR REV+AscI) were designed to amplify the entire pBBR1MCS-2 plasmid while introducing AvrII and AscI restriction sites into the termini of this 4,604-bp amplicon. The 2,258-bp HPRO-′lacZ fragment was amplified from pUJ-HPRO with primers (HPROLAC REV+AvrII and HPROLAC FOR+AscI) that introduced the same restriction sites into the termini of this product. The pBBR1MCS-2 and HPRO-′lacZ amplicons were digested (AvrII/AscI) and ligated, resulting in the 6,878-bp reporter construct pHPRO+ LACZ+. The construct was verified by sequence analysis with primers HPRO FOR and HPROLAC REV+AvrII.
Two control plasmids were also generated with this reporter plasmid lacking either the hbpA promoter (pHPRO− LACZ+) or ′lacZ (pHPRO+ LACZ−). First, the hbpA promoter was removed from pHPRO+ LACZ+ by digestion (EcoRI/AscI) and was replaced with a 15-bp linker formed by hybridization of primers (LINK FOR+AscI and LINK REV+EcoRI) containing corresponding restriction sites. This 6,643-bp plasmid was confirmed by sequence analysis with primer HSEQ and was termed pHPRO− LACZ+. For the second control plasmid, ′lacZ was removed from pHPRO+ LACZ+ by digestion (AvrII/SpeI) and religation, resulting in pHPRO+ LACZ−, and this was confirmed by sequence analysis using HPRO FOR primer.
Electroporation-mediated transformation of B. quintana JK31.
Transformation of B. quintana JK31 was accomplished by methods similar to those we previously described for B. bacilliformis (7). Briefly, strain JK31 (in vitro passage 3) was harvested, washed in 10% glycerol, and diluted to 3 × 1010 cells/ml. A volume of 44 μl of this suspension was combined with 5.4 to 43.2 μg plasmid DNA in a 2-mm-gap electroporation cuvette (BTX, Holliston, MA) and pulsed with a GenePulser (Bio-Rad) at 2.5 kV, 25 μF, and 400 Ω. Kanamycin-resistant clones were verified as stable transformants by isolation of plasmid DNA and subsequent restriction fragment length polymorphism analysis.
Nucleotide sequencing and analysis.
DNA was sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction kit (ABI) and an automated DNA sequencer (ABI3130x1). Sequence analysis was accomplished with MacVector Software version 7.2.2 (Accelyrys, San Diego, CA).
qRT-PCR of hbp and ′lacZ transcripts.
The MyiQ Real-Time PCR Detection System (Bio-Rad) was used with One-Step RT-PCR Mastermix, Multiscribe, and RNase inhibitor reagents (ABI), where each reaction included 0.7 ng template RNA, 67 ng probe, and 167 ng of each primer in a 25-μl volume in a 96-well format. Thermal cycling was 50°C for 30 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Raw data were analyzed by Optical System Software version 1.0 (Bio-Rad). Calculation of fold differences in hpb or lacZ mRNA transcript levels between two environmental conditions was accomplished by using the comparative cycle threshold (CT) method (4, 26) (part #4371095; ABI). Specifically, triplicate qRT-PCRs using RNA derived from bacteria subjected to each condition were used to calculate the 2−ΔΔCt by normalizing to 16S rRNA. Independent determinations of fold differences were used to calculate standard deviation and demonstrate reproducibility of the results.
Two-dimensional electrophoresis and immunoblotting.
Bartonella organisms were harvested from culture plates with a sterile razor blade into HEPES buffer (20 mM HEPES, 50 mM NaCl, 4°C [pH 7.5]) supplemented with Complete Mini Protease Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany) per the manufacturer's instructions. After washing the bacteria three times in this solution (using centrifugations of 4,620 × g for 10 min at 4°C), cell lysis was achieved by three passes through a French Press Cell Disrupter (Thermo Electron Corp., Waltham, MA) at 12,000 lb/in2. The preparation was cleared of cellular debris by centrifugation (10,000 × g, 15 min, 4°C), and total protein was quantified with a bicinchoninic acid kit (Pierce, Rockford, IL). Ultracentrifugation (100,000 × g, 1.5 h, 4°C) was used to enrich for insoluble outer-membrane proteins when isolating protein spots for identification by mass spectrometry. A range of 60 to 200 μg of protein was precipitated with 3 volumes of acetone supplemented with 13.3% trichloroacetic acid (Sigma, St. Louis, Mo.) and 0.05% 2-mercaptoethanol (2-ME) (Fisher, Fair Lawn, NJ) and incubating for 1.5 h at −20°C. Proteins were pelleted by centrifugation (1,310 × g, 15 min, 4°C) and washed with acetone containing 0.07% 2-ME. The pellet was air dried for 2 min at 25°C and resuspended in 200 μl rehydration buffer (7 M urea, 2 M thiourea, 4% Triton X-100, 0.62% dl-dithiothreitol, 0.2% Bio-Lyte 3/10 ampholyte [Bio-Rad], 0.002% bromophenol blue, 0.2 mM Tris-HCl). The rehydration buffer was prepared with Ultra Pure H2O (Ambion), and all reagents were PlusOne grade (Amersham, Piscataway, NJ), with the exception of the ampholyte. Samples were then vortexed (2 min), incubated at 25°C (50 min), and centrifuged (16,000 × g, 10 min, 25°C) to pellet insoluble debris. The supernatant was loaded into the isoelectric focusing (IEF) tray of a Protean IEF Cell (Bio-Rad) followed by a Ready Strip IPG strip (11 cm, pH 3 to 10 nonlinear; Bio-Rad) and finally overlaid with mineral oil (Bio-Rad). Focusing was achieved by the following cycles: active rehydration for 12 h (50 V, 20°C), 250 V for 15 min, 8,000 V for 2.5 h, and 8,000 V until a total of 35,000 V · h was reached. Following a brief equilibration of the strips (per the manufacturer's instructions), a standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (12.5% [wt/vol] acrylamide) was used to separate the focused proteins. A Silver Quest stain kit (Invitrogen) was used to visualize spots, except when mass spectrometry followed separation, where staining was accomplished with 0.1% (wt/vol) Coomassie brilliant blue (CBB).
