Abstract
Sarcocystis neurona causes serious neurological disease in horses and other vertebrates in the Americas. Based on epidemiological data, this parasite has recently emerged. Here, the genetic diversity of Sarcocystis neurona was evaluated using the amplified fragment length polymorphism (AFLP) method. Fifteen S. neurona taxa from different regions collected over the last 10 years were used; six isolates were from clinically diseased horses, eight isolates were from wild-caught opossums (Didelphis virginiana), and one isolate was from a cowbird (Molothrus ater). Additionally, four outgroup taxa were also fingerprinted. Nine primer pairs were used to generate AFLP patterns, with a total number of amplified fragments ranging from 30 to 60, depending on the isolate and primers tested. Based on the presence/absence of amplified AFLP fragments and pairwise similarity values, all the S. neurona isolates tested were clustered in one monophyletic group. No significant correlation could be found between genomic similarity and host origin of the S. neurona isolates. AFLP revealed significant intraspecific genetic variations, and S. neurona appeared as a highly variable species. Furthermore, linkage disequilibrium analysis suggested that S. neurona populations within Michigan have an intermediate type of population structure that includes characteristics of both clonal and panamictic population structures. AFLP is a reliable molecular technique that has provided one of the most informative approaches to ascertain phylogenetic relationships in S. neurona and its closest relatives, allowing them to be clustered by relative similarity using band matching and unweighted pair group method with arithmetic mean analysis, which may be applicable to other related protozoal species.
Sarcocystis neurona is the main causal agent of equine protozoal myeloencephalitis (EPM) in horses and of similar neurological disease in other vertebrates. EPM is the most commonly diagnosed neurological disease of horses in the Americas (5). Recently, outbreaks of neurological disease in several marine mammals through a waterborne route have highlighted the wide dispersion of this apicomplexan cyst-forming coccidian protozoan (8). The parasite is heteroxenous, requiring two hosts to complete its life cycle. Recent studies have shown that the domestic cat (Felis domesticus) (6), the striped skunk (Mephitis mephitis) (3), the nine-banded armadillo (Dasypus novemcinctus) (2), and the raccoon (Procyon lotor) (7) can serve as intermediate hosts of S. neurona. In North America, the parasite utilizes the Virginia opossum (Didelphis virginiana) as its definitive host (17). The sporocyst stage of the parasite develops in the small intestine of the opossum and is passed in feces (4). Equids are infected when they consume feed or water contaminated with sporocysts from opossum feces (6), and recent evidence suggests they also may act as intermediate hosts (23). The propensity to cause neurological disease in a wide range of intermediate hosts suggested that isolates identified as S. neurona could have significant diversity, which would be unusual for species within the genus Sarcocystis.
As an initial and important step towards an appropriate strategy for management of EPM infections, the genetic diversity of S. neurona populations must be assessed. Understanding the clinical role of S. neurona genetic variants in causing EPM is greatly hampered by the difficulty involved in their isolation, cultivation, and identification (12). Additionally, the molecular identification and typing of S. neurona associated with equine diseases and that isolated from opossums by current published methods are hampered by a lack of resolution at the inter- or intraspecific level (12). There have been a few studies that used molecular methods for the identification of S. neurona. These included use of random amplified polymorphic DNA (RAPD) (31), sequencing analysis of SSU rRNA (9) and SAG1 genes (10), and restriction analysis of PCR amplicons of a diagnostic marker (12, 31). These methods differ in their discriminatory power, reproducibility, and ease of interpretation. Additionally, in all of the above-mentioned typing methods, only a very limited part of the parasite genome is covered through specific targeting of one or more DNA markers.
Accurate measurement of genetic relationships requires a neutral marker system that distributes randomly throughout the parasite genome, such as amplified fragment length polymorphism (AFLP). The AFLP method is based on (i) digestion of whole-genomic DNA with two restriction enzymes, (ii) ligation of double-stranded oligonucleotide adapters to the restriction half sites, and (iii) selective amplification of the modified restriction fragments with adapter-specific primers that have an extension of one or more bases at their 3′ end (19, 32). AFLP analysis is a sensitive whole-genome coverage fingerprinting technique which generates highly reproducible results and has superior discriminatory power (28). Also, AFLP provides an unbiased estimate of total genomic variance and is sufficiently abundant to minimize errors due to sampling variance (30). A recent review highlighted the value of this technique for parasite genetic studies (22). Although AFLP analysis has been widely used for taxonomic, diagnostic, epidemiological, and source-tracking applications (28), the feasibility of using AFLP fingerprinting to genotype S. neurona isolates or any other cyst-forming coccidians has not been investigated.
