Abstract
Bone morphogenetic proteins (BMPs) are pleiotropic growth and differentiation factors belonging to the transforming growth factor-β (TGF-β) superfamily. Signals of the TGF-β-like ligands are propagated to the nucleus through specific interaction of transmembrane serine/threonine kinase receptors and Smad proteins. GCCGnCGC has been suggested as a consensus binding sequence for Drosophila Mad regulated by a BMP-like ligand, Decapentaplegic. Smad1 is one of the mammalian Smads activated by BMPs. Here we show that Smad1 binds to this motif upon BMP stimulation in the presence of the common Smad, Smad4. The binding affinity is likely to be relatively low, because Smad1 binds to three copies of the motif weakly, but more repeats of the motif significantly enhance the binding. Heterologous reporter genes (GCCG-Lux) with multiple repeats of the motif respond to BMP stimulation but not to TGF-β or activin. Mutational analyses reveal several bases critical for the responsiveness. A natural BMP-responsive reporter, pTlx-Lux, is activated by BMP receptors in P19 cells but not in mink lung cells. In contrast, GCCG-Lux responds to BMP stimulation in both cells, suggesting that it is a universal reporter that directly detects Smad phosphorylation by BMP receptors.
INTRODUCTION
Bone morphogenetic proteins (BMPs) were originally identified as inducers of bone and cartilage formation in ectopic tissues (Reddi, 1997). More than 20 BMPs have been isolated and constitute the largest subfamily in the transforming growth factor-β (TGF-β) superfamily of growth and differentiation factors. Like other members of the superfamily, BMPs exert pleiotropic biological effects ranging from the regulation of early developmental processes to organogenesis (Hogan, 1996; Reddi, 1997; Kawabata et al., 1998a). Characterization of BMP homologues in invertebrates has greatly contributed to the elucidation of the signaling pathway of the TGF-β superfamily (Padgett et al., 1998; Whitman, 1998). Decapentaplegic (Dpp) identified in Drosophila is structurally similar to mammalian BMP-2 and -4. Dpp regulates the establishment of the dorsoventral axis in the Drosophila embryo and is required for gut morphogenesis and outgrowth and patterning of imaginal disks (Padgett et al., 1997). In Xenopus, BMPs induce ventral mesoderm, whereas activins, constituting another subfamily of the TGF-β superfamily, direct formation of dorsal mesoderm (Whitman, 1998). Gene disruption of BMP-4 or BMP type IA receptor (BMPR-IA) in mice results in early embryonal death associated with a defect in gastrulation (Kawabata et al., 1998a). BMPs induce bone in ectopic tissues, and Dpp exerts the same effect (Sampath et al., 1993). BMP-4 compensates a developmental defect in flies with a mutation in the dpp gene (Padgett et al., 1993). Thus these factors are functionally interchangeable.
BMPs bind to two types of transmembrane receptors denoted type I and type II with serine/threonine kinase activity (Kawabata et al., 1998a). The BMP receptors identified so far include three type II receptors; BMP type II (BMPR-II), activin type II and activin type IIB receptors, and three type I receptors: BMPR-IA, BMP type IB receptor (BMPR-IB), and activin receptor-like kinase 2 (ten Dijke et al., 1994; Kawabata et al., 1998a; Macías-Silva et al., 1998). Upon ligand binding, type II receptors phosphorylate the juxtamembrane region denoted the GS domain of type I receptors. The activated type I receptors then phosphorylate downstream substrates.
Genetic analyses in Drosophila and Caenorhabditis elegans resulted in the identification of Mad and Sma as signaling molecules downstream of receptors for BMP-like ligands in each organism, respectively (Padgett et al., 1998; Whitman, 1998). Eight homologues of Mad and Sma have been identified in mammals and are generically denoted Smad (Heldin et al., 1997; Massagué, 1998). Smads are grouped into three classes based on structure and function. Smads exist as monomers in the absence of ligand stimulation (Kawabata et al., 1998b). Receptor-regulated Smads (R-Smads) are directly phosphorylated by type I receptors and then associate with common Smads (Co-Smads) essential to distinct signaling pathways. The heteromeric complexes translocate to the nucleus, where they regulate transcription of target genes in concert with other nuclear proteins. Inhibitory Smads antagonize signaling by R-Smads and Co-Smads. Smads 1, 5, and presumably 8 propagate BMP signals and are structurally related to Mad that acts downstream of Dpp, a BMP homologue in Drosophila. Smads 2 and 3 are activated by TGF-βs or activins, and dSmad2 has been identified as the counterpart of Smad2/3 in Drosophila (Brummel et al., 1999; Das et al., 1999). Smad4 is the only Co-Smad in mammals, and Medea acts as a common Smad in flies (Whitman, 1998). Smad6 and Smad7 in mammals and Dad in Drosophila belong to inhibitory Smads.
Smads share two conserved regions denoted the Mad homology 1 (MH1) and MH2 domains (Heldin et al., 1997; Massagué, 1998). The MH1 domain of certain Smads directly binds to DNA, whereas the MH2 domain possesses intrinsic transactivation activity. The C-terminal end of R-Smads contains the SSXS motif, two serines of which serve as the phosphorylation sites by type I receptors. In the absence of ligand stimulation, the MH1 and MH2 domains repress the function of each other through intramolecular interaction. Receptor-induced phosphorylation of R-Smads releases this mutual inhibition. Upon entry to the nucleus, Smads form complexes containing sequence-specific DNA-binding proteins and transcriptional coactivators or corepressors (Derynck et al., 1998; Wotton et al., 1999). Smads can directly bind to DNA; however, the affinity is relatively low, and interaction with sequence-specific DNA-binding proteins is critical for the formation of a stable DNA-binding complex (Derynck et al., 1998). Extensive efforts have been directed toward the isolation of nuclear partners for Smads. DNA-binding proteins such as FAST-1, FAST-2, and AP-1 have been identified as interacting proteins for Smad2/3 and implicated in transactivation of various genes (Chen et al., 1996; Labbéet al., 1998; Zhang et al., 1998; Zhou et al., 1998; Liberati et al., 1999). Hoxc-8 interacts with Smad1, and Smad1 acts as a derepression factor for Hoxc-8 (Shi et al., 1999). STAT3 interacts with Smad1 indirectly via p300, and the complex is involved in transactivation of the glial fibrillary acidic protein gene, a marker for astrocyte differentiation (Nakashima et al., 1999). Transactivation partners that directly interact with Smad1/5/8 or Mad remain to be identified. Tinman could be one of such candidates, although direct interaction with Smads has not been demonstrated (Xu et al., 1998).