Immunoblots were prepared by transferring proteins separated by SDS-PAGE to Nitropure nitrocellulose membranes (0.45-μm pore size; Osmonics, Minnetonka, Minn.) by the methods of Towbin et al. (46). Immunoblots were probed with rabbit anti-HbpA antibody (10) at a 1:6,666 dilution and developed using goat-anti-rabbit:horseradish peroxidase and 4-chloro-1-naphthol (Sigma) using standard procedures (5).
Protein identification by mass spectrometry.
A total of 200 μg of an outer-membrane enriched fraction was focused, separated, and stained with CBB as described above. Spots corresponding to the predicted molecular weight and pI of the Hbps were excised from the gel, transferred to siliconized microcentrifuge tubes, destained with 50% acetonitrile-25 mM NH4HCO3 (at 25°C until colorless), and dried in a speedvac. Sequencing Grade Modified Trypsin (Promega) was prepared per the manufacturer's instructions and diluted to 12.5 ng/μl in 25 mM NH4HCO3. Dried gel fragments were reswelled in the trypsin solution (4°C, 20 min), resuspended in 25 mM NH4HCO3, and digested for 16 h at 37°C. Peptides were extracted from the gel fragments with 0.1% trifluoracetic acid (TFA)-60% methanol, dried in a speedvac, and resuspended in 4 μl 2.5% TFA. Finally, peptides were cleaned and concentrated using Omix C18 pipette tips (Varian, Palo Alto, CA) per the manufacturer's instructions, analyzed using the Voyager-DE PRO MALDI-TOF BioSpectrometery Workstation (ABI), and fingerprinted with MASCOT software (http://www.matrixscience.com) (38).
RESULTS
Growth of B. quintana at “louse-like temperature” results in a dramatic increase in hbpC.
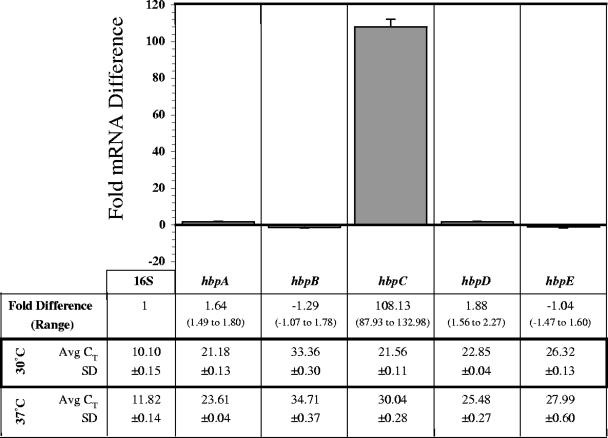
To determine if the hbp transcript profiles were influenced by temperature, we compared the effects of growth at louse-like temperature (30°C) (27, 29) to those at human bloodstream temperature (37°C). B. quintana strain JK31 was grown on BA-H (0.15 mM hemin) at 37°C and 30°C. RNA was prepared from bacteria cultivated at each temperature, and qRT-PCR was performed as described above. To clarify the methods and compound calculations of the comparative method of qRT-PCR (4) using this first experiment, a brief description follows. The threshold cycle (CT) is the cycle number at which the fluorescence signal generated by PCR crosses a threshold just above background and begins exponential increase. The detection system (MyiQ; Bio-Rad) statistically determines the threshold value during each run using a series of computations based on real-time output of fluorescent signal (data not shown). The average of three CT values (Avg CT) was determined for each target (16S as well as hbpA to hbpE) using RNA preparations from 30°C and 37°C and is listed in Fig. 1. The Avg CT value represents the quantity of a specific target transcript in a given RNA sample, where the lower the Avg CT the more abundant the specific transcript. In this experiment, the 16S transcript is the most common and hbpB is the least common in both 30°C and 37°C RNA samples. Considering only the hbp genes, the hierarchy of transcripts from the 30°C RNA sample is hbpA > hbpC > hbpD > hbpE > hbpB, and from the 37°C preparation it is hbpA > hbpD > hbpE > hbpC > hbpB.
FIG. 1.
Fold differences in hbp mRNA following growth at louse-like temperature (30°C) relative to that at human temperature (37°C). At 96 h, the amount of target hbp mRNA transcript in 30°C preparations was normalized to the amount of 16S rRNA and is relative to the quantity of that particular hbp mRNA transcript in 37°C preparations. A representative experiment is shown, where CT values were determined in triplicate. SD bars were generated by comparing fold differences to those obtained from a second independent triplicate determination.
The Avg CT values are useful for determination of transcript quantity in a single sample. However, when determination of relative quantities between RNA samples is necessary, the Avg CTs and corresponding standard deviations (SDs) are used to calculate the fold difference using the comparative method. The fold difference is the amount of target mRNA normalized to an endogenous reference and relative to a calibrator. In this experiment, the amount of a specific hbp mRNA transcript in the 30°C preparations (target) was normalized to the amount of 16S rRNA (endogenous reference) and is relative to the quantity of that particular hbp mRNA transcript in the 37°C preparations (calibrator). Fold differences calculated by this method for each hbp transcript are derived from the given Avg CTs and corresponding SDs and are listed with resultant range limits (Fig. 1). To illustrate the reproducibility of these transcript profile determinations, the bar graph portion of Fig. 1 depicts fold differences calculated from the given data set (Avg CTs and corresponding SDs) together with a second independently derived data set where error bars show the SD between these independent fold difference determinations.
