Abstract
The plasma membrane ATPase, encoded by PMA1, is delivered to the cell surface via the secretory pathway. Previously, we characterized a temperature-sensitive pma1 mutant in which newly synthesized Pma1-7 is not delivered to the plasma membrane but is mislocalized instead to the vacuole at 37°C. Several vps mutants, which are defective in vacuolar protein sorting, suppress targeting-defective pma1 by allowing mutant Pma1 to move once again to the plasma membrane. In this study, we have analyzed trafficking in the endosomal system by monitoring the movement of Pma1-7 in vps36, vps1, and vps8 mutants. Upon induction of expression, mutant Pma1 accumulates in the prevacuolar compartment in vps36 cells. After chase, a fraction of newly synthesized Pma1-7 is delivered to the plasma membrane. In both vps1 and vps8 cells, newly synthesized mutant Pma1 appears in small punctate structures before arrival at the cell surface. Nevertheless, biosynthetic membrane traffic appears to follow different routes in vps8 and vps1: the vacuolar protein-sorting receptor Vps10p is stable in vps8 but not in vps1. Furthermore, a defect in endocytic delivery to the vacuole was revealed in vps8 (and vps36) but not vps1 by endocytosis of the bulk membrane marker FM 4-64. Moreover, in vps8 cells, there is defective down-regulation from the cell surface of the mating receptor Ste3, consistent with persistent receptor recycling from an endosomal compartment to the plasma membrane. These data support a model in which mutant Pma1 is diverted from the Golgi to the surface in vps1 cells. We hypothesize that in vps8 and vps36, in contrast to vps1, mutant Pma1 moves to the surface via endosomal intermediates, implicating an endosome-to-surface traffic pathway.
INTRODUCTION
In mammalian cells, entry of newly synthesized hydrolases into the lysosomal pathway is mediated by a sorting receptor at the trans-Golgi complex (Kornfeld and Mellman, 1989). Analogously, in Saccharomyces cerevisiae, Vps10 is a sorting receptor that recycles between the trans-Golgi and the endosome, recognizing a signal within soluble hydrolases and thereby effecting their delivery to the vacuole (Marcusson et al., 1994; Cooper and Stevens, 1996). Genetic screens to identify yeast mutants defective in delivery of the vacuolar marker carboxypeptidase Y (CPY) have resulted in >40 VPS genes required for proper vacuolar protein sorting, revealing the complexity of the vacuole biosynthetic pathway (Rothman and Stevens, 1986; Robinson et al., 1988). Because biosynthetic traffic to the vacuole or lysosome intersects with endocytic traffic at the endosome, a subset of the vps mutants also displays defects in endocytosis (Davis et al., 1993; Munn and Riezman, 1994; Singer-Kruger et al., 1994).
The general organization of the endocytic pathway has been especially well characterized in mammalian cells. Specifically, extracellular molecules and plasma membrane proteins travel through the endocytic pathway to the lysosome via two sequential intermediates, early and late endosomes. Similarly, in yeast, two populations of endosomes have been defined morphologically as well as biochemically (Singer-Kruger et al., 1993; Hicke et al., 1997; Mulholland et al., 1999). Nevertheless, a molecular understanding of endosome function remains far from complete (Gruenberg and Maxfield, 1995; Mellman, 1996; Riezman et al., 1997). In mammalian cells, a recycling pathway is well established by which certain internalized proteins are delivered back to the plasma membrane (Gruenberg and Maxfield, 1995; Mellman, 1996). In contrast, in yeast, the existence of a direct traffic pathway from the endosome to the cell surface has not been established to date, although a number of recent studies suggest such a pathway (Harsay and Bretscher, 1995; Yuan et al., 1997; Ziman et al., 1998). Indeed, two populations of Golgi-derived secretory vesicles have been isolated from yeast; the finding that preventing endocytosis results in the accumulation of only one of these populations is consistent with the idea that one of these classes of vesicles may derive from endocytic recycling (Harsay and Bretscher, 1995).
We were prompted to examine whether an endosome-to-surface traffic pathway exists in yeast by studies of PMA1, which encodes the plasma membrane ATPase. Normally, Pma1 reaches the cell surface via the secretory pathway (Brada and Schekman, 1988; Chang and Slayman, 1991). However, in the temperature-sensitive pma1-7 mutant, newly synthesized Pma1 is defective for targeting to the plasma membrane at 37°C and instead is delivered to the vacuole via the endosome (Chang and Fink, 1995; Luo and Chang, 1997). Although the molecular basis for vacuolar delivery of Pma1-7 is unknown, we have considered the possibility that there is a post-endoplasmic reticulum quality control mechanism that recognizes and targets mutant Pma1 into the endosomal/vacuolar system (Chang and Fink, 1995; Hong et al., 1996; Li et al., 1999). Several vps mutants, which are defective in vacuolar protein sorting, have been identified that cause rerouting of mutant Pma1 to the plasma membrane (Luo and Chang, 1997). By disrupting the recycling of a Golgi-based quality control receptor, these vps mutants might allow Pma1-7 to travel directly from the Golgi to the cell surface. With this in mind, we have compared trafficking pathways of mutant Pma1 in vps1, vps8, and vps36. Remarkably, the data suggest that in vps8 and vps36 cells, Pma1-7 moves to the plasma membrane only after it has entered the endosomal system.
MATERIALS AND METHODS
Media and Strains
Standard yeast media and genetic manipulations were as described (Sherman et al., 1986). Yeast transformations were performed by the lithium acetate method (Gietz et al., 1992). Strains used in this study are listed in Table 1. All strains except those marked with asterisks are isogenic with L3852. vps8-Δ1::LEU2 and vps36-Δ1 were isolated as suppressors of pma1-7 after insertional mutagenesis (Luo and Chang, 1997). ACY76 was generated in a one-step gene replacement by transformation of L3852 with pPS83, a HIS3-marked VPS8 disruption construct (Horazdovsky et al., 1996) provided by B. Horazdovsky (Texas Southwestern Medical Center, Dallas, TX). ACY33 was generated in a one-step gene replacement by transformation of L3852 with pKJH2, a LEU2-marked VPS27 disruption construct (Piper et al., 1995) provided by T. Stevens (University of Oregon, Eugene). WLY65 was generated in a one-step gene replacement by transformation of L3852 with pBS-YPT51-LYS2 (Singer-Kruger et al., 1994) provided by B. Singer-Kruger (University of Stuttgart, Germany). WLX20-5D is an ascospore from a cross between WLX16-1A and WLX4-2A (MATa lys2Δ201 leu2-3,112 ura3-52 ade2 vps27-Δ1::LEU2). Integration of MET-HA-PMA1 and MET-HA-pma1-7 at ura3-52 was accomplished by transforming yeast with pWL10 and pWL9 linearized with NcoI. ACY72 was constructed by pop-in, pop-out gene replacement of STE3 by transformation of ACX66-2D (MATα his3Δ200 leu2-3,112 ura3-52 ade2 trp1Δ63 GAL+) with pSL1904 (provided by N. Davis, Wayne State University, Detroit, MI), resulting in replacement of the STE3 promoter with a GAL1,10 promoter. ACY81 was constructed by transformation of ACY72 with pPS83, a vps8::HIS3 disruption construct. ACY84 and ACY85 were constructed by transformation of ACY72 and ACY81 with pAS173, a pep4::hisG-URA3-hisG disruption construct (Chang and Fink, 1995), to disrupt PEP4.
Table 1.