Several DNA-binding motifs for Smads have been identified. PCR-based screening of random sequences identified palindromic GTCTAGAC (GTCT motif) as a Smad-binding motif (Zawel et al., 1998). Close examination of TGF-β responsive genes has revealed a sequence containing CAGACA (CAGA motif) as a Smad3-binding motif (Dennler et al., 1998; Jonk et al., 1998). The first demonstration that Smads can directly bind to DNA was reported in Drosophila (Kim et al., 1997). Vestigial, labial, and Ultrabithorax (Ubx) are Dpp-responsive genes. Mad was shown to directly bind to the enhancer of these genes, and GCCGnCGC (GCCG motif) was identified as the consensus binding site. The same motif was found as a Mad- and Medea-binding motif in the tinman gene (Xu et al., 1998). We investigated whether mammalian BMP-regulated Smads can bind to the GCCG motif. Smad1 bound to the sequence with Smad4 in a BMP-dependent manner. We constructed a heterologous reporter gene containing multiple copies of this motif and found that BMP stimulation activated the reporter, whereas TGF-β or activin stimulation did not. Thus the GCCG motif is a common BMP-responsive motif both in invertebrates and vertebrates. Furthermore, we showed that the reporter gene provides a useful detection system of BMP signals in a cell type-independent manner.
MATERIALS AND METHODS
Plasmid Construction
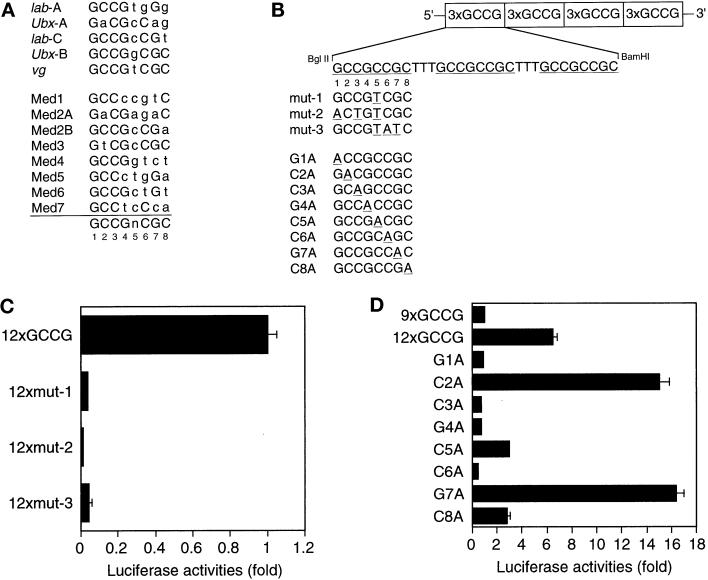
Reporters containing multiple copies of the GCCG motif were constructed as follows. 90COLXLUC (a gift from S. Harada, Merck Research Laboratories, West Point, PA) contains the core promoter of the mouse collagen X gene (−90 to +59) between the BglII and HindIII sites of pGL2-Basic (Promega, Madison, WI) (Harada et al., 1997). Various numbers of oligonucleotides containing three copies of the wild-type or mutant GCCG motif were inserted into the BglII site of 90COLXLUC. The sequences of the oligonucleotides are 5′-GATCTGCCGCCGCTTTGCCGCCGCTTTGCCGCCGCG-3′ (sense) and 5′-GATCCGCGGCGGCAA-AGCGGCGGCAAAGCGGCGGCA-3′ (antisense) for the wild-type, 5′-GATCTGCCGTCGCTTTGCCGTCGCTTTGCCGTCGCG-3′ (sense) and 5′-GATCCGCGACGGCAAAGCGACGGCAAAG-CGACGGCA-3′ (antisense) for 12xmut-1, 5′-GATCTACTGTCGCTTT-ACTGTCGC-TTTACTGTCGCG-3′ (sense) and 5′-GATCCGCGAC-ATAAAGCGACAGTAAAGCGACAGTA-3′ (antisense) for 12xmut-2, and 5′-G-ATCTGCCGTATCTTTGCCGTATCTTTGCCGTATCG-3′ (sense) and 5′-GATCCGATACGGCAAAGATACGGCAAAGATA CGGCA-3′ (anti-sense) for 12xmut-3. Another series of mutant reporters (G1A to C8A) were constructed by inserting oligonucleotides with one base replacement to A between the NheI and BglII sites of 90COLXLUC containing nine copies of the GCCG motif.
Cell Culture and Plasmid Transfection
COS-7 cells and R mutant mink lung epithelial cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma, St. Louis, MO) containing 10% FBS, 100 U/ml penicillin, and 100 μl/ml streptomycin. P19 murine embryonal carcinoma cells (a gift from T. Momoi, National Institute of Neuroscience, Tokyo, Japan) were cultured in α-minimal essential medium (Life Technologies, Gaithersburg, MD) containing 10% FBS and antibiotics. C3H10T1/2 mesenchymal progenitor cells and ATDC5 chondrogenic cells were obtained from the RIKEN Cell Bank (Tsukuba, Japan). C3H10T1/2 cells were cultured in DMEM containing 10% FBS and antibiotics. ATDC5 cells were grown in medium consisting of a 1:1 mixture of DMEM and Ham's F-12 medium (Life Technologies) containing 5% FBS and antibiotics. The cells were maintained in humidified atmosphere with 5% CO2 at 37°C. DNA transfection was performed using FuGENE 6 (Boehringer Mannheim, Mannheim, Germany) according to the manufacturer's protocol.