The present study was carried out to test the hypothesis that AFLP markers reveal the phylogenetic relationships between S. neurona isolates at the species and subspecies level. AFLP was used to analyze 15 S. neurona isolates and four outgroup taxa. We demonstrate here the potential of the AFLP technique for inferring phylogenetic relationships and providing information on infraspecific genetic variation in S. neurona isolates mainly from horses and opossums. This information provides a robust methodology and advances understanding of S. neurona population structure.
MATERIALS AND METHODS
Parasite taxa.
A total of 15 S. neurona taxa were collected from six naturally infected horses, eight opossums (D. virginiana), and one brown-headed cowbird from various geographic locations within the state of Michigan. Four closely related taxa were also included in the study as outgroup species. The parasite taxa, their geographical origin, year of isolation, and host sources are shown in Table 1. Methods for isolation and cultivation of S. neurona isolates from horses were previously described (21). Samples of S. neurona oocysts/sporocysts were collected from small intestines of naturally infected opossums and purified by centrifugation of the mucosal scrapings on a potassium bromide discontinuous density gradient as described elsewhere (11). Sporocysts were processed and introduced into cell cultures as described previously (24). In instances where this method failed, sporocysts were bioassayed in gamma interferon gene knockout mice as described previously (10). Parasite growth and tissue culture conditions, described elsewhere (11, 21), were used to obtain viable clonal isolates in the asexual stage of the life cycle. Briefly, equine dermal cells were inoculated with parasites and incubated until cell death due to parasite proliferation was evident and merozoites were present in high numbers. At this time, merozoites were harvested and cultures were passaged to fresh equine dermal cells. Harvest and purification of the culture-derived parasites were performed using PD-10 columns as described previously (14). This method produced high quantities of purified parasites free of detectable host cell contamination.
TABLE 1.
Origin, source, and year of isolation of taxa used in this study
| Species | Designation | State | Yr of isolation | Host | Publication source |
|---|---|---|---|---|---|
| S. neurona | MIH2 | Michigan | 1997 | Horse (Equus caballus)a | 21 |
| S. neurona | MIH5 | Michigan | 1997 | Horse (E. caballus)a | 21 |
| S. neurona | MIH6 | Michigan | 1998 | Horse (E. caballus)a | 21 |
| S. neurona | MIH7 | Michigan | 1998 | Horse (E. caballus)a | 21 |
| S. neurona | MIH8 | Michigan | 1998 | Horse (E. caballus)a | 21 |
| S. neurona | MIH9 | Michigan | 1998 | Horse (E. caballus)a | 21 |
| S. neurona | MIOP1 | Michigan | 1996 | Opossum (D. virginiana)b | 21 |
| S. neurona | MIOP4 | Michigan | 1997 | Opossum (D. virginiana)b | 21 |
| S. neurona | MIOP7 | Michigan | 1998 | Opossum (D. virginiana)b | 21 |
| S. neurona | MIOP17 | Michigan | 2002 | Opossum (D. virginiana)b | 13 |
| S. neurona | MIOP19 | Michigan | 2002 | Opossum (D. virginiana)b | 13 |
| S. neurona | MIOP20 | Michigan | 2002 | Opossum (D. virginiana)b | 13 |
| S. neurona | MIOP29 | Michigan | 2002 | Opossum (D. virginiana)b | 13 |
| S. neurona | MIOP30 | Michigan | 2002 | Opossum (D. virginiana)b | 13 |
| S. neurona | MSUOP/CB5 | Michigan | 1999 | Cowbird (Molothrus ater) | L. S. Mansfield, unpublished |
| S. falcatula | Sfs | New York | 2002 | Cowbird (Molothrus ater) | ATCC |
| T. gondii | RH | USA | Homo sapiens | D. S. Lindsay, unpublished | |
| N. caninum | Nc-1 | USA | Dog | D. S. Lindsay, unpublished | |
| B. darlingi | MIBD1 | Michigan | 2002 | Opossum | H. M. Elsheikha, unpublished data |
Horse samples all came from horses with neurological signs of equine protozoal myeloencephalitis.
Opossums were picked up as road kill or donated from animal control officers and humanely euthanatized in compliance with an approved animal use form according to the Michigan State University institutional animal care and use committee.
DNA extraction and AFLP reactions.