Yeast strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| L3852 | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 | Fink lab collection |
| WLX16-1A | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 vps8-Δ1::LEU2 | Luo and Chang, 1997 |
| WLX12-7C | MATα lys2Δ201 leu2-3,112 ura3-52 vps36-Δ1::LEU2 | Luo and Chang, 1997 |
| ACX58-3C | MATa his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 vps1Δ::LEU2 | Luo and Chang, 1997 |
| ACY33 | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 vps27Δ::LEU2 | Luo and Chang, 1997 |
| WLX20-5D | MATα lys2Δ201 leu2-3,112 ura3-52 ade2 vps8-Δ1::LEU2 vps27-Δ1::LEU2 | This study |
| ACY76 | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 vps8Δ::HIS3 | This study |
| WLY65 | MATα his3Δ200 lys2Δ201 leu2-3,112 ura3-52 ade2 ypt51::LYS2 | This study |
| ACY72* | MATα his3Δ200 leu2-3, ura3-52 trp1-Δ63 ade2 GAL1-STE3 | This study |
| ACY81* | MATα his3Δ200 leu2-3,112 ura3-52 trp1-Δ63 ade2 vps8::HIS3 GAL1-STE3 | This study |
| ACY84* | MATα his3Δ200 leu2-3,112 ura3-52 trp1-Δ63 ade2 pep4::URA3 GAL1-STE3 | This study |
| ACY85* | MATα his3Δ200 leu2-3,112 ura3-52 trp1-Δ63 ade2 vps8::HIS3 pep4::URA3 GAL1-STE3 | This study |
| WLY152 | MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 pep4Δ ura3-52::MET-HA-pma1-7 | This study |
| WLY153 | MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 pep4Δ ura3-52::MET-HA-PMA1 | This study |
| WLY103 | MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 ura3-52::MET25-HA-pma1-7 | This study |
| WLY104 | MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 ura3-52::MET25-HA-PMA1 | This study |
| WLY127 | MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 vps8-Δ1::LEU2 ura3-52::MET25-HA-pma1-7 | This study |
| WLY128 | MATα his3Δ200 lys2Δ201 leu2-3,112 ade2 vps8-Δ1::LEU2 ura3-52::MET25-HA-PMA1 | This study |
| WLY144 | MATa his3Δ200 lys2Δ201 leu2-3,112 ade2 vps1Δ::LEU2 ura3-52::MET25-HA-pma1-7 | This study |
| WLY145 | MATa his3Δ200 lys2Δ201 leu2-3,112 ade2 vps1Δ::LEU2 ura3-52::MET25-HA-PMA1 | This study |
| WLY156 | MATα lys2Δ201 leu2-3,112 vps36-Δ1::LEU2 ura3-52::MET25-HA-pma1-7 | This study |
| WLY157 | MATα lys2Δ201 leu2-3,112 vps36-Δ1::LEU2 ura3-52::MET25-HA-PMA1 | This study |
Molecular Biology
Plasmids with HA-tagged pma1-7 and PMA1 under the control of the MET25 promoter were constructed as follows. With the use of XhoI (polylinker) and BstEII sites, a 1.7-kilobase (kb) fragment from the 5′ region was removed from pAC7 and pAC4 bearing 4.5-kb HindIII–HindIII pma1-7 and PMA1 inserts, respectively (Chang and Fink, 1995). The fragment was replaced with a 750-base pair (bp) fragment from pFT4 (provided by C. Slayman, Yale University, New Haven, CT), which has a HindIII site introduced at −27 bp from the start codon, generating pWL1 and pWL2. A 4.2-kb HindIII–HindIII fragment bearing the PMA1 coding sequence was excised from pWL1 and pWL2 and placed after the MET25 promoter of FB1521 (Mumberg et al., 1994). The 4.6-kb fragments containing MET-pma1-7 and MET-PMA1 were excised with the use of SacI–XhoI polylinker sites and placed into pRS306, a URA3-marked YIp, generating pWL5 and pWL6, respectively. To introduce an HA epitope, the plasmid pXZ28 (containing PMA1 with an HA epitope introduced after the second amino acid; provided by J. Haber, Brandeis University, Waltham, MA) was used as a template for PCR. A fragment of 0.8 kb was amplified with the use of the oligonucleotide TCCCCCGGGAGCTAGTTAAAGAAAATC to introduce a SmaI site at −67 bp from the start codon and the oligonucleotide CCTTCACCTCTCTTAACA. After cutting with SmaI and BstEII, the PCR fragment was used to replace the corresponding fragments in pWL5 and pWL6 to generate pWL9 and pWL10, respectively.
Protein Induction
To detect newly synthesized Pma1, plasmids were used in which HA-tagged mutant or wild-type Pma1 was placed under the control of the MET25 promoter. Cells were grown under repressing conditions in minimal medium containing 600 μM methionine. To induce synthesis of Pma1, cells were washed once with water and resuspended in methionine-free medium. At the same time, cells were shifted to 37°C. Synthesis of HA-tagged Pma1 was shut off by adding 2 mM methionine alone or in the presence of 100 μg/ml cycloheximide.
To study Ste3, cells were grown to midlog phase at 30°C in synthetic complete minus uracil medium with 2% galactose. Glucose (3%) was added to stop synthesis of Ste3. For detection of Ste3 by Western blot, anti-Ste3 mAb (provided by G. Sprague, University of Oregon) was used. For Ste3 detection by indirect immunofluorescence, cells were transformed with a GAL1-STE3 construct in which a c-myc epitope is fused to the carboxyl terminus of STE3 (pSL2015; provided by N. Davis, Wayne State University).
Indirect Immunofluorescence, Cell Fractionation, Western Blotting, and Metabolic Labeling
For indirect immunofluorescence, cells were spheroplasted with oxalyticase (Enzogenetics, Corvallis, OR) and permeabilized with methanol and acetone, as described (Rose et al., 1990). Cells were stained with anti-HA (BABCO, Berkeley, CA) or anti-myc mAb (Santa Cruz Biotechnology, Santa Cruz, CA) followed by Cy3-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA). Pulse-chase experiments were visualized with the use of an Olympus (Lake Success, NY) IX70 microscope, and the images were collected digitally (with the same exposure for each time point within an experiment) and adjusted at the same settings with Adobe Photoshop 4.0 (Adobe Systems, Mountain View, CA). All other fluorescent microscopy experiments were photographed with the use of a Zeiss Axiophot microscope (Carl Zeiss, Thornwood, NY).
Cell fractionation on Renografin (gift of L. Marsh, Albert Einstein College of Medicine) density gradients was performed essentially as described (Schandel and Jenness, 1994; Jenness et al., 1997). Harvested samples were placed on ice in the presence of 10 mM azide. Cell lysates were prepared by vortexing cells with glass beads in the presence of a protease inhibitor cocktail including 1 mM PMSF (Chang and Slayman, 1991). After centrifugation at 400 × g for 5 min to remove unbroken cells, lysate (0.5 ml) was mixed with 0.5 ml of Renografin-76, placed at the bottom of a centrifuge tube, and overlaid with 1 ml of 34, 30, 26, and 22% Renografin solutions. For pulse-chase experiments, samples from different time points were loaded on gradients after normalization to lysate protein by the Bradford assay (Bio-Rad Laboratories, Hercules, CA). Gradients were centrifuged in an SW50.1 rotor overnight at 150,000 × g at 4°C. Fourteen fractions (350 μl) were collected from the top of each gradient. To prevent Renografin from interfering with subsequent Western blotting, membranes were diluted with Tris-EDTA buffer and pelleted by centrifugation at 100,000 × g for 1 h. Intracellular membranes and plasma membrane were consistently contained in fractions 6–7 and 10–11, respectively. Therefore, quantitation of newly synthesized Pma1 was performed by pooling fractions 1–8 and 9–14. Distribution of mutant Pma1 after induction and chase was determined by Western blotting and calculated after subtraction of the background signal obtained at time 0.
For Western blotting of cell lysate, samples were prepared as described previously (Chang and Slayman, 1991) and normalized to lysate protein. After separation by SDS-PAGE, proteins were transferred to nitrocellulose. Anti-HA Western blotting was performed with the use of mAb. Antibodies against Kex2, Pep12, and Gas1 were provided by S. Nothwehr (University of Missouri, Columbus), H. Pelham (Medical Research Council Laboratories, Cambridge, UK), and T. Doering (Washington University, St. Louis, MO), respectively. Immune complexes were visualized by chemiluminescence detection reagents (ECL Western blotting detection system; Amersham, Arlington Heights, IL) or 125I-protein A (Amersham). Quantitation of Western blots with the use of 125I-protein A was performed with the use of a Molecular Dynamics (Sunnyvale, CA) phosphorimager.