Preparation of Glutathione S-Transferase (GST)-Smad Fusion Proteins
GST-Smad fusion proteins were prepared essentially as described (Nishihara et al., 1998). Briefly, GST-Smad fusion proteins were expressed in Escherichia coli (DH5α) in the presence of 1 mM of isopropyl-d-thiogalactopyranoside. After sonication of the bacteria, the fusion proteins were purified with glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech, Uppsala, Sweden), washed, eluted, and dialyzed against dialysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl).
Gel Mobility Shift Assay
Gel mobility shift assays were performed as described (Kawabata et al., 1998b). Whole-cell extracts from COS-7 cells transfected with an appropriate combination of plasmids or GST-Smad fusion proteins were used. Probes were labeled with [α-32P]dCTP using Klenow enzyme. Three micrograms of cell lysates or 1 μg of GST fusion proteins were added to premix solution containing poly(dI-dC) and 4 × 104 cpm of the labeled probe. Complexes were resolved on a 4% polyacrylamide gel and analyzed by autoradiography. The sequences of the probes used are 5′-CGCGTTTCTGGACTGGCGTCAGCGCCGGCGCTTCCAGCTGCCAAATTGCTGCTTTATTAG-CTGCGTAAGTGC-3′ (sense) and 5′-TCGAGCACTTACGCAGCT-AATAAAGCAGCAATTTGGCAGCTGGAAGCGCCGGCGCTGA-CGCCAGTCCAGAAA-3′ (antisense) for Ubx, and 5′-CGCGTACG-GGCTGCCGTGGGGAGACACCAGAGCTGTGTAGCAAGAATC-GTATCGAACGGCGGCCAC-3′ (sense) and 5′-TCGAGTGCC-GCCGTTCGATACGATTCTTGCTACACAGCTCTGGTGTCTCC-CCACGGCAGCCCGTA-3′ (antisense) for labial. The sequences of the oligonucleotides for the 3xGCCG probe are the same as used in the construction of the reporter gene. The template for 9xGCCG was prepared by ligation and gel selection of the 3xGCCG DNA.
Luciferase Assay
Luciferase assays with various reporters were carried out using R mutant mink lung epithelial cells or P19 cells. Cells were transiently transfected with an appropriate combination of the reporter, receptor, or Smad expression plasmids and pcDNA3 (Invitrogen, San Diego, CA). Total amounts of the transfected DNAs were the same throughout the experiments, and luciferase activities were normalized using the sea pansy luciferase activity under the control of the thymidine kinase promoter.
RESULTS
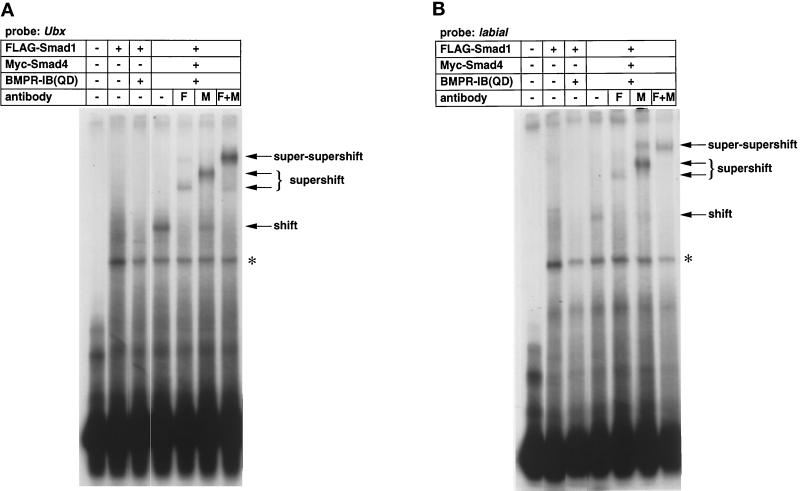
Mad was shown to bind to the enhancers of vetigial, labial, and Ubx that are responsive to Dpp (Kim et al., 1997). Kim et al. (1997) used the GST fusion proteins of Mad and showed that full-length Mad does not bind to the enhancer, and removal of the MH2 domain resulted in DNA binding of Mad. The result indicates that the MH2 domain inhibits DNA binding of the MH1 domain. Phosphorylation of the SSXS motif by type I receptors presumably induces conformational change that releases the inhibition of DNA binding by the MH2 domain. However, direct evidence that Dpp induces DNA binding of Mad has not been presented. We first tested whether mammalian Smad1 and Smad4 bind to the enhancer of Ubx upon BMP stimulation (Figure 1A). Smad1 and/or Smad4 were expressed in COS cells in the absence or presence of a constitutively active form of BMPR-IB, BMPR-IB(QD). Smad1 did not bind to DNA in either the presence or absence of BMPR-IB(QD). When Smad1 and Smad4 were coexpressed in the presence of BMP stimulation, DNA-binding complex appeared. Addition of antibodies against Smad1 or Smad4 supershifted the complex, and simultaneous addition of the two antibodies caused super-supershift of the DNA-binding complex. The results suggest that Smad1 and Smad4 bind to the Ubx enhancer in the presence of BMP stimulation. We used the enhancer of another Dpp-responsive gene, labial (Figure 1B), and obtained an identical result.
Figure 1.
Binding of Smad1 and Smad4 to Dpp-responsive genes. (A) A gel mobility shift assay using the Ubx probe was conducted. COS cells were transfected with the indicated combinations of plasmids, and the cell lysates were mixed with radiolabeled probe. DNA-binding complexes were resolved by gel electrophoresis. The first lane does not contain COS cell lysates, showing the free probe. The asterisk represents a nonspecific band. F and M, anti-FLAG and anti-Myc antibodies, respectively. (B) A gel mobility shift assay was performed using labial as the probe. The first lane does not contain COS cell lysates, showing the free probe.