The more than 108-fold difference in hbpC transcript quantity at 30°C is the largest fold difference of any environmental condition tested in this study, and it shows that hbpC transcription is temperature regulated. Furthermore, this significant increase in hbpC expression at 30°C suggests that HbpC function may be important in the louse. The fold difference of the other four hbp transcripts is statistically insignificant, suggesting that the expression of these hbp genes is not temperature regulated and not necessarily specific to the louse.
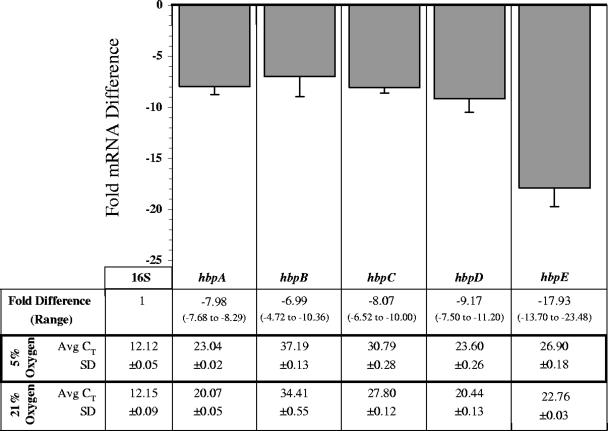
Growth at “bloodstream” oxygen levels results in down-regulation of all hbp genes.
As the combination of O2 and heme (or iron) can result in the formation of toxic reactive oxygen species (ROS) (6, 18), we were curious to see if hbp transcript profiles are influenced by oxygen. B. quintana strain JK31 was grown on BA-H (0.15 mM hemin) at 37°C in an environment containing 5% O2 or 21% O2. RNA was prepared from bacteria cultivated at each O2 concentration, and qRT-PCR was performed as described above. Average CTs and corresponding SDs are listed in Fig. 2, where the hierarchy of hbp transcripts is the same at both O2 concentrations (hbpA > hbpD > hbpE > hbpC > hbpB). Fold differences calculated from these Avg CT values are listed in Fig. 2 with corresponding range limits. In this experiment, the amount of a specific hbp mRNA transcript derived from 5% O2-grown bacteria (target) was normalized to the amount of 16S rRNA (endogenous reference) and is relative to the quantity of that particular hbp mRNA transcript from a 21% O2-grown preparation (calibrator). The bar graph portion of Fig. 2 shows the fold differences calculated from the given data set together with a second independently derived data set, where error bars show the SD between these independent fold difference determinations. The fold differences of all hbp transcripts are significantly decreased when bacteria are grown in a 5% O2 environment relative to 21% O2, indicating that hbp expression is influenced by oxygen and/or ROS.
FIG. 2.
Fold differences in hbp mRNA following growth at human bloodstream oxygen concentration (5% O2) relative to that of the routine in vitro culture environment (21% O2). At 96 h, the amount of target hbp mRNA transcript in 5% O2 preparations was normalized to the amount of 16S rRNA and is relative to the quantity of that particular hbp mRNA transcript from 21% O2 preparations. A representative experiment is shown, where CT values were determined in triplicate. SD bars were generated by comparing fold differences to those obtained from a second independent triplicate determination.
Variable hemin levels reveal expression subgroups of the hbp genes.
B. quintana must survive and replicate in environments containing markedly different heme concentrations throughout the infectious cycle. Humans maintain very low free heme levels by sequestering it within hemopexin, albumin, and hemoglobin complexes (6). On the contrary, body lice feed several times per day and digest their blood meal within minutes, exposing B. quintana to very high levels of potentially toxic heme with each meal (9, 47). Thus, we were interested in examining hbp expression at low (human) and high (louse) heme levels.
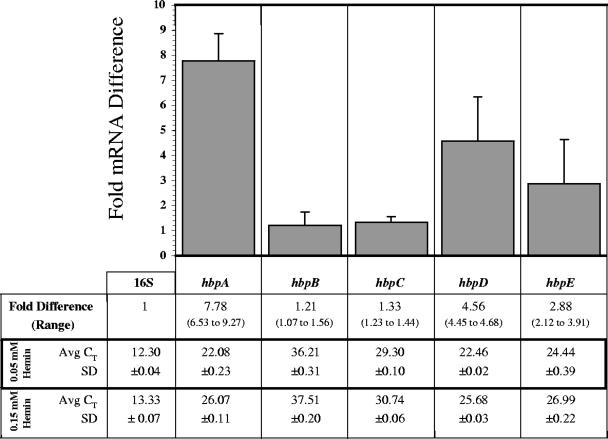
B. quintana strain JK31 was grown at 37°C on BA-H supplemented with a range of hemin concentrations (0.0, 0.035, 0.04, 0.05, 0.15, 1.0, 2.5, 5.0, and 8.0 mM), and RNA was prepared from the cultures. The control (0.15 mM), low (0.05 mM), and high (1.0, 2.5, 5.0 mM) hemin concentrations were empirically determined by relative growth rates and 16S Avg CT values. Growth was significantly affected in the extremes of this range, where growth did not occur (0.0, 8.0 mM heme) or a slower growth rate combined with a high 16S Avg CT value (0.035, 0.04 mM heme) eliminated these conditions from our experiment (data not shown).
qRT-PCR and fold difference calculations were performed as described above. Figure 3 shows the effect on hbp transcript profiles at low hemin concentration (0.05 mM) compared to the control concentration (0.15 mM). The average CT hierarchy at low hemin (hbpA > hbpD > hbpE > hbpC > hbpB) is similar to that of the control (hbpD > hbpA > hbpE > hbpC > hbpB), where hbpA and hbpD quantities are nearly equal. However, fold differences indicate that expression of hbpA, hbpD, and hbpE are significantly increased at low hemin concentration, whereas hbpC and hbpB remain relatively unchanged. This demonstrates that hbpA, hbpD, and hbpE respond to low hemin levels and suggests that HbpA, HbpD, and HbpE may play a more significant role in the human environment relative to HbpC and HbpB. Finally, these data infer that two subgroups exist within the hbp family: subgroup I (hbpC and hbpB) and subgroup II (hbpA, hbpD, and hbpE).