Metabolic labeling was performed with the use of cultures grown to midlog phase in synthetic complete medium without methionine and cysteine. Cells were resuspended at 1 OD600/ml, labeled with Expre35S35S (New England Nuclear, Boston, MA) (2 mCi/25 OD600 cells) for 5 min at room temperature, and chased in the presence of 10 mM methionine and cysteine. At various times during the chase, aliquots were placed on ice in the presence of 10 mM Na azide. Lysates were prepared and resuspended in RIPA buffer (10 mM Tris, pH 7.5, 150 nM NaCl, 2 mM EDTA, 1% NP40, 1% deoxycholate, 0.1% SDS) for immunoprecipitation.
RESULTS
Inducible Synthesis of Mutant Pma1
To visualize movement of newly synthesized Pma1, we used constructs in which wild-type PMA1 and mutant pma1-7 were tagged with an HA epitope and placed under the control of the MET25 promoter. Although a low level of synthesis is detected under repressing conditions, the Western blot in Figure 1 shows a large increase in synthesis of Pma1 upon activation of the MET25 promoter (by removal of methionine from the medium). Quantitation reveals that the level of wild-type Pma1 upon induction is approximately fivefold greater than that of mutant Pma1, probably because of concurrent degradation of the mutant protein. After stopping synthesis, degradation of newly synthesized mutant Pma1 is readily apparent after 90 min, whereas wild-type Pma1 remains stable (Figure 1).
Figure 1.
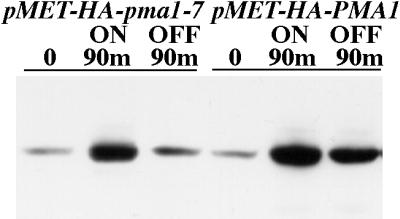
Stability of newly synthesized wild-type and mutant Pma1. Pma1 levels in wild-type cells bearing either pMET-HA-pma1-7 or pMET-HA-PMA1 (WLY103 and WLY104). Exponentially growing cells were washed, transferred to methionine-free medium to induce synthesis of epitope-tagged protein, and shifted to 37°C. After 90 min, methionine was added to prevent further Pma1 synthesis, and incubation continued for an additional 90 min. Lysates were prepared and analyzed by Western blot with anti-HA antibody. Note that newly synthesized wild-type Pma1 is stable, whereas mutant Pma1 is degraded during chase.
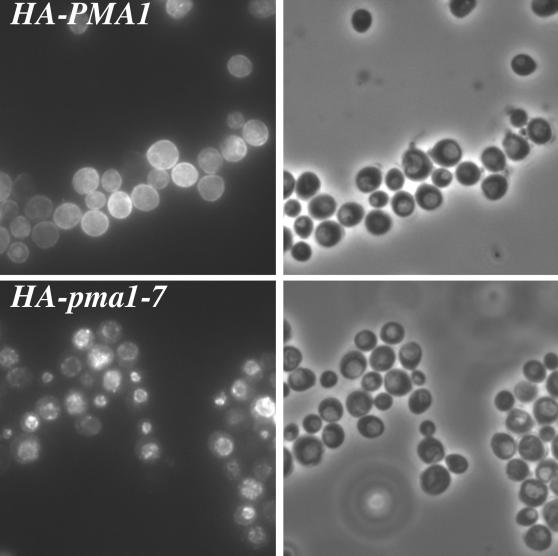
Previously, we demonstrated that degradation of mutant Pma1 occurs at 37°C upon trafficking of the newly synthesized protein to the vacuole (Chang and Fink, 1995; Luo and Chang, 1997). To confirm that epitope-tagged Pma1 behaves in the same manner, indirect immunofluorescence localization was performed in pep4 cells defective in vacuolar protease activity. Synthesis of HA-tagged Pma1 was induced for 1 h at 37°C, and the cells were then stained with anti-HA antibody. As shown in Figure 2, newly synthesized Pma1-7 is accumulated at the vacuole (bottom left), which is recognized as pale regions under phase contrast optics (bottom right). In contrast, staining of cells expressing wild-type Pma1 is at the cell surface (top).
Figure 2.
Vacuolar delivery of mutant Pma1. Indirect immunofluorescence localization of newly synthesized Pma1. pep4 cells carrying pMET-HA-pma1-7 (WLY152) or pMET-HA-PMA1 (WLY153) were washed free of methionine to induce synthesis of epitope-tagged Pma1 and shifted to 37°C for 1 h. Samples were then fixed, spheroplasted, and permeabilized for indirect immunofluorescence staining with anti-HA antibody followed by Cy3-conjugated secondary antibody. Left panels show indirect immunofluorescence images; right panels show the corresponding phase images. Newly synthesized wild-type Pma1 is localized at the cell surface, whereas mutant Pma1 is delivered to the vacuole.
Newly Synthesized Mutant Pma1 Reaches the Prevacuolar Compartment of vps36 Cells before Arrival at the Cell Surface
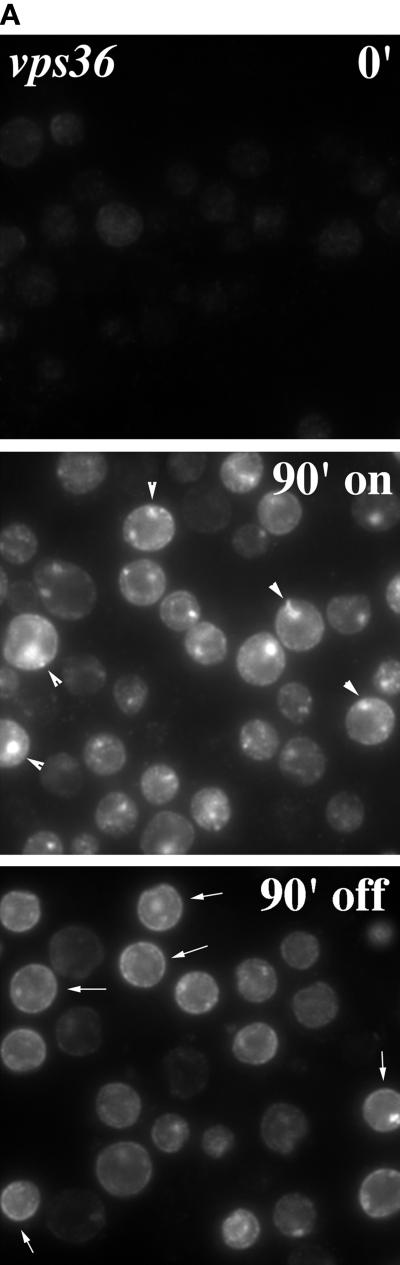
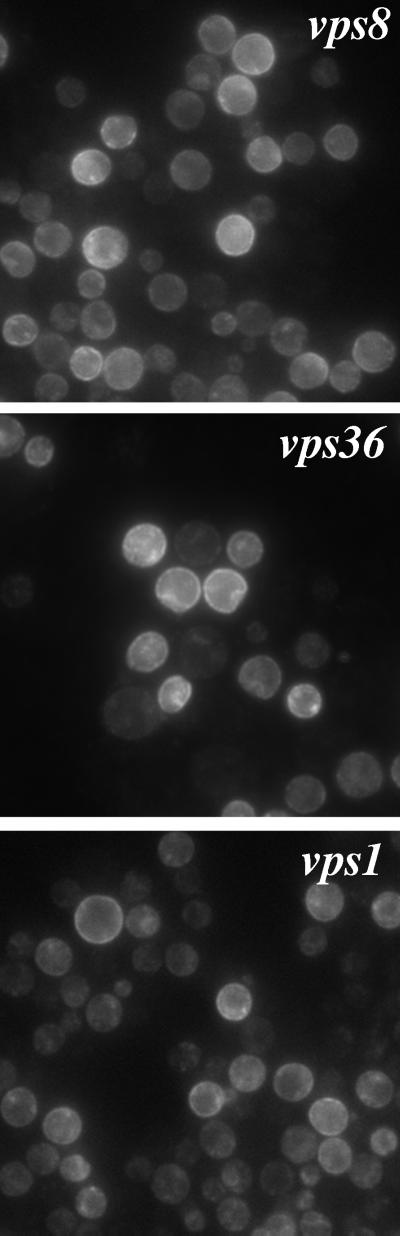
Mutant Pma1 is delivered to the cell surface in two class E vps mutants, vps36 and vps27 (Luo and Chang, 1997). Class E vps mutants are characterized by defective trafficking from the endosome to the vacuole as well as from the endosome back to the Golgi (Piper et al., 1995). In both vps36 and vps27 mutants, some proteins traversing endocytic and biosynthetic pathways are trapped in a novel prevacuolar compartment that is visualized as a characteristic large spot next to the vacuole; on the other hand, newly synthesized CPY is missorted in these cells and travels directly from the Golgi to the cell surface (Raymond et al., 1992; Piper et al., 1995). To determine the route by which mutant Pma1 travels to the plasma membrane in the class E vps mutants, the localization of newly synthesized Pma1 was determined by staining with anti-HA antibody. As shown in Figure 3A (0′, top), no staining is seen in vps36Δ cells before induction of mutant Pma1 synthesis. After a 90-min induction period, staining of newly synthesized Pma1-7 appears predominantly in large perivacuolar spots; little cell surface staining is apparent (Figure 3A, middle, arrowheads). Previously, we established that this staining pattern is characteristic of mutant Pma1 accumulating in the prevacuolar compartment (Luo and Chang, 1997). The same pattern of Pma1-7 accumulation was also seen in vps27Δ cells (data not shown). These observations confirm that newly synthesized mutant Pma1 enters the endosomal system in class E vps mutants. In contrast, wild-type Pma1 does not detectably accumulate in the prevacuolar compartment or other intracellular compartments (Figure 4).