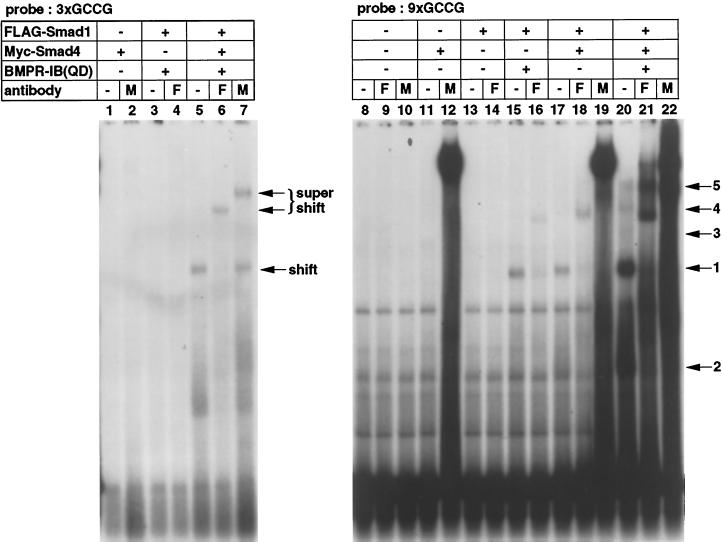
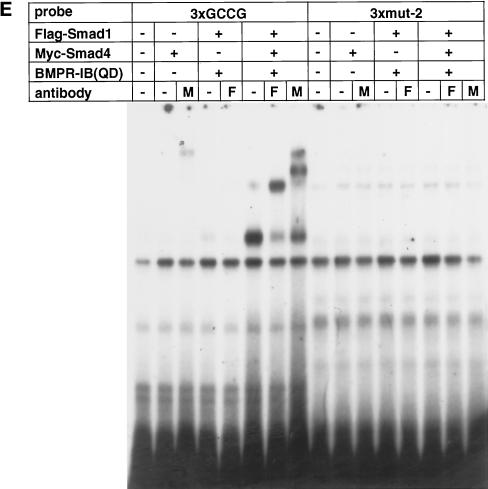
The consensus Mad-binding motif identified among various Dpp-responsive genes is GCCGnCGC (Kim et al., 1997). Smads directly bind to DNA, but the affinity is likely to be relatively low (Derynck et al., 1998). We thus multimerized the GCCG motif and examined the binding of Smad1 and Smad4 to three (3xGCCG) or nine (9xGCCG) copies of the GCCG motif (Figure 2). The same radioactive counts of the 3xGCCG and 9xGCCG probes were used. Free 3xGCCG probe ran out of the gel under the condition used. Smad1 and Smad4 bound to 3xGCCG in the presence of BMPR-IB(QD) (lane 5), and antibodies against each Smad supershifted the band (lanes 6 and 7). Notably, Smad1 alone or Smad4 alone did not bind to 3xGCCG (lanes 1–4). When 9xGCCG was used as a probe, much more strong binding of Smad1 and Smad4 was observed in the presence of BMP stimulation (lanes 20–22). Other weaker bands with slower or faster mobility were also detected (lane 20, bands 2–5). These bands could represent various forms of DNA-binding complexes with different modes of DNA–protein interaction because of the use of multimerized binding sites. We consistently observed enhancement of DNA binding by anti-Myc antibody (lanes 12, 19, and 22). The antibody may stabilize the DNA binding of Smad proteins. Expression of Smad1 alone in the presence of BMPR-IB(QD) (lanes 15 and 16), but not in the absence of the receptor (lanes 13 and 14), caused DNA binding of Smad1. This DNA-binding complex might incorporate endogenous Smad4 in COS cells. Coexpression of Smad1 and Smad4 resulted in DNA binding even in the absence of BMP stimulation (lanes 17–19), although the intensities of the bands are remarkably weaker than those in the presence of receptor stimulation. Thus simple overexpression of Smad1 and Smad4 may force DNA binding of the Smad proteins to some extent. We obtained similar results with Smad5 and Smad8 (our unpublished results).
Figure 2.
Binding of Smad1 and Smad4 to the GCCG motif. 3xGCCG (left panel) and 9xGCCG (right panel) probes were used in a gel mobility shift assay. The same radioactive counts of probes were used. All of the samples were run in the same gel, but the 3xGCCG probe ran out of the gel. The numbers above the gel indicate the numbers of the lanes. The numbers on the right represent various forms of DNA-binding complexes detected in lane 20. Band 1 is the major DNA-binding complex. F and M, anti-FLAG and anti-Myc antibody, respectively.
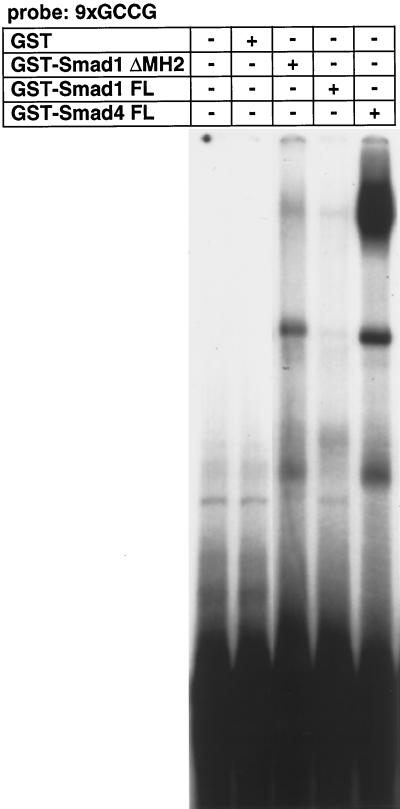
We next studied whether Smad1 or Smad4 directly binds to the GCCG motif as Mad or Medea. GST fusion proteins of full-length Smad4 efficiently bound to the 9xGCCG probe (Figure 3), whereas those of full-length Smad1 did not. When the MH2 domain of Smad1 was removed, the truncted Smad1 fusion proteins bound to the GCCG probe. These results indicate that both Smad1 and Smad4 can directly bind to the GCCG motif. In addition, the MH2 domain of Smad1 interferes with the DNA binding of the protein. The same amounts of GST-Smad proteins were used for Smad1 and Smad4, suggesting that the DNA-binding affinity of Smad1 may be lower than that of Smad4.