FIG. 3.
Fold differences in hbp mRNA following growth at a low hemin concentration (0.05 mM) relative to the control concentration (0.15 mM). At mid-log phase, the amount of target hbp mRNA transcript from 0.05 mM preparations was normalized to the amount of 16S rRNA and is relative to the quantity of that particular hbp mRNA transcript from 0.15 mM preparations. A representative experiment is shown, where CT values were determined in triplicate. SD bars were generated by comparing fold differences to those obtained from a second independent triplicate determination.
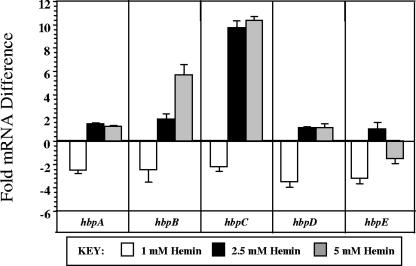
The subgroup hypothesis is further supported by analyzing the fold differences at high hemin levels (1.0, 2.5, 5.0 mM) relative to the control (0.15 mM) and is illustrated in Fig. 4. As the hemin concentration is elevated to 1.0 mM, a decrease is evident in all hbp transcript quantities and implies that hemin is saturating but not yet toxic, suggesting that this may be the ideal hemin concentration for B. quintana strain JK31. Increasing concentrations further (2.5 to 5 mM) results in an elevation of subgroup I (hbpC and hbpB), whereas subgroup II (hbpA, hbpD, and hbpE) remains relatively unchanged. This demonstrates that hbpC and hbpB respond to concentrations of free hemin that are approaching toxicity, an environment that would most likely be encountered in the louse gut. These results, combined with the >100-fold increase seen in hbpC transcript at louse-like temperature (Fig. 1), strongly suggest that HbpC function is critical in the louse.
FIG. 4.
Fold differences in hbp mRNA at high heme levels from mid-log-phase cells. The amount of target hbp mRNA transcript from cultures grown in 1, 2.5, and 5 mM heme was normalized to the amount of 16S rRNA and is relative to the quantity of that particular hbp mRNA transcript from 0.15 mM preparations. A representative experiment is shown, where CT values were determined in triplicate. SD bars were generated by comparing fold differences to those obtained from a second independent triplicate determination.
Finally, hbp transcript quantity was not significantly affected by cultivation at several different free iron concentrations or media pH backgrounds, indicating that these environmental cues do not influence expression (data not shown). Due to the inherent peroxidase activity of hemin, it was impossible to investigate the oxidative stress response by cultivation in an H2O2-rich environment.
Synthesis of subgroup II proteins correlates with expression data.
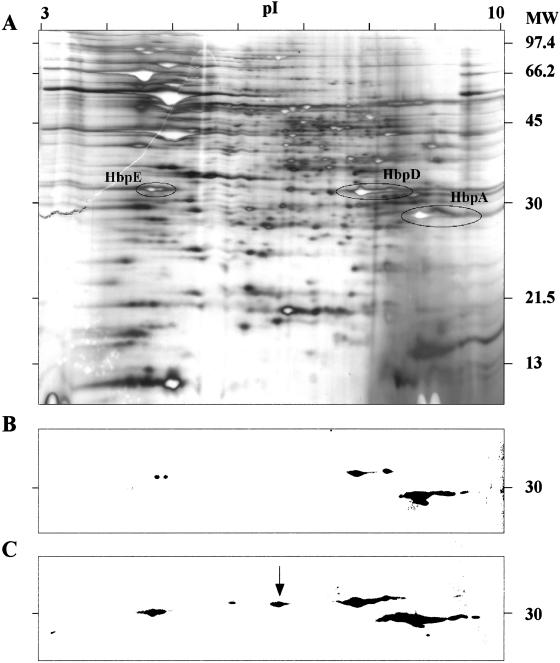
To determine if hbp transcript profiles correspond to protein profiles, we analyzed whole-cell lysates by two-dimensional electrophoresis, immunoblotting, and matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) mass spectroscopy (MS). B. quintana strain JK31 was grown at 37°C on BA-H supplemented with low (0.05 mM) and control (0.15 mM) hemin concentrations. The image in Fig. 5A shows a silver-stained two-dimensional gel of a lysate prepared from B. quintana organisms grown on control (0.15 mM) hemin plates with spots identified by MALDI-TOF MS corresponding to group II proteins (HbpA, HbpD, and HbpE). Immunoblots were prepared and probed with anti-HbpA antibody. It is obvious, from the intensity and size of the immunolabeled spots, that group II proteins are more abundant when bacteria are grown at low hemin (Fig. 5C) relative to control hemin (Fig. 5B). The arrow indicates the size of the HbpD spot, which was confirmed by MALDI-TOF MS to extend into a lower pI range. Previously, we described our rabbit anti-HbpA antibody as monospecific, based on single-dimension immunoblots and knowledge of only a single member of the Hbp family (10). This experiment clearly demonstrates that anti-HbpA antibody cross-reacts with HbpD and HbpE, the other members of subgroup II. We were unable to locate HpbC or HbpB by MALDI-TOF MS or by cross-reactivity with anti-HbpA from cultures grown at low hemin concentration.
FIG. 5.