Genomic DNA was isolated from each isolate according to the manufacturer's recommendations using a DNeasy tissue kit (QIAGEN). AFLP analysis was performed using the Perkin-Elmer AFLP plant mapping kit (PE Applied Biosystems, Foster City, CA). Details regarding the adapter and primer sequences are available in the kit documentation. The restriction and ligation steps of the AFLP reaction were performed simultaneously in a total reaction volume of 11 μl by adding 200 to 400 ng of DNA, 1 U of MseI, 5 U of EcoRI, and 1 U of T4 DNA ligase (all enzymes from New England BioLabs, Beverly, MA) to a reaction mixture containing 1× T4 DNA ligase buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, and 25 μg ml−1 bovine serum albumin [New England BioLabs]), 0.55 M NaCl, 0.2 μM EcoRI adapter, and 2 μM MseI adapter (PE Applied Biosystems). The reaction mixtures were incubated in the dark overnight at room temperature in room air and then diluted by adding 189 μl of TE0.1 buffer (20 mM Tris-HCl, 0.1 mM EDTA; pH 8.0). Preselective amplification reactions were performed following the manufacturer's protocol. Successful amplification was verified by running 10 μl of the PCR products on a 1.5% agarose gel stained with ethidium bromide and visualizing this using UV illumination. The remaining product from the preselective amplification (10 μl) was then diluted with 190 μl TE0.1 buffer and used for subsequent steps. For the selective amplifications, 64 primer combinations were initially tested and 9 primer sets were selected for later use. The selected primer pairs produced the most polymorphic and reproducible fingerprints. In each case, one EcoRI primer, labeled with a fluorescent dye, was paired with one MseI primer. The AFLP patterns obtained using primers of the AFLP plant mapping kit (PE applied Biosystems) were few. Therefore, primers from the AFLP microbial fingerprinting kit (PE Applied Biosystems) were also tested. These primers were identical to those designed for the plant samples but contained fewer selective nucleotides at the 3′ end of the primer. Selective amplification was performed as directed in the kit documentation in a total reaction volume of 20 μl. Selective primer pairs designed for these studies are shown below in Table 2.
TABLE 2.
Selected primer combinations and polymorphism rates for AFLP analysis of 15 Sarcocystis neurona isolates
| Primer combination (plus adapter)
|
Total no. of bandsa | No. of polymorphic bands | Polymorphism (%) | |
|---|---|---|---|---|
| EcoRI | MseI | |||
| T | CAG | 51 | 20 | 39 |
| C | CTG | 91 | 62 | 68 |
| C | CAG | 64 | 34 | 53 |
| —b | CTG | 62 | 33 | 53 |
| A | CTC | 77 | 73 | 95 |
| C | CTC | 53 | 36 | 68 |
| C | CAT | 33 | 17 | 51 |
| C | CTT | 44 | 18 | 41 |
| A | CAT | 73 | 66 | 90 |
| Mean | 60.9 | 39.9 | 65.5 | |
The total number of bands per line per primer pair.
—, no selective nucleotide in this primer combination.
AFLP technique optimization.
To test the reproducibility of both the DNA extraction method and AFLP methodology, we conducted repeated measures of AFLP on two well-characterized Sarcocystis isolates (S. neurona MIH1 isolate and S. falcatula) (21). DNA extraction and restriction-ligation and AFLP reactions were performed on aliquots of ∼1 × 107 merozoites of each cloned isolate on three separate occasions. Additionally, 64 primer combinations from the AFLP microbial fingerprinting kit and the AFLP small-genome plant mapping kit were screened. Primer combinations producing patterns comprising 30 to 60 amplified fragments throughout the size range of 50 to 500 bp were selected to test all of the isolates. An iterative process was used to select the primer combinations. When the AFLP patterns were too complex, the complexity was reduced by the addition of an additional selective nucleotide to the primers. However, if the selective nucleotides were numerous, the AFLP patterns were comprised of only a few low-molecular-weight fragments. Therefore, to obtain a greater number of amplified fragments for detection of polymorphism, stringency was reduced by using primers with fewer numbers of selective nucleotides. Since a preamplification step could preferentially enrich heterosite fragments (fragments with different adapters at each end) following adapter ligation (32), the two-step PCR amplification procedure was chosen: a preselective reaction (without any selective nucleotide on primer) followed by selective amplification reaction.
AFLP fragment detection and analysis.
Fluorescent dye-labeled amplification products were separated using a 5% (wt/vol) Long Ranger gel (J.T. Baker, Toronto, Ontario, Canada) for 3.5 h and an ABI 3100 DNA sequencer (Applied Biosystems). Detection mixtures consisted of 2 μl of PCR products diluted 1:20 in sterile water, 2 μl of deionized formamide, 0.3 μl of internal lane size standard labeled with tetramethyl carboxyrhodamine dye (GeneScan 500 TAMRA; Applied Biosystems, Foster City, Calif.), and 0.7 μl of loading buffer (supplied with the size standard). Data collection, processing, fragment sizing, and pattern analysis were done by using ABI PRISM GeneScan 2.0.2 software (PE Applied Biosystems) to obtain fragment sizes and their fluorescent intensities. For the purpose of numerical analysis, the background level was subtracted from the raw AFLP data with GeneScan. Data outputs were obtained both as electropherograms and tabular data. The number of polymorphic bands (scored on the basis of presence/absence) was determined for the fingerprints of all of the parasite taxa. Only fragments in the range from 50 to 500 bp were considered, because the resolving capability of the sequencing gel functions best in this size range. Bands that appeared predominantly in more than one isolate but were absent from or less frequent in the other isolates were considered to be discriminatory bands. For each primer pair, the number of polymorphic fragments was divided by the total number of fragments to calculate a percentage of polymorphism.