Figure 3.
Direct binding of Smad1 and Smad4 to the GCCG motif. GST fusion proteins of Smad1 or Smad4 were used in a gel mobility shift assay with the 9xGCCG probe. The MH2 domain is removed in Smad1 ΔMH2. FL, full-length. The same amounts of GST fusion proteins (1 μg) were used.
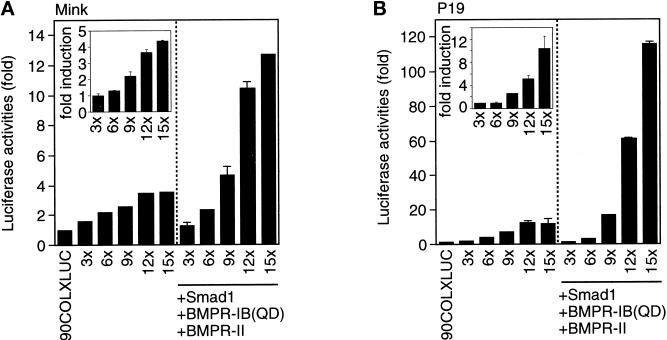
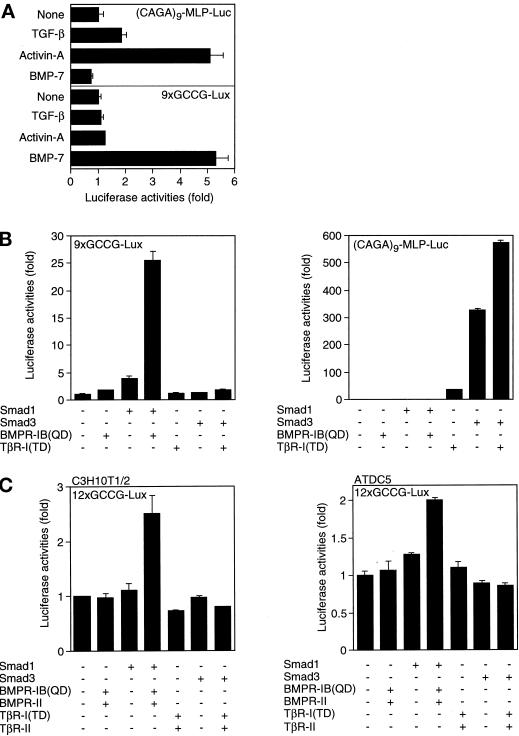
To investigate whether the DNA binding of Smad1 and Smad4 correlates with transactivation, we constructed heterologous luciferase reporter genes with different copies of the GCCG motif. We tested two different promoters to drive transcription. We observed some transactivation of the luciferase gene with the SV40 promoter, but the response was much higher with the collagen X (COLX) promoter (our unpublished results). Therefore, we conducted the following experiments using reporter genes with the COLX promoter. We first used R mutant mink lung epithelial cells (Figure 4A). Reporter genes with six or fewer copies of the GCCG motif was almost unresponsive to BMP stimulation. In contrast, a reporter gene with nine copies of the GCCG motif (9xGCCG-Lux) responded to BMP stimulation, and increase in the repeats of the motif further enhanced the response. We next used P19 mouse embryonal carcinoma cells (Figure 4B). Essentially the same result was obtained with P19 cells, although the induction upon BMP stimulation was more conspicuous.
Figure 4.
Transactivation through the GCCG motif. (A) A luciferase reporter assay was performed using R mutant mink lung epithelial cells. Reporter genes with various numbers of the GCCG motif were transfected into the cells in the absence or presence of BMP stimulation. Luciferase activity was normalized against the cotransfected sea pansy luciferase activity. Experiments were done in duplicate, and the SDs are shown in vertical lines. The values represent fold induction as determined by comparing with the basal 90COLXLUC activity. The fold induction of each reporter activity upon BMP stimulation is shown in the inset. (B) P19 mouse embryonal carcinoma cells were used in a luciferase assay. The fold induction of each reporter activity upon BMP stimulation is shown in the inset. Note the difference of the scale between A and B.
The GCCG motif has been suggested as a Mad- or Medea-binding element (Figure 5A) (Kim et al., 1997; Xu et al., 1998). To determine which bases within the GCCG motif are essential in response to BMP, we generated a series of reporters with 12 copies of mutant sequences (Figure 5B). The nonconserved fifth base was replaced with T from C in all of the three mutants. In addition, the first and third bases were changed in 12xmut-2, and the sixth and seventh bases were changed in 12xmut-3. Both 12xmut-2 and 12xmut-3 lost responsiveness to BMP (Figure 5C). Unexpectedly, alteration of the fifth base (12xmut-1) also abrogated the responsiveness. The fifth base is not highly conserved among Dpp-responsive genes, and the GCCGTCGC sequence in vestigial has high affinity to Mad (Kim et al., 1997). In the Medea- and Mad-binding sites in tinman, however, the fifth base is relatively conserved as C (Figure 5A) (Xu et al., 1998). Thus this position may have some base preference. We also tested the SP-1-binding motif (GGGGCGGGGC), and it did not respond to BMP (our unpublished results). To evaluate the importance of every base of the GCCG motif in response to BMP, we took advantage of the BMP responsiveness of 9xGCCG-Lux. We added three repeats of each mutant sequence (G1A to C8A, as in Figure 5B) to the 9xGCCG sequence and compared the luciferase activity with that of the reporter gene with 12 copies of the GCCG motif (12xGCCG-Lux; Figure 5D). Mutation of the first G, third C, fourth G, or sixth C did not show enhancement of the responsiveness as was observed with 12xGCCG-Lux, suggesting that these four bases are critical in response to BMP. Mutation of the fifth C or eighth C to A only minimally affected the enhancement. In contrast, alteration of the second C or seventh G to A rather augmented the enhancement. Intriguingly, these bases are A in some of the binding sites listed in Figure 5A. To evaluate the correlation between transcriptional responsiveness and the Smad-binding ability, we compared the wild-type probe (3xGCCG) and a mutant probe (3xmut-2) in a gel shift assay. As in Figure 5E, the mutant probe failed to bind activated Smad1, suggesting that the unresponsiveness of the mutant sequence to BMP stimulation is due to lack of Smad binding.