A. Two-dimensional SDS-PAGE gel of a whole-cell lysate of B. quintana grown for 96 h at the control hemin concentration (0.15 mM), with positions of HbpA, HbpD, and HbpE circled. B. Corresponding immunoblot of two-dimensional SDS-PAGE developed with anti-HbpA antiserum reacting with subgroup II Hbps. C. Immunoblot of two-dimensional SDS-PAGE of B. quintana grown to mid-log phase at a low hemin concentration (0.05 mM) developed with anti-HbpA showing significant increases in HbpA, HbpD, and HbpE. Identity of the Hbps was verified by MALDI-TOF MS, and the spot indicated with the arrow is additional HbpD not seen at the 0.15 mM heme level. The pI range of the IEF is shown at the top, and the molecular weight (MW) is given to the right in thousands.
Expression of ′lacZ by the hbpA promoter parallels hbpA transcription.
Data presented thus far demonstrate that hbp transcription is affected by physiologically relevant environmental conditions. We were curious to determine if transcription factors were involved in regulation of transcript quantity. To this end, we constructed a reporter plasmid, pHPRO+ LACZ+, consisting of the hbpA promoter region, HPRO, fused to a truncated lacZ gene (′lacZ). A diagram of this reporter plasmid is shown in Fig. 6A, along with two control constructs lacking either HPRO, pHPRO− LACZ+ (Fig. 6B), or ′lacZ, pHPRO+ LACZ− (Fig. 6C). These three constructs were electroporated into low-passage B. quintana strain JK31, and transformation was confirmed by restriction fragment length polymorphism analysis and sequencing. The transformation efficiency was approximately 0.8 transformants per μg DNA.
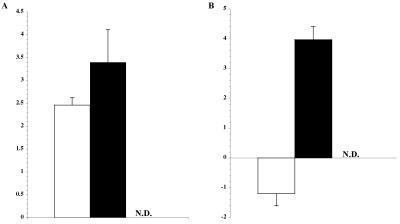
The resulting strains (JK31-pHPRO+ LACZ+, JK31-pHPRO− LACZ+, and JK31-pHPRO+ LACZ−) were then analyzed by qRT-PCR in the two environments where hbpA expression is most affected: low hemin (0.05 relative to 0.15 mM) and low oxygen (5% relative to 21%). Fold differences were calculated for ′lacZ transcripts and are shown in Fig. 7. Although ′lacZ appears to be transcribed from a cryptic promoter in the control plasmid lacking HPRO (black bars), where relative amount of transcript (more than 3.5 to 4.0) is not influenced by the environment, data in Fig. 7B clearly show that without HPRO, repression is lost under low oxygen. ′lacZ is not detectable in strains lacking this truncated open reading frame (ORF), showing that B. quintana does not have a genomic copy of lacZ. hbpA expression in these strains and under the two conditions is similar to previous results in Fig. 2 and 3 (data not shown). These data suggest that a transcriptional regulator(s) is influencing the expression of hbpA, at least under conditions of low O2, and that the hbpA promoter region contains a cis-acting regulatory element.
FIG. 7.
Fold differences in ′lacZ mRNA in B. quintana containing the reporter construct pHPRO+ LACZ+ (white) or the control plasmids pHPRO− LACZ+ (black) and pHPRO+ LACZ+ (N.D., not detectable), under conditions of (A) low heme (0.05 mM) or (B) low O2 (5%) relative to normal conditions (0.15 mM heme or 21% O2, respectively). A representative experiment is shown, where CT values were determined in triplicate. SD bars were generated by comparing fold differences to those obtained from a second independent triplicate determination. All strains were grown in the environments indicated, and mRNA preparations were made at mid-log phase.
DISCUSSION
Bartonella species require extraordinarily high hemin concentrations for growth, yet an explanation for this remains unknown. Several researchers have hypothesized that heme is essential as a source of porphyrin (34) and/or iron (10, 42) or as a hydrogen peroxide-detoxifying agent (33). Recently published genomic sequences (1) provide data that address these hypotheses: (i) neither B. quintana nor B. henselae is capable of de novo heme synthesis, as genes for nearly all porphyrin biosynthetic enzymes are missing; (ii) ORFs encoding uptake systems for heme (HutABC/HmuV), iron-siderophore (FatBCD/CeuD), and free iron (YfeABCD) are found in both species; (iii) genes coding for catalase, peroxidase, bacterioferritin, or ferritin-like heme or iron storage molecules and iron detoxification proteins (Dps) are absent in both genomes.
A survey of over 100 genomes using the KEGG database (http://www.genome.ad.jp/kegg/pathway.html) demonstrates that bacteria lacking the enzymes to perform de novo heme synthesis are quite rare. Although the absence of heme biosynthesis in B. quintana and B. henselae describes why this molecule is essential, it does not explain the extraordinary quantity that is necessary for routine culture. Compared to other pathogens that also lack this capability, the heme supplement for Bartonella is approximately 100-fold greater than that of Porphyromonas gingivalis and 1,000-fold greater than that of Haemophilus influenzae in aerobic iron-replete conditions (23, 25, 34, 49). One explanation assumes that more heme and/or iron is required as a nutrient by Bartonella to synthesize a relatively larger number of enzymes that utilize heme prosthetic groups and/or iron. However, while genomic sequence annotations for B. quintana and B. henselae (1) describe a number of enzymes that use iron as a cofactor, only two utilize heme prosthetic groups, succinate dehydrogenase and cytochrome O oxidase.
