Abstract
Some genes on the inactive X chromosome escape silencing. One possible escape mechanism is that heterochromatization during X inactivation can be blocked by boundary elements. DNA insulators are candidates for blocking because they shield genes from influences of their chromosomal environment. To test whether DNA insulators can act as boundaries on the X chromosome, we inserted into the mouse X-linked Hprt locus a GFP transgene flanked with zero, one, or two copies of a prototypic vertebrate insulator from the chicken β-globin locus, chicken hypersensitive site 4, which contains CCCTC binding factor binding sites. On the active X chromosome the insulators blocked repression of the transgene, which commences during early development and persists in adults, in a copy number-dependent manner. CpG methylation of the transgene correlated inversely with expression, but the insulators on the active X chromosome were not methylated. On the inactive X chromosome, insulators did not block random or imprinted X inactivation of the transgene, and both the insulator and transgene were almost completely methylated. Thus, the chicken hypersensitive site 4 DNA insulator is sufficient to protect an X-linked gene from repression during development but not from X inactivation.
Keywords: CCCTC binding factor
Eukaryotic genomes contain interspersed domains of transcriptionally active euchromatin and inactive heterochromatin. Heterochromatin can encroach into transcriptionally active euchromatin and silence adjoining genes, as illustrated by position effect variegation in Drosophila (1, 2). Maintaining these chromosomal domains and preventing the spread of heterochromatin implies the existence of boundary elements that act as barriers (3). One class of boundary element candidates consists of DNA insulators because they can block the positive effects of an adjacent enhancer and protect a gene from negative position effects (4, 5).
One of the best-characterized vertebrate DNA insulators is from the chicken β-globin locus. In chicken erythrocytes, DNaseI hypersensitive site 4 upstream of the β-globin locus marks the transition between a euchromatic region that contains the β-globin genes and upstream heterochromatin (6, 7). Chicken hypersensitive site 4 (cHS4), a 1.2-kb fragment that encompasses hypersensitive site 4, has both insulator properties: it can block an enhancer in an enhancer blocking assay, and it has barrier activity in a position effect assay (8). A binding site for the transcription factor CTCF (CCCTC binding factor) within cHS4 is necessary and sufficient for its enhancer blocking activity, but is not necessary for its barrier function (9, 10). Flanking copies of cHS4 without the CTCF binding site can still protect randomly integrated transgenes from position effects. In a position effect assay the cHS4 insulator itself and transgenes flanked with it are associated with acetylated histones, histone H3 methylated at lysine 4, and reduced CpG methylation relative to the methylation of uninsulated transgenes (11–13).
A specialized form of heterochromatinization occurs in mammals during X inactivation and results in one of the two X chromosomes in female somatic cells becoming largely transcriptionally inactive (14–16). The silencing of genes on the inactive X chromosome (Xi) is patchy. Ten to 20% of the genes on the human Xi escape inactivation; however, only seven genes on the mouse Xi are known to escape inactivation (17, 18). In humans at least, this escape appears to operate at the level of chromosomal domains possibly demarcated by boundary elements (19, 20). The role for chromosomal domains in escape from X inactivation is supported by the demonstration that a 17-kb chicken transferrin gene randomly integrated as multiple copies on the X chromosome is expressed whether the transgene is on the Xi or the active X chromosome (Xa) (21). However, in two separate transgenic studies, neither the human β-globin locus control region, nor a functional DNA fragment containing matrix attachment regions, resisted silencing on the Xi (22, 23), indicating that these boundary elements are not sufficient to form escape domains on the Xi. A recent report (24) characterized a DNA fragment with insulator properties and CTCF binding sites at the 5′ end of three X-linked genes (mouse Jarid1c and Eif2s3x and human EIF2S3) that escape inactivation although they are adjacent to genes that are inactivated, and suggested that CTCF may play a role in maintaining escape domains.
The experiments we describe here show that copies of the cHS4 DNA insulator, which contains a CTCF binding site, flanking a transgene inserted adjacent to the X-linked Hprt gene, block in a copy number-dependent manner the partial repression of the transgene that occurs during development and persists in adults. However, random and imprinted X inactivation of the transgene are not blocked by cHS4.
Results and Discussion
Transgenes Were Targeted in Mouse ES Cells to Hprt Locus.
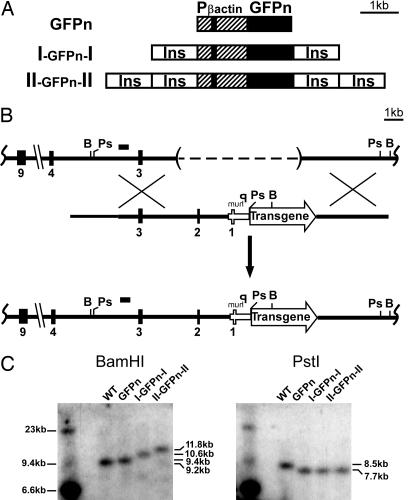
We chose the hypersensitive site 4 from the chicken β-globin locus, a well characterized vertebrate DNA insulator with a CTCF binding site, to test the effects of flanking insulators on an X-linked transgene. To monitor X inactivation, we used a GFP gene with a nuclear localization signal (GFPn) driven by a ubiquitously active 1.3-kb human β-actin promoter (25). This reporter transgene, flanked with zero, one, or two copies of the 1.2-kb cHS4 insulator sequence (8) (Fig. 1A), was inserted into the X-linked Hprt locus of mouse ES cells by using the single-copy chosen site integration method described by Bronson et al. (26) (Fig. 1B). It was advantageous to insert the reporter transgene into the X-linked Hprt locus because we could measure the effect an insulator has on expression when the X chromosome is transcriptionally active or inactive. Southern blot analyses confirmed that the transgenes were integrated as single copies in targeted ES cells (Fig. 1C).