Figure 5.
Effect of mutations in the GCCG motif. (A) Various Mad (Kim et al., 1997) and Mad/Medea (Xu et al., 1998) binding sites are compared. Lowercase letters indicate bases different from the consensus sequence. (B) The sequences of various reporter mutants are presented. The structure of 12xGCCG-Lux is schematically drawn as an example. (C) The luciferase activities of 12xGCCG-Lux, 12xmut-1, 12xmut-2, and 12xmut-3 were compared in the presence of BMP stimulation in P19 cells. All 12 GCCG sequences were mutated in 12xmut-1, 12xmut-2, and 12xmut-3. The values represent relative luciferase activity of the reporters. (D) The responses of mutants with a single-base replacement were compared in the presence of BMP stimulation in P19 cells. Three copies of the mutated sequence were added to the 5′ part of 9xGCCG-Lux. The values represent fold induction as determined by comparing with the activity of 9xGCCG-Lux. (E) A gel mobility shit assay was performed to compare the Smad binding ability of the wild sequence (3xGCCG) and a mutant one (3xmut-2).
We next examined whether the GCCG motif is specifically responsive to BMP. The CAGA motif was identified as a TGF-β-responsive Smad-binding site in the PAI-1 and junB promoters (Dennler et al., 1998; Jonk et al., 1998). (CAGA)9-MLP-Luc contains nine repeats of the CAGA motif (Dennler et al., 1998). P19 cells were transfected with (CAGA)9-MLP-Luc or 9xGCCG-Lux and treated with various ligands (Figure 6A). Both TGF-β and activin activated (CAGA)9-MLP-Luc, whereas BMP-7 did not. In contrast, 9xGCCG-Lux responded only to BMP-7. Thus 9xGCCG-Lux responds specifically to BMP stimulation. We then tested the response of the reporters to activated Smads in P19 cells (Figure 6B). The combination of Smad1 and BMPR-IB(QD) activated 9xGCCG-Lux, whereas that of Smad3 and constitutively active form of TGF-β type I receptor, TβR-I(TD), did not. The latter combination strongly activated (CAGA)9-MLP-Luc. Because the GCCG reporter uses murine collagen X promoter, we tested two cell lines that are osteogenic or chondrogenic (Figure 6C). C3H10T1/2 cells are mesenchymal progenitor cells that are osteogenic, chondrogenic, or adipogenic depending on the culture condition (Bachner et al., 1998). ATDC5 cells are murine teratocarcinoma cells that are chondrogenic (Shukunami et al., 1996). In C3H10T1/2 cells, the GCCG reporter responded to BMP stimulation but not to TGF-β stimulation. Similarly, the reporter was activated by BMP stimulation but not by TGF-β stimulation in ATDC5 cells. Taken together, the GCCG motif is specifically responsive to BMPs among the three major ligands in the TGF-β superfamily.
Figure 6.
Response specificity of the GCCG motif. (A) The responses of (CAGA)9-MLP-Luc and 9xGCCG-Lux to various ligands were compared in P19 cells. Ten ng/ml TGF-β, 100 ng/ml activin, and 1000 ng/ml BMP-7 were used. The values represent fold induction as determined by comparing with the basal activity of each reporter. (B) The responses of (CAGA)9-MLP-Luc (right panel) and 9xGCCG-Lux (left panel) to Smad1 or Smad3 were compared in mink lung cells. The values represent fold induction as determined by comparing with the basal activity of each reporter. Transactivation of (CAGA)9-MLP-Luc by BMP stimulation is too low to see (see the values of the fold induction). (C) Transactivation of 12xGCCG-Lux was tested in C3H10T1/2 (left panel) and ATDC5 (right panel) cells. The values represent fold induction as determined by comparing with the basal activity of the reporter.
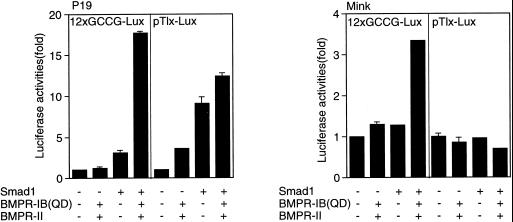
Tlx-2 is a BMP-responsive gene in mouse embryos (Tang et al., 1998). pTlx-Lux is a luciferase reporter gene derived from Tlx-2. Both 12xGCCG-Lux and pTlx-Lux responded to BMP stimulation in P19 cells (Figure 7, left panel). In contrast to the result with 12xGCCG-Lux, pTlx-Lux did not respond to BMP in mink lung cells (Figure 7, right panel). Thus 12xGCCG-Lux may be a more universal reporter for the detection of BMP signals than pTlx-Lux.
Figure 7.
Comparison of the GCCG reporter and the Tlx-2 reporter. The activities of 12xGCCG-Lux and pTlx-Lux were compared in P19 (left panel) and mink lung cells (right panel). The values represent fold induction of the luciferase activity of each reporter.