An alternative explanation for the high requirement is that heme molecules are not just metabolic nutrients per se but are also mediators of metabolic homeostasis, where heme may (i) function as a defense mechanism against ROS (ii) or exogenously generate a decreased oxygen environment for the bacterium. Although there is one report suggesting that B. quintana can respire anaerobically (17), it is generally accepted that the Bartonella species are aerobic (8). During aerobic respiration, superoxide (O2−) and hydrogen peroxide (H2O2) are naturally generated, and superoxide dismutase and catalase/peroxidase are normally employed, respectively, for intracellular detoxification (30). Although genes encoding superoxide dismutase can be found in both B. quintana and B. henselae, there are no genes encoding catalase or peroxidase (1). The apparent absence of a method for endogenous H2O2 detoxification suggests that either these bacteria possess an uncharacterized mechanism for its degradation, or they do not respire aerobically. First, since almost all catalases and peroxidases utilize a heme prosthetic group, it is possible that one or more of the hypothetical genes encode a novel heme enyzme for H2O2 detoxification and the concurrent requirement for high-level heme uptake. Second, although in vitro growth of B. quintana can be accomplished under atmospheric oxygen, the results of this study demonstrate that a microaerophilic environment of 5% O2 is sufficient for replication. It has been demonstrated biochemically that B. quintana does not produce H2O2 and is catalase negative, two metabolic traits shared with clostridia and lactobacteria that respire anaerobically, even in the presence of oxygen (34). This suggests that B. quintana is not an aerobe and may not require much oxygen for metabolism. Determination of the heme requirements of B. quintana at very low O2 concentrations is currently being examined to help address this hypothesis.
The second nonnutritional method whereby heme can mediate metabolic homeostasis is by creating a decreased oxygen microenvironment. First, Bartonella are members of the order Rhizobiales, along with several human (Brucella spp.) and plant (Agrobacterium spp.) pathogens. Many rhizobia form a symbiotic relationship with their legume host plant by fixing atmospheric nitrogen in root nodules. For nitrogen fixation to occur, a microaerophilic environment must be established for the bacteria. This is accomplished by plant-generated leghemoglobin (a molecule similar to hemoglobin) binding to the rhizobial surface, effectively shielding the bacteria, and O2-labile nitrogenase, from oxygen (3). Considering the close relationship of Bartonella and rhizobia, it is tempting to speculate that heme binding is a common strategy used by members of this order to decrease oxygen in the environment. In addition, orthologues of the Hbps can be found in Brucella and Agrobacterium (13, 32). Second, P. gingivalis stores heme dimers on its surface to both exclude oxygen from the cell (44) and function as an antioxidant by the intrinsic peroxidase activity of heme (43). Interestingly, the hemin blotting technique that we first used to identify the Hbp family proteins (10) relies on this intrinsic peroxidase activity and shows that the heme bound to the Hbps might act as H2O2 detoxifiers.
The human body louse has been implicated as the insect vector for three major human diseases: epidemic typhus (Rickettsia prowazekii), relapsing fever (Borrelia recurrentis), and trench fever (B. quintana) (15). Body lice live between the skin and clothing of humans, where the temperature is approximately 30°C (27, 29). Lice imbibe human blood several times per day and hemolyze erythrocytes almost immediately (9, 47), resulting in waves of potentially toxic heme, iron, and ROS with each meal. It has been well established that B. quintana multiplies extracellularly in the gut of the louse (14, 19) and is thus repeatedly exposed to these toxic molecules and digestive enzymes.
Our data show that hbpC transcript increases >100-fold at louse-like temperature (Fig. 1) and 10-fold at high hemin concentration (Fig. 4). Together, these results strongly suggest that HbpC would be preferentially synthesized in the louse gut. Considering the nutritive and nonnutritive explanations for the high heme requirement given above, the most straightforward hypothesis is that HbpC functions to bind heme on the surface, creating an antioxidant barrier. Heme and iron detoxification and storage are also challenges for hematophagous arthropod vectors, as hemoglobin is a major protein source (35, 37). This implies that B. quintana must compete with the louse for heme and iron molecules and suggests that HbpC could also function as a heme storage site. An intriguing parallel occurs in the flea-borne agent of bubonic plague, Yersinia pestis, where (i) the outer membrane is the primary site of exogenous heme storage (39), (ii) the storage phenotype is most evident at the temperature of the flea (40), and (iii) heme storage is required for colonization of the flea proventriculus and subsequent transmission (16). Finally, louse fecal matter is the most common vehicle for B. quintana transmission to humans, where B. quintana can survive up to a year (21). Each environmental cue of fecal matter (21% O2, low temperature, and high heme) resulted in an increased expression of hbpC, suggesting that HbpC may contribute to survival in fecal matter. In nature, these conditions are experienced simultaneously and may have a different effect on the hbp transcript profile than the individual in vitro simulations presented in this study. We propose that HbpC is required for survival of B. quintana in the louse and/or fecal matter and are presently addressing this hypothesis with an hbpC mutant and our established body louse colony.
B. quintana is exposed to a very different environment in the human. Available heme is scavenged by hemopexin, hemoglobin, and serum albumin, and free iron is chelated by a number of molecules depending on whether the bacterium is intracellular or extracellular. As growth of B. quintana is dependent on heme availability, mechanisms must be employed for its binding and transport into the cell. Genes encoding uptake systems for heme, iron-siderophore, and free iron are found in both B. quintana and B. henselae (1), yet their functions have not been studied. To date, all Bartonella species examined also contain genes encoding the Hbp family proteins (originally termed “Pap” in B. henselae) (50). Our data demonstrate that the relative quantity of subgroup II (hbpA, hbpD, and hbpE) transcript is significantly upregulated at low hemin concentrations (Fig. 3), suggesting that subgroup II proteins are utilized in the human for acquisition of heme. This notion is supported by qRT-PCR analysis of the hbp transcript profile in a Rhesus macaque where the hierarchy (hbpD > hbpA > hbpE > hbpC > hbpB) is consistent with this in vitro low-heme condition (data not shown).