Clustering and phylogenetic analysis.
Only distinct, reproducible, well-resolved fragments were scored. The band pattern of each taxon was scored by visual inspection in the GeneScan program and converted to binary codes based on the presence or absence of the discriminatory bands (1 for presence and 0 for absence). To simplify data treatment, quantitative polymorphisms (i.e., variable band intensities and/or reproducible faint bands) were not taken into account. The 1/0 matrix was used to calculate (dis)similarity coefficients following the Jaccard coefficient for each primer pair separately and also for the data pooled over all nine primer combinations (26). The correlation between the different genetic similarity matrices, obtained from the nine separate primer pairs, was evaluated. The resulting distance matrices were used to construct an unweighted pair group method by using arithmetic averages (UPGMA) clustering method (29) as performed with the BioNumerics software package (version 3.0; Applied Maths, Kortrijk, Belgium). A dendrogram was used to depict graphically the genetic relationship and phylogeny of isolates (see Fig. 1, below). The reliability and robustness of the phylogenetic trees were tested by bootstrap analysis with 1,000 replications (16).
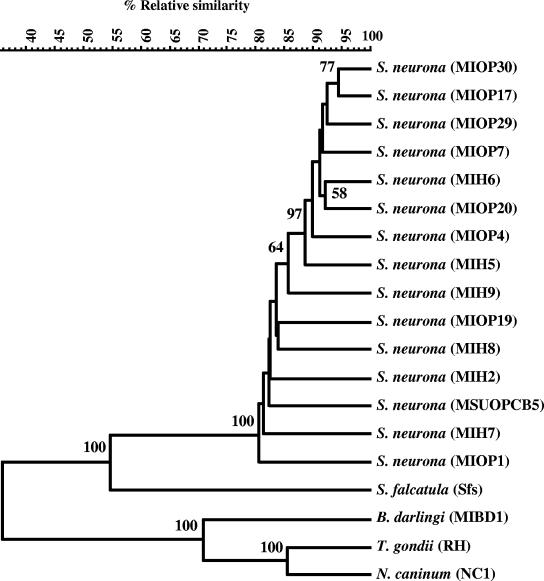
FIG. 1.
UPGMA dendrogram based on AFLP markers obtained with nine primer pairs. Numbers shown at different nodes represent percent confidence limits obtained in the bootstrap analysis. Nodes without numbers had bootstrap values of less than 50. The scale shown above is the measure of genetic similarity calculated according to the Jaccard similarity coefficient prepared from AFLP banding patterns of 15 S. neurona isolates. Taxon names and designations are indicated on the right side of the panel.
LD analysis.
Patterns of linkage disequilibrium (LD) were studied in 15 S. neurona populations and included a total of 434 alleles (two possibilities, presence/absence). Lewontin's D′ (pairwise linkage disequilibrium coefficient) was computed using the ETLINK program modified for binary data by Thomas S. Whittam (Microbial Evolution Laboratory, Michigan State University, East Lansing). All parsimonious uninformative characters were excluded from analysis.
RESULTS
The AFLP patterns generated were highly reproducible. When we tested the repeatability of DNA extraction and AFLP technique using identical samples from either the S. neurona MIH1 isolate or S. falcatula, identical fingerprint patterns were obtained from three replicate DNA samples, isolated, and processed on separate occasions from merozoites of each parasite. These three different attempts produced no visible differences in GeneScan for each of the parasites, which ensured the accuracy and reliability of the data. We used the AFLP technique to develop a convenient and reliable method for genetic differentiation of S. neurona at the inter- and intraspecific level. Sixty-four primer combinations were initially tested to identify the most appropriate pairs of selective primers that gave the most polymorphic and phylogenetically informative patterns. Nine primers were selected (Table 2), based on the number of fragments amplified and the polymorphism rate observed. These primer pairs were applied to the complete set of taxa listed in Table 1. Analysis of the 15 isolates with nine AFLP primer pairs identified a total of 548 fragments, of which 359 were polymorphic (65.5% polymorphism).
Correlation findings showed that AFLP markers obtained from different sets of amplification products provided complementary information. Even though each primer pair individually could have given an approximation of almost all the data, considerable differences existed between them. Hence, data obtained from all nine primer combinations were used for a more robust analysis.