DISCUSSION
The GCCG motif was identified as a consensus Mad-binding site in Dpp-responsive genes (Kim et al., 1997). The motif was also suggested as a Medea-binding site (Xu et al., 1998). We investigated whether the GCCG motif serves as a binding motif for mammalian R-Smads regulated by BMPs. Mad and Smad1 are highly similar in structure and propagate signals of the BMP family. Smad1 bound to the enhancers of Dpp-responsive genes in the presence of Smad4 and BMP stimulation. Smad1 also bound to multimerized copies of the consensus GCCG motif. The binding affinity was highly dependent on the number of the copies of the motif, suggesting that the DNA-binding affinity for a single GCCG motif is relatively low. The binding of Smad1 to Dpp-responsive genes with a single GCCG motif was detected perhaps because the natural sequence contains a binding site(s) for a nuclear partner(s) for Smad1 provided from the COS cell lysates, although direct evidence is not available at present. A gel shift assay using GST-Smad fusion proteins indicates that both Smad1 and Smad4 can directly bind to the GCCG motif, although the MH2 domain of Smad1 inhibits the DNA binding of the protein. BMP stimulation induced the activation of reporters with multiple copies of the GCCG motif. In accordance with the result of the gel shift assay, the activity of the reporters increased in proportion to the number of the GCCG motif. The result is similar to that obtained with TGF-β-responsive reporters containing the CAGA motif (Dennler et al., 1998). The GCCG reporter responded to BMP even in cells in which a natural BMP-responsive reporter, pTlx-Lux, was inert. Activation of R-Smads through phosphorylation by BMP receptors may be sufficient to transactivate the GCCG reporters. pTlx-Lux may contain only a low number of Smad1-binding sites and requires other factors to recruit stable DNA-binding complexes. P19 cells may contain such nuclear partners for BMP-stimulated R-Smads, whereas mink lung cells may not. The GCCG reporters did not respond to TGF-β or activin stimulation, indicating that the GCCG motif may be specific to BMP-regulated Smads.
Screening of random sequences resulted in the identification of the GTCT motif as a binding site for Smad3 and Smad4 (Zawel et al., 1998). The crystal structure of Smad3 bound to the GTCT motif was determined (Shi et al., 1998). In the same report, it was mentioned that Smad1 also binds to the GTCT motif, although data were not shown. Indeed, sequences with multiple copies of the GTCT motif were bound and transactivated by Smad1 and Smad4 (Johnson et al., 1999). CAGACA was identified as a Smad-binding sequence in TGF-β-responsive genes such as PAI-1 and junB (Dennler et al., 1998; Jonk et al., 1998). The GTCTAGAC and CAGACA sequences share AGAC. Detailed analysis of the CAGACA sequence in the junB promoter revealed that the GAC core sequence is critical for Smad4 binding (Jonk et al., 1998). A reporter containing the CAGACA sequences from the junB promoter responded to BMPs (Jonk et al., 1998), whereas the (CAGA)12-MLP-Luc did not respond to BMP stimulation (Dennler et al., 1998). Although the reason for this difference is not clear, BMP-regulated Smads did not bind to the CAGA motif (Dennler et al., 1998; Jonk et al., 1998). goosecoid is an activin-responsive gene. Smad2 activates transcription of goosecoid only in the presence of mouse FAST-2 (Labbéet al., 1998). In the goosecoid promoter, Smad3 and Smad4 were shown to bind to GC-rich sequences distinct from the GCCG motif and unrelated to the CAGA motif. It has been suggested that the DNA-binding affinity of Smads is relatively low, and that the sequence requirement for DNA binding may not be very strict (Derynck et al., 1998). Our mutational analyses of the GCCG motif also suggest that Smad1 can bind to sequences different from the consensus GCCGnCGC sequence.
Smad3 alone strongly bound to the CAGA motif but not to the GCCG motif upon TGF-β stimulation. However, Smad3 weakly bound to the GCCG motif in the presence of Smad4 upon TGF-β stimulation (our unpublished results). Smad3 may be tethered to the GCCG motif via Smad4, because Smad3 interacts with Smad4 upon activation by TGF-β. The discrepancy between this DNA binding and the unresponsiveness of the GCCG motif to Smad3 in a reporter assay is currently unknown.
Smad4 has been shown to play an essential role in distinct signaling pathways; however, the molecular basis for this requirement has not been fully understood. Smad4 alone does not translocate into the nucleus, and R-Smads are required for the nuclear translocation of Smad4 (Liu et al., 1997). Therefore, Smad4 is not required for nuclear translocation of Smads. R-Smads recruit transcriptional coactivators such as p300 and CREB-binding protein (CBP). The interaction of Smad4 with CBP was TGF-β dependent, suggesting that Smad4 may interact with CBP via R-Smads (Feng et al., 1998). The MH2 domain of Smad4 does not have significant transactivation activity (Wu et al., 1997). Hence, Smad4 may not be directly involved in transactivation. Smad2 does not directly bind to DNA, because it contains an extra sequence in the MH1 domain, which interferes with DNA binding (Dennler et al., 1999; Yagi et al., 1999). The Mix.2 gene contains two CAGA motifs adjacent to the FAST-1-binding site, and Smad4 is likely to tether Smad2 to the binding sites (Dennler et al., 1998). Indeed, Smad4 was shown to promote the binding of the Smad–FAST-1 complex to DNA (Liu et al., 1997). In addition, Smad4 may stabilize the structure of hetero-oligomeric Smad complexes (Kawabata et al., 1998b). Homo-oligomers of Smad3 bind to DNA upon TGF-β stimulation (Kawabata et al., 1998b). Thus Smad3 does not require Smad4 in DNA binding, although it is not known whether Smad3 homo-oligomers induce transcriptional activation. Our results indicate that Smad4 is required for Smad1 to bind to DNA in vivo, and this seems to be at least one of the crucial roles of Smad4 in transcriptional regulation by BMP. The DNA-binding affinity of Smad1 to the GCCG motif may be lower than that of Smad3 to the CAGA motif and may require Smad4 to stably bind to DNA.
Only a few BMP-responsive reporters have been reported (Harada et al., 1997; Chen et al., 1998; Jonk et al., 1998; Tang et al., 1998; Johnson et al., 1999). The activation of the Tlx-2 gene, for example, depends on the cell type (Figure 7). The junB and GTCT reporters respond to BMP stimulation as well as to TGF-β stimulation (Jonk et al., 1998; Johnson et al., 1999). The GCCG reporters are likely to respond specifically and directly to Smads phosphorylated by BMP receptors. Thus they may serve as a universal reporter to detect BMP signals and contribute to investigate BMP signaling in a variety of systems.
ACKNOWLEDGMENTS
We thank S. Harada for the COLX luciferase reporter plasmid, J.L. Wrana for pTlx-Lux, J.-M. Gauthier for (CAGA)9-MLP-Luc, T. Momoi for P19 cells, J. Massagué for R mutant mink lung cells, Y. Eto for activin A, and T.K. Sampath for BMP-7. This study was supported by grants-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture of Japan and special coordination funds for promoting science and technology from the Science and Technology Agency.