Figure 3.
Localization of newly synthesized mutant Pma1 in vps36 cells. Synthesis of HA-tagged mutant Pma1 was induced at 37°C, followed by a 90-min chase in the presence of cycloheximide, as described in MATERIALS AND METHODS. (A) Indirect immunofluorescence localization was performed before induction of synthesis (0′), after a 90-min induction (90′ on), and after a 90-min chase in vps36 cells (WLY156) (90′ off). Cells were stained with anti-HA antibody followed by Cy3-conjugated secondary antibody. In vps36 cells, newly synthesized mutant Pma1 is accumulated in the prevacuolar compartment after induction (middle panel, arrowheads). A slight increase in staining at the plasma membrane is apparent after chase (bottom panel, arrows). (B) Quantitative distribution of newly synthesized Pma1-7 in vps36. Distribution in intracellular and plasma membrane fractions was quantitated after fractionation on Renografin density gradients, as described in MATERIALS AND METHODS. Data are expressed as absolute arbitrary units.
Figure 4.
Localization of newly synthesized wild-type Pma1 in vps cells. Synthesis of HA-tagged wild-type Pma1 was induced at 37°C in vps1 (WLY145), vps8 (WLY128), and vps36 (WLY157), as described in MATERIALS AND METHODS. Indirect immunofluorescence staining was performed on cells after 90 min of induction. Cells were stained with anti-HA antibody followed by Cy3-conjugated secondary antibody.
To determine whether mutant Pma1 can move to the cell surface from the prevacuolar compartment, vps36Δ cells were reexamined after an additional 90-min chase period. Cycloheximide was included during the chase to ensure that no additional Pma1 synthesis could occur. As shown in Figure 3A (bottom), staining of the prevacuolar compartment declines after the chase. Much of the decrease is likely due to degradation in the proteolytically active prevacuolar compartment (Cereghino et al., 1995). In addition, staining of the plasma membrane appears to increase slightly (Figure 3A, bottom, arrows). A similar pattern was observed in vps27Δ cells (data not shown). Quantitation of newly synthesized Pma1-7 at the cell surface was performed by fractionation on Renografin density gradients, which efficiently separate plasma membranes from intracellular membranes (Jenness et al., 1997) (see below). Figure 3B shows the results of such an experiment in which the level of mutant Pma1 in intracellular and plasma membranes in vps36 cells was quantitated after induction and chase and plotted in arbitrary units. In this experiment, the plasma membrane fraction contains 31% of total Pma1-7 present during the induction period; after chase, the plasma membrane fraction represents 41% of Pma1-7 remaining. In three independent experiments, an ∼9% fractional increase at the cell surface was observed after chase. Thus, relocation of Pma1-7 from the endosomal system to the cell surface appears to occur, albeit inefficiently.
Cell Surface Delivery of Newly Synthesized Mutant Pma1 in vps1 and vps8
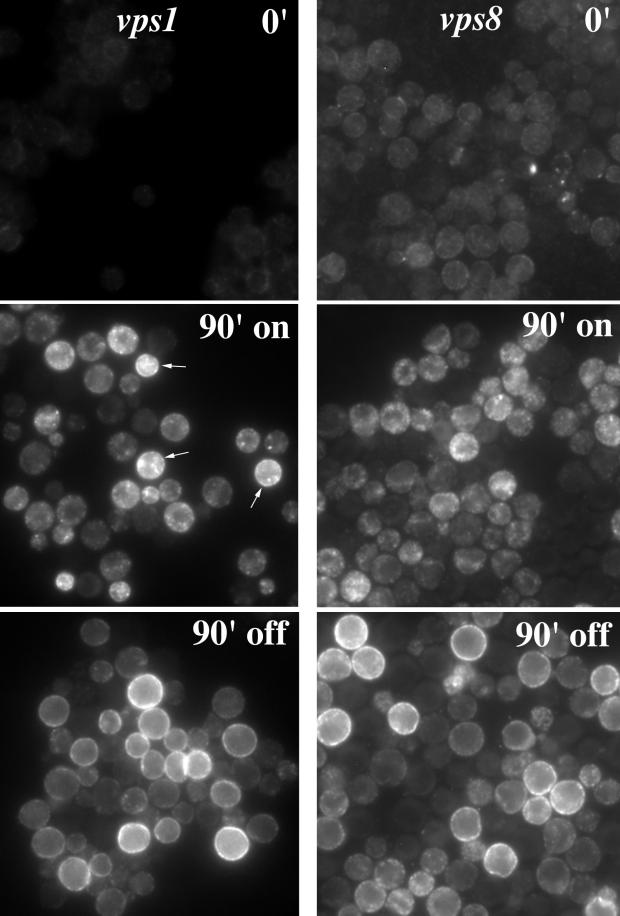
To compare a possible endosome-to-surface route with a Golgi-to-surface route, mutant Pma1 trafficking was followed in vps1 cells. Vps1 is a dynamin-like protein required for formation of endosome-bound vesicles from the Golgi, and all endosome-directed traffic is diverted to the cell surface in vps1 mutants (Nothwehr et al., 1995). Figure 5 (left middle panel) shows mutant Pma1 localization in vps1Δ cells after a 90-min induction period. Both bright punctate cytoplasmic staining and some surface staining (arrows) are apparent. After 90 min of chase (left bottom panel), the punctate staining has disappeared and there is exclusive staining of the cell surface. These observations are consistent with direct transport of newly synthesized mutant Pma1 from the Golgi to the plasma membrane.
Figure 5.