The AFLP banding patterns were analyzed, and genetic relationships among the isolates were indicated in an evolutionary tree constructed by the UPGMA method from a similarity matrix of the Jaccard coefficient using the BioNumerics software. Figure 1 shows the UPGMA dendrogram of relatedness (Jaccard similarity coefficient) based on a total of 842 AFLP markers. The dendrogram shows that there is a close relationship between S. neurona isolates. At the intraspecific level, AFLP fingerprints of S. neurona were grouped by numerical analysis in one main cluster, allowing a clear separation of S. neurona from S. falcatula, Neospora caninum, Toxoplasma gondii, and Besnoitia darlingi. This clustering revealed a single monophyletic group of S. neurona species with a high (100%) confidence value at 82 to 94% genomic similarity. At this level of similarity, S. neurona did not form separate monophyletic groups, despite the fact that they originated from different hosts. S. neurona isolates were distantly separated from the outgroup taxa S. falcatula at a similarity level of 56% and separated from N. caninum, T. gondii, and B. darlingi at a similarity level of 41%. The similarity level between individual fingerprints of each pair of (sub)species, calculated by the Jaccard coefficient, ranged from 80 to 94%, indicating an overall low level of genetic heterogeneity among the isolates.
The results show that AFLP does not provide characteristic markers to differentiate S. neurona isolates recovered from different host species in this sample set. In general, Fig. 1 did not show any discrete clustering in the S. neurona clade except for a few subgroups. For example, S. neurona isolates MIOP30 and MIOP17 from opossums clustered together with 94% similarity; likewise, the isolates MIH6 and MIOP20 clustered together at 93% similarity. These two clusters and MIOP7 and MIOP14 from opossums clustered together with an overall similarity of 92%. The UPGMA analysis demonstrated that S. neurona isolates from various hosts showed considerable genetic variability and demonstrated no correlation between the isolates and their hosts, suggesting that the distinct host origins are not a constraint in the relatedness between the S. neurona isolates.
Among a total of 434 AFLP bands, 217 were parsimony informative for determining phylogenetic relationships between S. neurona isolates. A total of 93,744 comparisons were made. The distribution of Lewontin's D′ (pairwise linkage disequilibrium coefficient) had a peak around 0, which can be a result of recombination and/or parallel mutation to generate all four possible haplotypes. However, these analyses could not document sufficient shuffling by recombination to get to linkage equilibrium. This can be seen in the variance of the mismatch distribution, which is inflated over the randomized data (observed standard deviation, 20.2; expected standard deviation for randomized data, 7.04).
DISCUSSION
The present study was conducted to evaluate the utility of whole-genome AFLP technology as a new method for DNA fingerprinting of S. neurona in order to study the phylogenetic relationships to the (sub)species level of S. neurona isolated from clinically diseased horses and from opossums. Documentation of genetic relatedness of Sarcocystis spp. has been a significant impediment to advances in studying these pathogens. To our knowledge, this is the first time that AFLP has been used to characterize genomic variability of S. neurona species or any other cyst-forming coccidian taxa.
AFLP accessed multiple independent sites within the genome and allowed a better definition of the phylogenetic relatedness of different S. neurona isolates. The results demonstrated the usefulness of the AFLP technique as a reliable taxonomic tool for the differentiation of S. neurona and other related species. It served as a powerful method for the characterization of infraspecific polymorphisms among populations of clinical and environmental isolates of S. neurona. To optimize the method, it was necessary to do several repeat measures on two different isolates on three separate occasions to ensure reproducibility. Additionally, criteria for comparisons were established as nine primer pairs that yielded 30 to 60 DNA fragments after the PCR, within the size range of 50 to 500 bp. These were used to screen all of the isolates. The results of various primer pairs conducted to check the robustness of the dendrogram/estimates of phylogeny clearly establish that polymorphism revealed by AFLP is not only abundant but also statistically reliable. Moreover, these results demonstrate that genetic resolution provided by AFLP is likely amenable to phylogenetic analysis not only of closely related Sarcocystis species as shown earlier but even of highly diverse genera, such as T. gondii, N. caninum, and B. darlingi.
The high sensitivity of the AFLP technique and its rich banding pattern (30 to 60 bands for each pair of selective primers) provided more information about polymorphisms between S. neurona isolates, allowing them to be clustered based on percent similarity. Genotyping techniques based on a single genetic locus can lead to erroneous conclusions if other methods or approaches are not employed. Sarcocystis neurona isolates from opossums and horses from the state of Michigan could not be distinguished based on SSU rRNA sequences, because all of them showed absolute sequence homology (9). These results suggest that the SSU rRNA region has limited value as a marker within S. neurona populations, since there was not enough variation at that level. Also, 25/396 sequence data also demonstrated low nucleotide divergence within S. neurona isolates from horses and opossums (12). The AFLP method has a higher multiplex ratio (i.e., number of loci simultaneously analyzed per experiment) than single-gene analysis. In general, the AFLP method has been a very useful and reliable technique for detecting polymorphisms, and its reproducibility is reported to be very high (33). A noteworthy result of the present study is the finding that AFLP displayed a higher rate of polymorphisms among S. neurona isolates than that obtained with PCR-RFLP or sequencing of SAG1 or SSU rRNA genes in previous studies (9, 27).