REFERENCES
- Bachner D, Ahrens M, Schroder D, Hoffmann A, Lauber J, Betat N, Steinert P, Flohe L, Gross G. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2) Dev Dyn. 1998;213:398–411. doi: 10.1002/(SICI)1097-0177(199812)213:4<398::AID-AJA5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Brummel T, Abdollah S, Haerry TE, Shimell MJ, Merriam J, Raftery L, Wrana JL, O'Connor MB. The Drosophila activin receptor Baboon signals through dSmad2 and controls cell proliferation but not patterning during larval development. Genes Dev. 1999;13:98–111. doi: 10.1101/gad.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rubock MJ, Whitman M. A transcriptional partner for MAD proteins in TGF-β signaling. Nature. 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- Chen Y-G, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massagué J. Determinants of specificity in TGF-β signal transduction. Genes Dev. 1998;12:2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P, Inoue H, Baker JC, Beppu H, Kawabata M, Harland RM, Miyazono K, Padgett RW. Drosophila dSmad2 and Atr-I transmit activin/TGFβ signals. Genes Cells. 1999;4:123–134. doi: 10.1046/j.1365-2443.1999.00244.x. [DOI] [PubMed] [Google Scholar]
- Dennler S, Huet S, Gauthier J-M. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18:1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J-M. Direct binding of Smad3 and Smad4 to critical TGF β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng X-H. Smads: transcriptional activators of TGF-β responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- Feng X-H, Zhang Y, Wu R-Y, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S, Sampath TK, Aubin JE, Rodan GA. Osteogenic protein-1 up-regulation of the collagen X promoter activity is mediated by a MEF-2-like sequence and requires an adjacent AP-1 sequence. Mol Endocrinol. 1997;11:1832–1845. doi: 10.1210/mend.11.12.0022. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Johnson K, Kirkpatrick H, Comer A, Hoffmann FM, Laughon A. Interaction of Smad complexes with tripartite DNA-binding sites. J Biol Chem. 1999;274:20709–20716. doi: 10.1074/jbc.274.29.20709. [DOI] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin C-H, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Imamura T, Miyazono K. Signal transduction by bone morphogenetic proteins. Cytokine Growth Factor Rev. 1998a;9:49–61. doi: 10.1016/s1359-6101(97)00036-1. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998b;17:4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen H-J, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Labbé E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF β-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Liberati NT, Datto MB, Frederick JP, Shen X, Wong C, Rougier-Chapman EM, Wang X-F. Smads bind directly to the Jun family of AP-1 transcription factors. Proc Natl Acad Sci USA. 1999;96:4844–4849. doi: 10.1073/pnas.96.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Pouponnot C, Massagué J. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 1997;11:3157–3167. doi: 10.1101/gad.11.23.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macías-Silva M, Hoodless PA, Tang SJ, Buchwald M, Wrana JL. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J Biol Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nishihara A, Hanai J-i, Okamoto N, Yanagisawa J, Kato S, Miyazono K, Kawabata M. Role of p300, a transcriptional coactivator, in signaling of TGF-β. Genes Cells. 1998;3:613–623. doi: 10.1046/j.1365-2443.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- Padgett RW, Das P, Krishna S. TGF-β signaling, Smads, and tumor suppressors. Bioessays. 1998;20:382–390. doi: 10.1002/(SICI)1521-1878(199805)20:5<382::AID-BIES5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Padgett RW, Savage C, Das P. Genetic and biochemical analysis of TGF β signal transduction. Cytokine Growth Factor Rev. 1997;8:1–9. doi: 10.1016/s1359-6101(96)00050-0. [DOI] [PubMed] [Google Scholar]
- Padgett RW, Wozney JM, Gelbart WM. Human BMP sequences can confer normal dorsal-ventral patterning in the Drosophila embryo. Proc Natl Acad Sci USA. 1993;90:2905–2909. doi: 10.1073/pnas.90.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddi AH. Bone morphogenetic proteins: an unconventional approach to isolation of first mammalian morphogens. Cytokine Growth Factor Rev. 1997;8:11–20. doi: 10.1016/s1359-6101(96)00049-4. [DOI] [PubMed] [Google Scholar]
- Sampath TK, Rashka KE, Doctor JS, Tucker RF, Hoffmann FM. Drosophila transforming growth factor β superfamily proteins induce endochondral bone formation in mammals. Proc Natl Acad Sci USA. 1993;90:6004–6008. doi: 10.1073/pnas.90.13.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Yang X, Chen D, Chang Z, Cao X. Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J Biol Chem. 1999;274:13711–13717. doi: 10.1074/jbc.274.19.13711. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang Y-F, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Shukunami C, Shigeno C, Atsumi T, Ishizeki K, Suzuki F, Hiraki Y. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J Cell Biol. 1996;133:457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Hoodless PA, Lu Z, Breitman ML, McInnes RR, Wrana JL, Buchwald M. The Tlx-2 homeobox gene is a downstream target of BMP signaling and is required for mouse mesoderm development. Development. 1998;125:1877–1887. doi: 10.1242/dev.125.10.1877. [DOI] [PubMed] [Google Scholar]
- ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DL, Ichijo H, Heldin C-H, Miyazono K. Identification of type I receptors for osteogenic protein-1 and bone morphogenetic protein-4. J Biol Chem. 1994;269:16985–16988. [PubMed] [Google Scholar]
- Whitman M. Smads and early developmental signaling by the TGFβ superfamily. Genes Dev. 1998;12:2445–2462. doi: 10.1101/gad.12.16.2445. [DOI] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Lee S, Massagué J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- Wu R-Y, Zhang Y, Feng X-H, Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol Cell Biol. 1997;17:2521–2518. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Yin Z, Hudson JB, Ferguson EL, Frasch M. Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng X-H, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- Zhou S, Zawel L, Lengauer C, Kinzler KW, Vogelstein B. Characterization of human FAST-1, a TGF β and activin signal transducer. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]