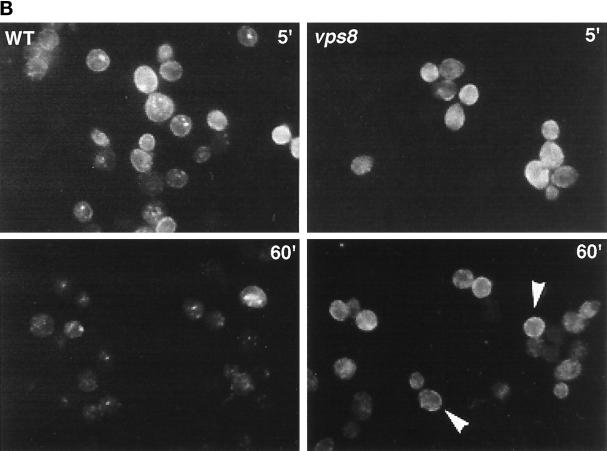
Localization of newly synthesized mutant Pma1 in vps1 and vps8 cells. Synthesis of HA-tagged mutant Pma1 was induced for 90 min at 37°C. Indirect immunofluorescence localization of mutant Pma1 was performed in vps1 (WLY144; left panel) and vps8 (WLY127; right panel) at time 0 (0′), after a 90-min induction (90′ on), and after a 90-min chase (90′ off). Upon induction, Pma1-7 is seen in small punctate structures in vps1 and vps8 cells. At this time in vps1, surface staining is also apparent (arrows). After chase, mutant Pma1 is distributed predominantly at the cell surface in vps1 and vps8 cells.
vps8 mutation also allows Pma1-7 to travel to the plasma membrane (Luo and Chang, 1997). Because vps8 cells accumulate the bulk membrane marker FM 4-64 in an endocytic intermediate compartment distinct from the prevacuolar compartment (Luo and Chang, 1997) (see below), it was of interest to examine the pathway to the cell surface taken by Pma1-7 in vps8. As shown in Figure 5, after induction for 90 min, mutant Pma1 localizes to punctate cytoplasmic structures (right middle panel). After a 90-min chase, staining is predominantly at the plasma membrane, indicating that newly synthesized Pma1-7 moves to the cell surface (right bottom panel). Movement to the plasma membrane in both vps8 and vps1 cells is unaffected by the presence of cycloheximide (data not shown). Compared with vps36, cell surface delivery of mutant Pma1 appears more efficient in vps8 and vps1 (compare bottom panels of Figures 3A and 5).
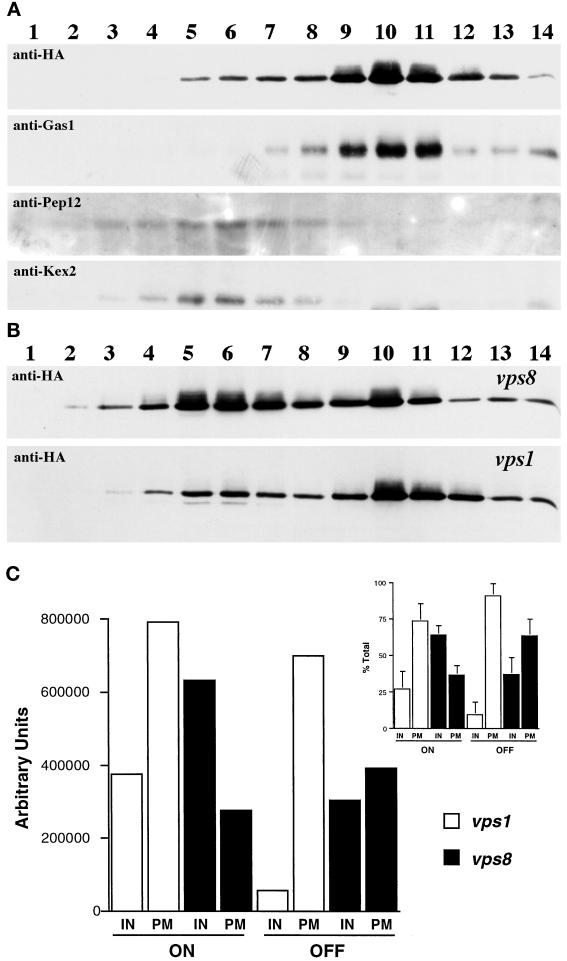
Although the immunofluorescence localization pattern of Pma1-7 in vps8Δ resembles that in vps1Δ cells, careful comparison suggests slightly more surface staining after induction in vps1 (Figure 5, left middle panel, arrows). To compare further vps1 and vps8 mutants, cell fractionation was performed on Renografin density gradients. Figure 6A shows the distribution of marker proteins after fractionation of wild-type cells on a Renografin gradient. The plasma membrane protein Gas1 is found predominantly in fractions 10 and 11. Newly synthesized wild-type Pma1 is also localized in these fractions. In contrast, Kex2 and Pep12, membrane proteins that recycle between the Golgi and the endosome (Wilcox et al., 1992; Becherer et al., 1996), are predominantly distributed in fractions 5–7. Figure 6B shows the fractionation pattern of Pma1-7 in vps1Δ and vps8Δ cells. After 90 min of induction at 37°C in vps1Δ cells, the majority of newly synthesized Pma1-7 cofractionates with plasma membrane in fractions 10 and 11. In contrast, in vps8Δ cells, a larger fraction of newly synthesized Pma1-7 is distributed in intracellular membrane fractions. Because there is distinct and consistent separation between intracellular and plasma membranes on Renografin density gradients, fractions 1–8 and 9–14 were pooled for analysis by quantitative Western blotting. As shown in Figure 6C, after 90 min of induction, >80% of mutant Pma1 has reached the cell surface in vps1, whereas in vps8, Pma1-7 is mostly intracellular. After 90 min of chase, the fraction of mutant Pma1 at the cell surface is increased in vps8Δ cells. These data reveal that Pma1-7 is delivered to the plasma membrane with different kinetics in vps1 and vps8.
Figure 6.
Kinetics of mutant Pma1 movement to the cell surface in vps1 and vps8 cells. Wild-type cells bearing pMET-HA-PMA1 and vps1 and vps8 cells bearing pMET-HA-pma1-7 were resuspended in methionine-free medium and shifted to 37°C for 90 min to induce synthesis of epitope-tagged Pma1. Lysates were prepared and fractionation on Renografin density gradients was performed as described in MATERIALS AND METHODS. Fractions were collected and assayed for marker proteins. (A) Western blot showing distribution of newly synthesized wild-type Pma1, the plasma membrane marker Gas1, and the Golgi/endosome markers Kex2 and Pep12 in wild-type cells (WLY104). (B) Gradient distribution of Pma1-7 in vps1 and vps8 cells (WLY144 and WLY127) . Synthesis of HA-tagged mutant Pma1 was induced for 90 min at 37°C. At this time in vps1 cells, newly synthesized Pma1 is predominantly in plasma membrane–containing fractions (lanes 10 and 11); in vps8, mutant Pma1 is found in intracellular fractions (lanes 6 and 7) as well as in plasma membrane–containing fractions (lanes 10 and 11). (C) Quantitative distribution of newly synthesized mutant Pma1 in vps1 and vps8 cells. Distribution of Pma1-7 in intracellular and plasma membrane fractions was quantitated after a 90-min induction at 37°C (on) and after a 90-min chase (off), as described in MATERIALS AND METHODS. Data from a representative experiment are expressed as absolute arbitrary units. There is a decrease in the total amount of Pma1 after the chase, likely attributable to a fraction of newly synthesized Pma1 moving to and being degraded in the vacuole. (Inset) Mutant Pma1 levels are expressed as a percentage of the total at each time point. Data are means ± SD of three or four independent experiments. Delivery to the plasma membrane is more rapid in vps1 cells than in vps8 cells.