At the specific level, the genetic relationships inferred from this study based on AFLP data are in agreement with SSU rRNA and SAG1 gene sequence data in that the S. neurona isolates were found to be genetically closer to each other and seemed to radiate from a common monophyletic origin. Indeed, the present study provides evidence suggesting that the S. neurona populations originated from a common ancestral species. This is apparent in the phyletic representation, wherein all the lineages in this species converge to a single point before separating from other related species and genera included as outgroup taxa. However, in the present study, the variability observed among S. neurona populations based on AFLP analysis suggests that the “species” S. neurona may be a composite group including sublineages. LD analysis of AFLP data suggests that S. neurona has an intermediate model of genetic structure and exhibits LD statistical values suggestive of both clonal and panamictic population structures. However, it is possible that the apparent linkage disequilibrium is a consequence of some geographic structuring, but the sample size used in this study was too limited geographically to infer a firm delineation.
We estimated that 30 S. neurona isolates might be the ideal minimum number of taxa required to precisely study the population structure of this organism. Additionally, these isolates should be diverse and collected from various animal hosts and from wide geographic regions. While more intrapopulation diversity was detected in S. neurona and its population structure was partly clarified with AFLP compared to other analyses based on a single gene/marker, one could argue that small sample sizes (e.g., 15 S. neurona isolates) such as ours could be responsible for these results. However, in our study we proceeded to analyze a smaller number of S. neurona isolates due to the great difficulty associated with isolating and culturing this parasite. The samples analyzed here represent 10 years of active work to obtain isolates from critically ill horses suffering from EPM. One major outcome of studies with small sample sizes is that low-frequency polymorphisms may go undetected. However, it is important to note that sampling error associated with use of small sample size for estimating population differentiation is greatly reduced through the use of large numbers of loci (25). Furthermore, studies on some other systems have been based on even more limited isolate sample sizes, and these clearly showed a significant genetic structuring for Cryptosporidium parvum (1) and for snakes (18).
Even though considerable genetic variation was revealed by AFLP between S. neurona populations, the cluster analysis did not reveal any correlation between genomic similarity and host origin of the populations. Some authors have suggested that AFLP may have little power to resolve complex and recent diversification (20). The facts that the evolutionary rate in the apicomplexan parasite genome is low (15) and that there was a lack of genetic structuring observed in S. neurona in this study both support the notion that AFLP markers alone may not be sufficient tools to investigate diversification of recently emerged organisms, such as has been suggested for S. neurona.
Even though the number of populations tested here is rather low and should be increased to strengthen the informative value of our data, this study nevertheless provides arguments for the hypothesis that S. neurona may not be indigenous throughout its current geographical distribution in the New World. The fact that S. neurona populations from horses and opossums are similar at the genome level suggests that they could share a common geographic center of origin. Additionally, the presence of populations with very low genomic similarity in the same region could be the result of random dissemination from a number of centers of origin and juxtaposition through agronomical practices rather than arising from extreme genetic drift from a common and local ancestor. The ability to accurately detect polymorphisms between S. neurona isolates is of major importance for the design of effective control strategies for this parasite. This study has established the AFLP conditions and primer combinations that permit the assessment of genetic diversity between S. neurona subspecies. It further demonstrated that the technique could be applied in Sarcocystis diversity studies, with potential use for the identification of intra- or interspecific genetic markers. Through the evaluation of large numbers of cloned and characterized field isolates, such AFLP fingerprinting may facilitate the identification of polymorphisms linked to parasite virulence factors and, thus, contribute to the understanding of host-parasite interactions at the molecular level. Therefore, further research focusing on the identification of molecular markers able to accurately discriminate at a number of levels (species, clones, populations, and pathotypes) should be encouraged.
In conclusion, the AFLP technique described in this paper was reproducible for analyzing genomic DNA from S. neurona isolates and isolates of related taxa and was effective for showing interspecific and intraspecific molecular variability between isolates. Although there was no obvious correlation between particular isolates and their host origin, this technique revealed close genetic similarity of S. neurona isolates obtained from horses, opossums, and a cowbird from the state of Michigan. Within the scope of subsequent ecological and epidemiological studies, AFLP fingerprinting could be further extended to a broad survey of populations present in different hosts and from different geographic regions in order to evaluate the impact of coevolution between host and S. neurona on the biodiversity. Future studies may also include the recovery and molecular cloning of AFLP marker bands for the identification of species-specific or type-specific S. neurona markers.