Oxygen, heme, and iron are molecules that are required by almost all living organisms to maintain metabolic homeostasis, yet each of these nutrients can become toxic if the concentration is too high (6, 18). Furthermore, heme and iron are capable of transforming oxygen into highly toxic ROS (O2−, H2O2, and OH−). Humans maintain their bloodstream O2 between 3 and 5%, much lower than the atmospheric 21% used for routine cultivation of B. quintana. Nothing is known about the O2 concentration in a louse gut, but we hypothesize that large amounts of ROS are present during blood meal digestion. Our data show that all of the hbp transcripts decrease significantly when B. quintana is grown at an O2 concentration that simulates the human bloodstream (Fig. 2). This finding strongly suggests that the Hbps respond to O2 and/or ROS. Again, the simplest explanation is that surface-bound heme functions as an antioxidant barrier, where bacteria grown at 5% oxygen encounter relatively less ROS and, thus, less Hbp is necessary to maintain metabolic homeostasis. Assuming that the Hbps also function in transport of hemin for nutritional purposes (50), it is also possible that enzymes which utilize porphyrin and/or iron are not required to the extent they are at 21% O2. In nature, one would expect relative hbp expression to be highest in louse fecal matter and lowest in the human bloodstream.
B. quintana is also unusual in that it apparently lacks mechanisms for iron storage (ferritin), heme storage (bacterioferritin), iron detoxification, and antioxidant defense (catalase and peroxidase). To prevent intracellular oxidative damage and iron toxicity, the regulation of genes involved in heme uptake and subsequent nutritive utilization must be tightly regulated (2, 45). It is obvious that hbpC transcription is temperature regulated and that oxygen and hemin also influence transcription of the hbp genes. We generated a ′lacZ reporter construct (Fig. 6) to determine if the hbpA promoter region could control transcription of an exogenous locus. Although the presence of the cryptic promoter may be affecting ′lacZ expression at low hemin concentrations (Fig. 7A), it is evident that repression of ′lacZ is occurring under low oxygen concentrations (Fig. 7B). This strongly suggests that one or more regulators are affecting transcription and that the hbpA promoter region contains a cis-acting regulatory element. Experiments are under way to identify the specific trans-acting regulators and the cognate promoter elements of the hbp genes.
In conclusion, the roles that the Hbp family play in essential heme acquisition and maintenance of metabolic homeostasis are unknown. We report here the differential and coordinated expression of hbp genes in response to environmental conditions that simulate the human host and louse vector. Based on differential expression patterns, we propose that there are two subgroups of hbp genes, subgroup I (hbpC and hbpB) and subgroup II (hbpA, hbpD, and hbpE). We are downplaying the role of hbpB for several reasons: (i) hbpB contains a ∼510-bp insert in the center of its ORF and recombinant HbpB does not bind hemin like other Hbps (J. A. Carroll and M. F. Minnick, unpublished data); and (ii) in all qRT-PCR studies, the relatively high average CT value suggests that very little hbpB transcript is produced. Continued work on this heme receptor gene family is expected to yield valuable clues regarding the extraordinary need for heme as well as the roles that these virulence factors play in the survival and pathogenesis of Bartonella.
Acknowledgments
We are grateful to Jane Koehler (UC Davis, San Fransisco, CA) for the generous contribution of B. quintana strain JK31. We thank Jesse Hay, Beverly Parker, and Holly Cox (University of Montana) as well as Matthew Chenoweth (National Cancer Institute) for their valuable technical assistance with cell disruption, MALDI-TOF, MASCOT fingerprinting analysis, and two-dimensional electrophoresis. Finally, we thank Patty McIntire (Murdock Sequencing Facility, University of Montana) for providing sequence data and James Gannon (University of Montana) and James Carroll (University of Pittsburgh) for helpful discussions on bacterial physiology and qRT-PCR, respectively.
This work was supported by Public Health Service grant R01 AI350111 from the National Institutes of Health.
Editor: D. L. Burns
REFERENCES
- 1.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Appleby, C. A. 1984. Leghemoglobin and Rhizobium respiration. Annu. Rev. Plant Physiol. 35:443-478. [Google Scholar]
- 4.Applied Biosystems. 2004. Guide to performing relative quantitation of gene expression using real-time quantitative PCR. [Online.] http://www.appliedbiosystems.com/support/tutorials/pdf/performing_rq_gene_exp_rtpcr.pdf. Accessed 13 February 2006.
- 5.Ausubel, F., et al. 1995. Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 6.Baker, H. M., B. F. Anderson, and E. N. Baker. 2003. Dealing with iron: common structural principles in proteins that transport iron and heme. Proc. Natl. Acad. Sci. USA 100:3579-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battisti, J. M., and M. F. Minnick. 1999. Development of a system for genetic manipulation of Bartonella bacilliformis. Appl. Environ. Microbiol. 65:3441-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birtles, R. J., T. G. Harrison, N. A. Saunders, and D. H. Molyneux. 1995. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int. J. Syst. Bacteriol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 9.Buxton, P. A. 1939. The louse: an account of the lice which infest man, their medical importance and control. Butler and Tanner, London, United Kingdom.