Trafficking of Newly Synthesized Vps10 in vps1 and vps8 Mutants
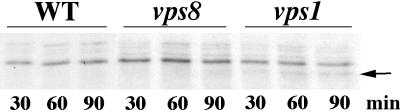
The kinetic differences in cell surface arrival is consistent with the possibility that Pma1-7 may take different routes to the plasma membrane in vps1 and vps8. To examine this possibility in greater detail, trafficking of Vps10 was compared in vps1 and vps8 mutants. In wild-type cells, Vps10, the CPY-sorting receptor, is a stable protein that recycles between the trans-Golgi and the endosomes (Marcusson et al., 1994; Cooper and Stevens, 1996). Previous reports have shown that recycling of trans-Golgi membrane proteins, including Vps10, is disrupted in vps1 mutants; Vps10 travels instead to the plasma membrane, where it undergoes rapid internalization, delivery to the vacuole, and degradation (Wilsbach and Payne, 1993; Cereghino et al., 1995; Nothwehr et al., 1995). Figure 7 shows analysis of newly synthesized Vps10 by pulse labeling of cells with [35S]methionine and [35S]cysteine followed by chase for various times. In vps1Δ cells, Vps10 undergoes proteolytic cleavage at 2 h of chase (arrow), whereas it appears stable in wild-type and vps8Δ cells. Similarly, vps1, but not vps8, perturbed the stability of the trans-Golgi membrane protein Kex2 (data not shown). These data support the idea that biosynthetic membrane traffic follows different routes in vps8 and vps1.
Figure 7.
Trafficking of Vps10 in vps1 and vps8 cells. Pulse-chase experiments were used to analyze the stability of Vps10 in wild-type (L3852), vps8 (WLX16-1A), and vps1 (ACX58-3C) cells. Cells were radiolabeled at room temperature with Expre35S35S for 5 min and chased for various times. Vps10 was immunoprecipitated and analyzed by SDS-PAGE and fluorography. Newly synthesized Vps10 undergoes degradation in vps1 (arrow) but remains stable in vps8 and wild-type cells.
Endocytosis of the Bulk Membrane Marker FM 4-64
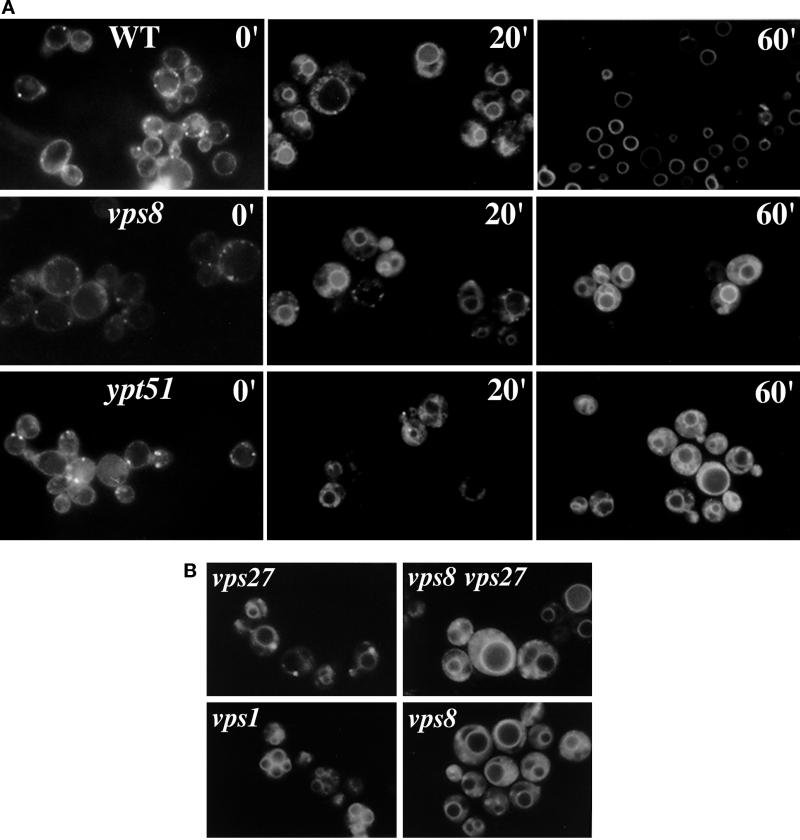
Of relevance to defining the trafficking route of Pma1-7 is the report that there is defective endocytosis of the fluorescent membrane marker FM 4-64 in vps8 (Luo and Chang, 1997). To characterize further the endocytic defect in vps8, a time course of FM 4-64 endocytosis was examined. Figure 8A shows that FM 4-64 staining occurs predominantly at the plasma membrane when wild-type or vps8 cells are incubated with the dye at 0°C, as described previously (Vida and Emr, 1995). (The small bright fluorescent spots that are also seen [Figure 8A, 0′] are likely endocytic structures formed during photography of the cells.) After warming the cells to permit endocytosis to proceed, cell surface staining disappears at similar rates in both wild-type and vps8 cells, indicating that internalization from the cell surface is not affected in vps8. At 20 min after warming, FM 4-64 is seen in the cytoplasm as well as in the vacuolar membrane in wild-type and vps8 cells. By 60 min after internalization, FM 4-64 has been cleared from the cytoplasm and is exclusively at the vacuole membrane in wild-type cells (Figure 8A, top right panel), whereas much of the internalized dye remains in vesicular intermediates in the cytoplasm of vps8Δ cells (Figure 8A, middle right panel).
Figure 8.
An early endocytic intermediate is accumulated in vps8 but not vps1 cells. (A) Time course of FM 4-64 endocytosis. Exponentially growing cells were incubated on ice for 30 min with 40 μM FM 4-64. Cells were then washed and incubated for an additional 20 or 60 min at 30°C. At 60 min in vps8 (WLX16-1A) and ypt51 (WLY65) cells, FM 4-64 is accumulated in an endocytic intermediate similar to that seen at 20 min in wild-type cells (L3852). (B) FM 4-64 endocytosis is delayed in vps8 at a step before the prevacuolar compartment. After labeling for 15 min with FM 4-64, cells were washed, resuspended, and incubated for 1 h at 30°C before visualization and photography. vps8 (ACY76), vps27 (ACY33), vps8 vps27 (WLX20-5D), and vps1 (ACX58-3C) cells are shown. The pattern of FM 4-64 accumulation in vps8 vps27 double mutants resembles that seen in vps8 cells.
In vps1 cells, FM 4-64 labels multiple vacuolar compartments (Figure 8B), reflecting the fragmented vacuolar morphology of the cells (Raymond et al., 1992). Nevertheless, no defect in endocytic delivery to the vacuole was detected, in agreement with previous reports (Wilsbach and Payne, 1993; Nothwehr et al., 1995).
FM 4-64 endocytosis was also performed in double mutants in which vps8 mutation was combined with a class E vps mutation. After 1 h of internalization in vps27 cells, accumulation of FM 4-64 in the prevacuolar compartment is seen as a large bright spot next to the vacuole (Figure 8B) (Vida and Emr, 1995). However, in vps8 vps27 double mutants, the pattern of FM 4-64 accumulation largely resembles that seen in vps8 cells, with much of the fluorescence signal in vesicular intermediates in the cytoplasm (Figure 8B). The same result was obtained in vps8 vps36 double mutants (data not shown). In contrast, in cells in which the class E vps mutation is combined with vps1, there is FM 4-64 accumulation in the prevacuolar compartment but not at an early endocytic step (data not shown). These data indicate that the endocytic defect seen in the vps8 mutant occurs before the prevacuolar compartment of class E vps cells.
Consistent with delayed transport through an early endocytic intermediate in vps8, a similar pattern of dye accumulation was observed in ypt51Δ cells (Figure 8A, bottom panels). Ypt51 is a small GTPase and homologue of mammalian Rab5, and previous work has shown that ypt51 cells accumulate internalized α factor in an early endocytic intermediate (Sambrook et al., 1989; Singer-Kruger et al., 1994, 1995).