Acknowledgments
We are grateful to Thomas S. Whittam for his assistance with statistical analyses and Seth Walk for his kind help with BioNumerics.
The study was supported by grant 61-6123 from the Grayson Jockey-Club Research Foundation.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Blears, M. J., N. J. Pokorny, R. A. Carreno, S. Chen, S. A. De Grandis, H. Lee, and J. T. Trevors. 2000. DNA fingerprinting of Cryptosporidium parvum isolates using amplified fragment length polymorphism (AFLP). J. Parasitol. 86:838-841. [DOI] [PubMed] [Google Scholar]
- 2.Cheadle, M. A., S. M. Tanhauser, J. B. Dame, D. C. Sellon, M. Hines, P. E. Ginn, R. J. MacKay, and E. C. Greiner. 2001. The nine-banded armadillo (Dasypus novemcinctus) is an intermediate host for Sarcocystis neurona. Int. J. Parasitol. 31:330-335. [DOI] [PubMed] [Google Scholar]
- 3.Cheadle, M. A., C. A. Yowell, D. C. Sellon, M. Hines, P. E. Ginn, A. E. Marsh, R. J. MacKay, J. B. Dame, and E. C. Greiner. 2001. The striped skunk (Mephitis mephitis) is an intermediate host for Sarcocystis neurona. Int. J. Parasitol. 31:843-849. [DOI] [PubMed] [Google Scholar]
- 4.Dubey, J. P., S. S. Black, L. G. Rickard, B. M. Rosenthal, D. S. Lindsay, S. K. Shen, O. C. Kwok, G. Hurst, and A. Rashmir-Raven. 2001. Prevalence of Sarcocystis neurona sporocysts in opossums (Didelphis virginiana) from rural Mississippi. Vet. Parasitol. 95:283-293. [DOI] [PubMed] [Google Scholar]
- 5.Dubey, J. P., D. S. Lindsay, W. J. Saville, S. M. Reed, D. E. Granstrom, and C. A. Speer. 2001. A review of Sarcocystis neurona and equine protozoal myeloencephalitis (EPM). Vet. Parasitol. 95:89-131. [DOI] [PubMed] [Google Scholar]
- 6.Dubey, J. P., W. J. Saville, D. S. Lindsay, R. W. Stich, J. F. Stanek, C. A. Speert, B. M. Rosenthal, C. J. Njoku, O. C. Kwok, S. K. Shen, and S. M. Reed. 2000. Completion of the life cycle of Sarcocystis neurona. J. Parasitol. 86:1276-1280. [DOI] [PubMed] [Google Scholar]
- 7.Dubey, J. P., W. J. Saville, J. F. Stanek, D. S. Lindsay, B. M. Rosenthal, M. J. Oglesbee, A. C. Rosypal, C. J. Njoku, R. W. Stich, O. C. Kwok, S. K. Shen, A. N. Hamir, and S. M. Reed. 2001. Sarcocystis neurona infections in raccoons (Procyon lotor): evidence for natural infection with sarcocysts, transmission of infection to opossums (Didelphis virginiana), and experimental induction of neurologic disease in raccoons. Vet. Parasitol. 100:117-129. [DOI] [PubMed] [Google Scholar]
- 8.Dubey, J. R., A. C. Rosypal, B. M. Rosenthal, N. J. Thomas, D. S. Lindsay, J. F. Stanek, S. M. Reed, and W. J. Saville. 2001. Sarcocystis neurona infections in sea otter (Enhydra lutris): evidence for natural infections with sarcocysts and transmission of infection to opossums (Didelphis virginiana). J. Parasitol. 87:1387-1393. [DOI] [PubMed] [Google Scholar]
- 9.Elsheikha, H. M., D. W. Lacher, and L. S. Mansfield. 2005. Phylogenetic relationships of Sarcocystis neurona isolates of horses and opossums to other cyst-forming coccidia deduced from SSU rRNA gene sequences. Parasitol. Res. 97:345-357. [DOI] [PubMed] [Google Scholar]
- 10.Elsheikha, H. M., and L. S. Mansfield. 2004. Sarcocystis neurona major surface antigen gene 1 (SAG1) shows evidence of having evolved under positive selection pressure. Parasitol. Res. 94:452-459. [DOI] [PubMed] [Google Scholar]
- 11.Elsheikha, H. M., A. J. Murphy, S. D. Fitzgerald, L. S. Mansfield, J. P. Massey, and M. A. Saeed. 2003. Purification of Sarcocystis neurona sporocysts from opossum (Didelphis virginiana) using potassium bromide discontinuous density gradient centrifugation. Parasitol. Res. 90:104-109. [DOI] [PubMed] [Google Scholar]
- 12.Elsheikha, H. M., A. J. Murphy, and L. S. Mansfield. 2005. Phylogenetic congruence of Sarcocystis neurona Dubey et al., 1991 (Apicomplexa: Sarcocystidae) in the United States based on sequence analysis and restriction fragment length polymorphism (RFLP). Syst. Parasitol. 61:191-202. [DOI] [PubMed] [Google Scholar]