- 10.Carroll, J. A., S. A. Coleman, L. S. Smitherman, and M. F. Minnick. 2000. Hemin-binding surface protein from Bartonella quintana. Infect. Immun. 68:6750-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, C. C., B. B. Chomel, R. W. Kasten, V. Romano, and N. Tietze. 2001. Molecular evidence of Bartonella spp. in questing adult Ixodes pacificus ticks in California. J. Clin. Microbiol. 39:1221-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delpino, M. V., J. Cassataro, C. A. Fossati, F. A. Goldbaum, and P. C. Baldi. 19. January 2006, posting date. Brucella outer membrane protein Omp31 is a haemin-binding protein. Microbes Infect. [Online.] http://www.elsevier.com/wps/find/journaldescription.cws_home/601557/description#description. [DOI] [PubMed]
- 14.Fournier, P. E., M. F. Minnick, H. Lepidi, E. Salvo, and D. Raoult. 2001. Experimental model of human body louse infection using green fluorescent protein-expressing Bartonella quintana. Infect. Immun. 69:1876-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, P. E., J. B. Ndihokubwayo, J. Guidran, P. J. Kelly, and D. Raoult. 2002. Human pathogens in body and head lice. Emerg. Infect. Dis. 8:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 17.Huang, K. Y. 1967. Metabolic activity of the trench fever rickettsia, Rickettsia quintana. J. Bacteriol. 93:853-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imlay, J. A. 2002. How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46:111-153. [DOI] [PubMed] [Google Scholar]
- 19.Ito, S., and J. W. Vinson. 1965. Fine structure of Rickettsia quintana cultivated in vitro and in the louse. J. Bacteriol. 89:481-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koehler, J. E. 1996. Bartonella infections, p. 1-27. In S. C. Aronoff, W. T. Hughes, S. Kohl, W. T. Speck, and E. R. Wald (ed.), Advances in pediatric infectious diseases, vol. 11. Mosby-Year Book, Inc., Chicago, Ill. [PubMed] [Google Scholar]
- 21.Kostrzewski, J. 1950. The epidemiology of trench fever. Med. Dosw. Mikrobiol. 11:233-263. (In Polish.) [PubMed] [Google Scholar]
- 22.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, Jr., and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 23.Kusaba, A., T. Ansai, S. Akifusa, K. Nakahigashi, S. Taketani, H. Inokuchi, and T. Takehara. 2002. Cloning and expression of a Porphyromonas gingivalis gene for protoporphyrinogen oxidase by complementation of a hemG mutant of Escherichia coli. Oral Microbiol. Immunol. 17:290-295. [DOI] [PubMed] [Google Scholar]
- 24.La, V. D., L. Tran-Hung, G. Aboudharam, D. Raoult, and M. Drancourt. 2005. Bartonella quintana in domestic cat. Emerg. Infect. Dis. 11:1287-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, X., T. Olczak, H. C. Guo, D. W. Dixon, and C. A. Genco. 2006. Identification of amino acid residues involved in heme binding and hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect. Immun. 74:1222-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 27.Marsh, F., and Buxton, P. A. 1937. Measurements of the temperature and humidity between clothes and body. J. Hyg. 37:254-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maurin, M., and D. Raoult. 1996. Bartonella (Rochalimaea) quintana infections. Clin. Microbiol. Rev. 9:273-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellannby, K. 1932. The conditions of temperature and humidity of the air between the skin and shirt of man. J. Hyg. 32:268-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messner, K. R., and J. A. Imlay. 1999. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J. Biol. Chem. 274:10119-10128. [DOI] [PubMed] [Google Scholar]
- 31.Minnick, M. F. 2001. Bartonella, p. 2115-2136. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, London, United Kingdom.
- 32.Minnick, M. F., K. N. Sappington, L. S. Smitherman, S. G. Andersson, O. Karlberg, and J. A. Carroll. 2003. Five-member gene family of Bartonella quintana. Infect. Immun. 71:814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myers, W. F., L. D. Cutler, and C. L. Wisseman, Jr. 1969. Role of erythrocytes and serum in the nutrition of Rickettsia quintana. J. Bacteriol. 97:663-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myers, W. F., J. V. Osterman, and C. L. Wisseman. 1972. Nutritional studies of Rickettsia quintana: nature of the hematin requirement. J. Bacteriol. 109:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichol, H., J. H. Law, and J. J. Winzerling. 2002. Iron metabolism in insects. Annu. Rev. Entomol. 47:535-559. [DOI] [PubMed] [Google Scholar]
- 36.Ohl, M. E., and D. H. Spach. 2000. Bartonella quintana and urban trench fever. Clin. Infect. Dis. 31:131-135. [DOI] [PubMed] [Google Scholar]
- 37.Oliveira, M. F., J. R. Silva, M. Dansa-Petretski, W. de Souza, U. Lins, C. M. Braga, H. Masuda, and P. L. Oliveira. 1999. Haem detoxification by an insect. Nature 400:517-518. [DOI] [PubMed] [Google Scholar]
- 38.Perkins, D. N., D. J. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 39.Perry, R. D., T. S. Lucier, D. J. Sikkema, and R. R. Brubaker. 1993. Storage reservoirs of hemin and inorganic iron in Yersinia pestis. Infect. Immun. 61:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perry, R. D., M. L. Pendrak, and P. Schuetze. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929-5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rolain, J. M., M. Franc, B. Davoust, and D. Raoult. 2003. Molecular detection of Bartonella quintana, B. koehlerae, B. henselae, B. clarridgeiae, Rickettsia felis, and Wolbachia pipientis in cat fleas, in France. Emerg. Infect. Dis. 9:338-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander, A., S. Kretzer, W. Bredt, K. Oberle, and S. Bereswill. 2000. Hemin-dependent growth and hemin binding of Bartonella henselae. FEMS Microbiol. Lett. 189:55-59. [DOI] [PubMed] [Google Scholar]
- 43.Smalley, J. W., A. J. Birss, and J. Silver. 2000. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the mu-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol. Lett. 183:159-164. [DOI] [PubMed] [Google Scholar]
- 44.Smalley, J. W., J. Silver, P. J. Marsh, and A. J. Birss. 1998. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the mu-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem. J. 331:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 46.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaughan, J. A., and A. F. Azad. 1993. Patterns of erythrocyte digestion by bloodsucking insects: constraints on vector competence. J. Med. Entomol. 30:214-216. [DOI] [PubMed] [Google Scholar]
- 48.Vinson, J. 1966. In vitro cultivation of the rickettsia agent of trench fever. Bull. W. H. O. 35:155-164. [PMC free article] [PubMed] [Google Scholar]
- 49.White, D. C., and S. Granick. 1963. Hemin biosynthesis in Haemophilus. J. Bacteriol. 85:842-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmermann, R., V. A. Kempf, E. Schiltz, K. Oberle, and A. Sander. 2003. Hemin binding, functional expression, and complementation analysis of Pap 31 from Bartonella henselae. J. Bacteriol. 185:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]