Endocytosis of the Mating Receptor Ste3
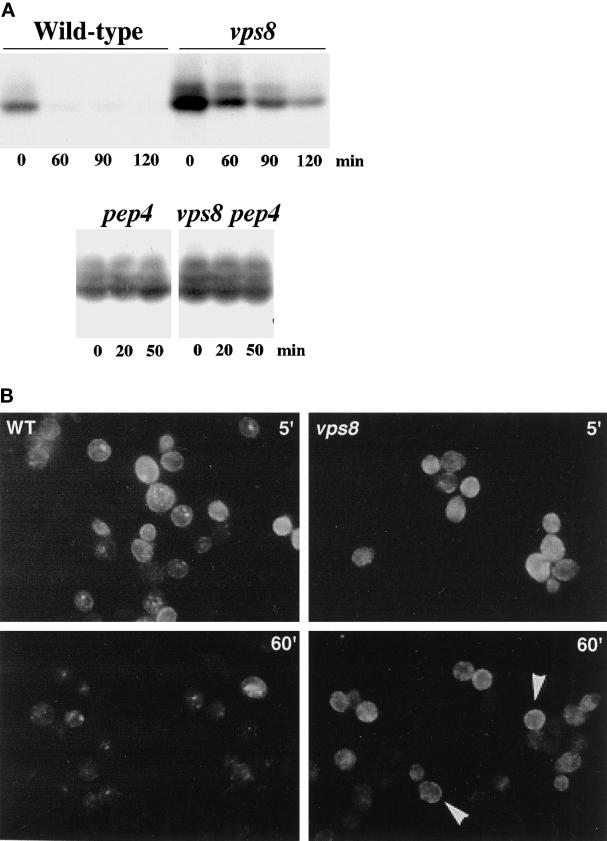
To analyze further the endocytic defect of vps8, the behavior of the cell surface receptor Ste3 was examined. In wild-type cells, Ste3 undergoes constitutive endocytosis and vacuolar degradation (Davis et al., 1993). To follow the fate of cell surface Ste3 in the absence of new receptor synthesis, STE3 was placed under the control of the GAL1 promoter. Figure 9A shows Western blot analysis of Ste3 stability at various times after glucose addition to repress STE3 expression. In wild-type cells, Ste3 is rapidly degraded upon glucose addition. In contrast, in vps8, the steady-state level of Ste3 (at time 0) is increased and the rate of Ste3 degradation is decreased (Figure 9A). These results are in agreement with the kinetic delay in delivery to the vacuole observed by FM 4-64 endocytosis (Figure 8). The pep4 mutation stabilizes Ste3 in both wild-type and vps8 cells, indicating that Ste3 degradation is due to vacuolar delivery (Figure 9A) (Davis et al., 1993).
Figure 9.
Effect of vps8 on endocytosis of Ste3. Inhibition of vacuolar degradation occurs concomitantly with an increase in cell surface Ste3 in vps8 cells. (A) Western blot after preventing new synthesis of Ste3 in wild-type (ACY72) and vps8 (ACY81) cells. To stop new Ste3 synthesis, glucose (3%) was added to GAL1-STE3 cells exponentially growing at 30°C in the presence of galactose. At the indicated times after glucose addition, cells were harvested and lysate was prepared for analysis of Ste3 protein level (top). Time 0 represents the steady-state level of Ste3 before the addition of glucose. The bottom panel shows that degradation of Ste3 upon glucose addition is dependent on PEP4; stabilization of Ste3 is seen in pep4 (ACY84) and vps8 pep4 (ACY85) cells. (B) Indirect immunofluorescence localization of Ste3 after inhibition of new Ste3 synthesis. Wild-type (ACY72) and vps8 (ACY81) cells expressing a GAL1-STE3-myc plasmid (pSL2015) were grown to midlog phase in galactose-containing medium. At 5 and 60 min after the addition of glucose (3%), cells were fixed, permeabilized, and stained with anti-myc antibody followed by Cy3-conjugated secondary antibody. Photographs showing the 5- and 60-min time points represent 4- and 8-s exposures, respectively. Arrowheads indicate cell surface staining in vps8 cells.
Indirect immunofluorescence was used to observe Ste3 endocytosis (Figure 9B). Within 5 min after glucose addition, the distribution of Ste3 in wild-type and vps8 cells appears similar (Figure 9B, top panels). At this time, Ste3 is seen predominantly at the plasma membrane, although some Ste3 is also found in small intracellular spots, likely reflecting endocytic intermediates. By 1 h after stopping further receptor synthesis, Ste3 staining in wild-type cells is markedly diminished; although some punctate staining is visible, staining at the cell surface is not readily apparent. In contrast to that in wild-type cells, plasma membrane Ste3 staining persists in vps8 cells, consistent with receptor recycling to the plasma membrane.
DISCUSSION
We have analyzed trafficking pathways in the endosomal system by monitoring the movement of newly synthesized Pma1-7. In several vps mutants, a fraction of Pma1-7 is rescued from vacuolar degradation and delivered to the cell surface. We show that in vps1 cells, newly synthesized mutant Pma1 can move to the plasma membrane after appearing briefly in punctate intracellular compartment(s) (Figure 5). Consistent with previous reports (Wilsbach and Payne, 1993; Nothwehr et al., 1995), it seems likely that mutant Pma1 in vps1 cells is routed directly to the plasma membrane from the Golgi complex.
In class E vps mutants, newly synthesized Pma1-7 accumulates first in the prevacuolar compartment (Figure 3). After chase in the presence of cycloheximide to ensure that no further synthesis could occur, a small fraction (∼10%) of newly synthesized Pma1-7 was observed to move to the plasma membrane. These data support a model in which protein traffic can flow from the prevacuolar compartment to the plasma membrane, albeit inefficiently. Because there is defective retrograde transport from the prevacuolar compartment to the Golgi in class E vps mutants (Piper et al., 1995), it is unlikely that transport of mutant Pma1 to the cell surface occurs via a Golgi intermediate. It is possible, however, that movement of Pma1-7 in these cells from the prevacuolar compartment to the surface occurs via an early endosomal intermediate (Figure 10) (see below).
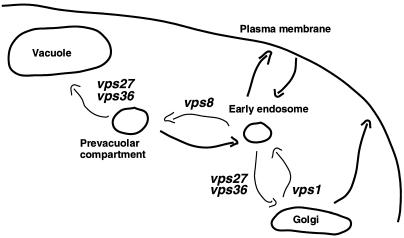
Figure 10.
Model showing trafficking step(s) impaired by vps mutations. Proposed impaired step(s) in transport from the Golgi to the vacuole are labeled with the corresponding vps mutations. Transport from the Golgi to the endosomal system is blocked in vps1Δ cells (Nothwehr et al., 1995); endosome-bound traffic, including Pma1-7, is redirected to the surface from the Golgi. In class E vps mutants, such as vps36 and vps27, there is defective anterograde transport from the prevacuolar compartment to the vacuole as well as defective retrograde transport to the Golgi (Piper et al., 1995; Nothwehr et al., 1996). In class E vps mutants, Pma1-7 is proposed to travel to the plasma membrane from the prevacuolar compartment via an early endosome. In vps8 cells, there is defective transport from the early endosome to the vacuole; movement of mutant Pma1 to the plasma membrane in vps8 is proposed to occur as a consequence of accumulation in an early endosome.
Our data have relevance to understanding the mechanism by which misfolded proteins are delivered to the vacuole. A model has been proposed in which vacuolar delivery of misfolded proteins occurs by receptor-mediated transport (Chang and Fink, 1995; Hong et al., 1996; Li et al., 1999). Our observation that mutant Pma1 enters the endosomal system of class E vps mutants argues that such a quality control receptor probably does not recycle routinely through the prevacuolar compartment.