- 13.Elsheikha, H. M., A. J. Murphy, and L. S. Mansfield. 2004. Prevalence of Sarcocystis species sporocysts in northern Virginia opossums (Didelphis virginiana). Parasitol. Res. 93:427-431. [DOI] [PubMed] [Google Scholar]
- 14.Elsheikha, H. M., B. M. Rosenthal, A. J. Murphy, D. B. Dunams, D. A. Neelis, and L. S. Mansfield. 2006. Generally applicable methods to purify intracellular coccidia from cell cultures and to quantify purification efficacy using quantitative PCR. Vet. Parasitol. 135:223-234. [DOI] [PubMed] [Google Scholar]
- 15.Escalante, A. A., and F. J. Ayala. 1995. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc. Natl. Acad. Sci. USA 92:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 17.Fenger, C. K., D. E. Granstrom, A. A. Gajadhar, N. M. Williams, S. A. McCrillis, S. Stamper, J. L. Langemeier, and J. P. Dubey. 1997. Experimental induction of equine protozoal myeloencephalitis in horses using Sarcocystis sp. sporocysts from the opossum (Didelphis virginiana). Vet. Parasitol. 68:199-213. [DOI] [PubMed] [Google Scholar]
- 18.Giannasi, N., R. S. Thorpe, and A. Malhotra. 2001. The use of amplified fragment length polymorphism in determining species trees at fine taxonomic levels: analysis of a medically important snake, Trimeresurus albolabris. Mol. Ecol. 10:19-426. [DOI] [PubMed] [Google Scholar]
- 19.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as an new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 20.Kim, K. S., H. W. Jeong, C. K. Park, and J. H. Ha. 2001. Suitability of AFLP markers for the study of genetic relationships among Korean native dogs. Genes Genet. Syst. 76:243-250. [DOI] [PubMed] [Google Scholar]
- 21.Mansfield, L. S., H. C. Schott, A. J. Murphy, M. G. Rossano, S. M. Tanhauser, J. S. Patterson, K. Nelson, S. L. Ewart, J. V. Marteniuk, D. D. Bowman, and J. B. Kaneene. 2001. Comparison of Sarcocystis neurona isolates derived from horse neural tissue. Vet. Parasitol. 95:167-178. [DOI] [PubMed] [Google Scholar]
- 22.Masiga, D. K., A. Tait, and C. M. Turner. 2000. Amplified restriction fragment length polymorphism in parasite genetics. Parasitol. Today 16:350-353. [DOI] [PubMed] [Google Scholar]
- 23.Mullaney, T., A. J. Murphy, M. Kiupel, J. A. Bell, M. G. Rossano, and L. S. Mansfield. 2005. Evidence to support horses as natural intermediate hosts for Sarcocystis neurona. Vet. Parasitol. 133:27-36. [DOI] [PubMed] [Google Scholar]
- 24.Murphy, A. J., and L. S. Mansfield. 1999. Simplified technique for isolation, excystation, and culture of Sarcocystis species from opossums. J. Parasitol. 85:979-981. [PubMed] [Google Scholar]
- 25.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 26.Rivera, I. G., M. A. R. Chowdhury, A. Huq, D. Jacobs, M. T. Martins, and R. R. Colwell. 1995. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholera O1, O139, and non-O1 strains. Appl. Environ. Microbiol. 61:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenthal, B. M., D. S. Lindsay, and J. P. Dubey. 2001. Relationships among Sarcocystis species transmitted by New World opossums (Didelphis spp.). Vet. Parasitol. 95:133-142. [DOI] [PubMed] [Google Scholar]
- 28.Savelkoul, P. H., H. J. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sneath, P. H., and R. R. Sokal. 1962. Numerical taxonomy. Nature 193:855-860. [DOI] [PubMed] [Google Scholar]
- 30.Spooner, D. M., J. Tivang, J. Nienhuis, J. T. Miller, D. S. Douches, and M. A. Contreras. 1996. Comparison of four molecular markers in measuring relationships among the wild potato relatives Solanum section Etuberosum (subgenus Potatoe). Theor. Appl. Genet. 92:532-540. [DOI] [PubMed] [Google Scholar]
- 31.Tanhauser, S. M., C. A. Yowell, T. J. Cutler, E. C. Greiner, R. J. MacKay, and J. B. Dame. 1999. Multiple DNA markers differentiate Sarcocystis neurona and Sarcocystis falcatula. J. Parasitol. 85:221-228. [PubMed] [Google Scholar]
- 32.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]