Mutant Pma1 is also routed to the cell surface in vps8 cells (Figure 6). Our immunofluorescence and cell fractionation experiments do not have sufficient resolution to prove unequivocally that Pma1-7 enters the endosomal system of vps8 mutants before reaching the surface. Nevertheless, several observations prompt us to hypothesize that mutant Pma1 moves to the plasma membrane from an early endosomal compartment in vps8 cells (Figure 10). First, cell surface arrival of mutant Pma1 occurs more slowly in vps8 compared with vps1 (Figure 6). Second, pulse-chase analysis shows that Vps10 undergoes rapid degradation in vps1 cells but not in vps8 cells. These data are consistent with vps8 and vps1 mutations having different effects on membrane traffic. Third, FM 4-64 endocytosis experiments indicate defective transport through an early endocytic compartment in vps8, in contrast to vps1 cells, which have no apparent endocytic defect (Figure 8). Finally, although plasma membrane internalization is not impaired (Figure 8), down-regulation of Ste3 from the cell surface is impaired by vps8 (Figure 9). These observations suggest persistent receptor recycling to the surface from an endosomal compartment. (Note that cell surface Ste3 is also increased in the class E vps mutant ren1/vps2, in which there is a block in traffic from the prevacuolar compartment to the vacuole [Davis et al., 1993].)
Enhanced endosome-to-surface trafficking in vps8 represents the simplest model that can account for all of the observations we have reported. Previous work on Vps8p reveals that it is a large membrane-associated protein containing a RING finger zinc-binding motif (Chen and Stevens, 1996; Horazdovsky et al., 1996). A role for Vps8 in the endosomal system is supported by genetic interactions between VPS8 and YPT51 (Horazdovsky et al., 1996) as well as between VPS8 and PEP5/END1 (Woolford et al., 1998). Nevertheless, we cannot formally rule out the possibility that mutant Pma1 moves to the surface directly from the Golgi in vps8 mutants. Similarly, we cannot exclude the possibility that mutant Pma1 undergoes retrograde transport from the endosome to the Golgi followed by delivery to the surface. (On the other hand, because CPY is missorted in vps8Δ cells [Chen and Stevens, 1996; Horazdovsky et al., 1996], recycling of Vps10 back to the Golgi is likely impaired.)
Our hypothesis is summarized in Figure 10, in which proposed impaired step(s) in transport from the Golgi to the vacuole are labeled with the corresponding vps mutations. We propose that in vps8 cells, there is an accumulation within early endosomes of proteins entering either from the cell surface (Ste3) or from the biosynthetic pathway (Vps10 and mutant Pma1). As a consequence, there is increased transport to the cell surface. Because plasma membrane delivery of Pma1-7 in class E vps mutants appears less efficient than in vps8 cells, it is possible that movement to the surface from the prevacuolar or late endosome compartment occurs via an early endosome intermediate.
In mammalian cells, an endosome-to-surface traffic pathway is taken constitutively by many internalized membrane proteins and lipids. In some cell types, the pathway is specialized for recycling synaptic vesicle components, antigen presentation, insulin-dependent control of glucose transport, and transcytosis. Indeed, some newly synthesized proteins are normally delivered to the plasma membrane via the endosome (Futter et al., 1995; Leitinger et al., 1995). Recent observations suggest that there is possibly analogous endosome-to-surface trafficking in yeast. Analysis of the chitin synthase Chs3 has led to the proposal that its distribution between the plasma membrane and the “chitosome,” an endosomal compartment, is regulated by recycling (Chuang and Schekman, 1996; Ziman et al., 1998). Work on copper entry into the yeast secretory pathway has suggested that copper loading of the surface enzyme ceruloplasmin occurs within an endosomal compartment (Yuan et al., 1997). Thus, selected plasma membrane proteins may pass through endosomes to fulfill specific processing requirements. Our finding that mutant Pma1 can move from the endosomal system to the plasma membrane represents a first step toward characterizing an endosome-to-surface trafficking pathway in yeast. Future work should focus on elucidating the physiological significance of this pathway.
ACKNOWLEDGMENTS
We thank Jim Haber, Nick Davis, George Sprague, Hugh Pelham, Steve Nothwehr, Tamara Doering, Birgit Singer-Kruger, Carolyn Slayman, and Scott Emr for strains, plasmids, antibodies, and advice. Thanks to Peter Arvan for reading the manuscript. This work was supported by grant GM58212 from the National Institutes of Health.
REFERENCES
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brada D, Schekman R. Coincident localization of secretory and plasma membrane proteins in organelles of the yeast secretory pathway. J Bacteriol. 1988;170:2775–2783. doi: 10.1128/jb.170.6.2775-2783.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Fink GR. Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Slayman CW. Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol. 1991;115:289–295. doi: 10.1083/jcb.115.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-J, Stevens TH. The VPS8 gene is required for localization and trafficking of the CPY sorting receptor in Saccharomyces cerevisiae. Eur J Cell Biol. 1996;70:289–297. [PubMed] [Google Scholar]
- Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis NG, Horecka JL, Sprague GF., Jr Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futter C, Connolly CN, Cutler DF, Hopkins CR. Newly synthesized transferrin receptors can be detected in the endosome before they appear on the cell surface. J Biol Chem. 1995;270:10999–11003. doi: 10.1074/jbc.270.18.10999. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Maxfield FR. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L, Zanolari B, Pypaert M, Rohrer J, Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol Biol Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Davidson AR, Kaiser CA. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horazdovsky BF, Cowles CR, Mustol P, Holmes M, Emr SD. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J Biol Chem. 1996;271:33607–33615. doi: 10.1074/jbc.271.52.33607. [DOI] [PubMed] [Google Scholar]
- Jenness DD, Li Y, Tipper C, Spatrick P. Elimination of defective α-factor pheromone receptors. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- Leitinger B, Hille-Rehfeld A, Spiess M. Biosynthetic transport of the asialoglycoprotein receptor H1 to the cell surface occurs via endosomes. Proc Natl Acad Sci USA. 1995;92:10109–10113. doi: 10.1073/pnas.92.22.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kane T, Tipper C, Spatrick P, Jenness DD. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol Cell Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W-j, Chang A. Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J Cell Biol. 1997;138:731–746. doi: 10.1083/jcb.138.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Konopka J, Singer-Kruger B, Zerial M, Botstein D. Visualization of receptor-mediated endocytosis in yeast. Mol Biol Cell. 1999;10:799–817. doi: 10.1091/mbc.10.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H+-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Yang H, Stevens TH. VPS27 controls vacuolar and endocytic traffic through prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. J Cell Biol. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H, Woodman PG, van Meer G, Marsh M. Molecular mechanisms of endocytosis. Cell. 1997;91:731–738. doi: 10.1016/s0092-8674(00)80461-4. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Hicks JB, Fink GR. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. pp. 523–585. [Google Scholar]
- Singer-Kruger B, Frank R, Crausaz F, Riezman H. Partial purification and characterization of early and late endosomes from yeast. J Biol Chem. 1993;268:14376–14386. [PubMed] [Google Scholar]
- Singer-Kruger B, Stenmark H, Dusterhoft A, Philippsen P, Yoo J-S, Gallwitz D, Zerial M. Role of three Rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer-Kruger B, Stenmark H, Zerial M. Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J Cell Sci. 1995;108:3509–3521. doi: 10.1242/jcs.108.11.3509. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbach K, Payne GS. Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 1993;12:3049–3059. doi: 10.1002/j.1460-2075.1993.tb05974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford CA, Bounoutas GS, Frew SE, Jones EW. Genetic interaction with vps8-200 allows partial suppression of the vestigial vacuole phenotype caused by a pep5 mutation in Saccharomyces cerevisiae. Genetics. 1998;148:71–83. doi: 10.1093/genetics/148.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan DS, Dancis A, Klausner RD. Restriction of copper export in Saccharomyces cerevisiae to a late Golgi or postGolgi compartment in the secretory pathway. J Biol Chem. 1997;272:25787–25793. doi: 10.1074/jbc.272.41.25787. [DOI] [PubMed] [Google Scholar]
- Ziman M, Chuang JS, Tsung M, Hamamoto S, Schekman R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1565–1576. doi: 10.1091/mbc.9.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]