Fig. 1.
Transgenic constructs and targeting scheme to insert transgenes into the X-linked Hprt locus. (A) The reporter gene (large solid black rectangle) encodes a GFPn. The GFPn gene is driven by a 1.3-kb human β-actin promoter (hatched box); the small solid black rectangle is the first exon of β-actin. Open boxes labeled Ins represent the 1.2-kb insulator from the chicken β-globin locus (cHS4). (B) (Top) The untargeted Hprt locus in the ES cell line (E14Tg2a). The dashed line within parentheses represents the ≈50-kb deletion that removes the Hprt promoter and exons 1 and 2. (Middle) The targeting construct. (Bottom) The Hprt locus after homologous recombination. The small black bar shows the location of the probe used for Southern blotting. (C) (Left) The Southern blot obtained when the ES cell DNA was digested with BamHI. The Hprt locus in untargeted, WT ES cells gave a 9.2-kb BamHI fragment. Targeted ES cells with the GFPn, I-GFPn-I, or II-GFPn-II transgene gave 9.4-, 10.6-, and 11.8-kb BamHI fragments, respectively. (Right) The Southern blot obtained when the ES cell DNA was digested with PstI. The Hprt locus in untargeted, WT ES cells gave a 8.5-kb PstI fragment. The targeted ES cells gave a 7.7-kb PstI fragment with all three transgenes. Ps, PstI; B, BamHI; Phum, human HPRT promoter.
cHS4 DNA Insulators Affect Changes in Expression During Early Development.
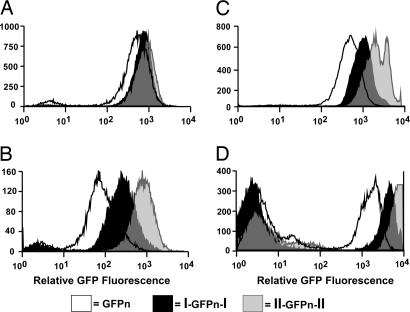
Our first experiments determined the effects of flanking DNA insulators on expression of the GFPn transgene during early development when the transgene is on an Xa. We performed FACS analysis on undifferentiated and differentiated ES cells to measure GFP fluorescence. Fig. 2A shows that the level of fluorescence was the same in undifferentiated male ES cells with zero, one, or two copies of cHS4 flanking the GFPn transgene. When ES cells were differentiated in vitro to embryoid bodies, the level of GFP fluorescence differed markedly among the three cell lines. Expression of the GFPn transgene was highest when flanked by two copies of cHS4, lowest when uninsulated, and intermediate when flanked by a single copy of cHS4 (Fig. 2B). Thus, the flanking insulators block in a copy number-dependent manner the partial repression of the GFPn transgene at the Hprt locus that occurs during development.
Fig. 2.
GFP fluorescence in undifferentiated and differentiated ES cells and in adult hemizygous male and heterozygous female mice. The levels of GFP fluorescence were measured by FACS analysis, and representative histograms are shown. Solid white, black, and gray histograms represent the GFPn, I-GFPn-I, and II-GFPn-II transgenes, respectively. (A) The histograms are from undifferentiated male ES cells grown on feeder layers. (B) Shown is fluorescence of ES cells 4 weeks after they were removed from feeders and allowed to form embryoid bodies with the FACS detector settings identical to those used for undifferentiated ES cells. (C) The histograms are from lymphocytes of male mice hemizygous for the GFPn transgenes. The mean fluorescence ± standard deviation for three male mice for each transgene are: GFPn, 520 ± 20; I-GFPn-I, 1,360 ± 270; and II-GFPn-II, 2,630 ± 610. (D) Shown is the fluorescence in splenocytes from heterozygous females. The mean fluorescence ± standard deviation for the GFP+ cells are: GFPn, 2,031 ± 340 (n = 4); I-GFPn-I, 4,076 ± 644 (n = 5); and II-GFPn-II, 7,511 ± 576 (n = 6). Note the presence of an approximately equal number of nonfluorescent cells in the females but none in males.
cHS4 DNA Insulators Modulate Expression in Adults.
We next determined the effects of flanking insulators on the expression of the transgene when on the Xa in adults. Peripheral WBCs were isolated from transgenic adult males and their GFP fluorescence was determined by FACS analysis. Fig. 2C shows representative histograms of the relative fluorescence in WBCs from transgenic males. When compared with expression from the uninsulated transgene (520 ± 20), expression from the transgene with one flanking copy of the insulators is ≈2-fold greater (1,360 ± 270) and expression from the transgene with two flanking copies is ≈4-fold greater (2,630 ± 610). A similar dose effect on the level of fluorescence was seen in GFP+ splenocytes from heterozygous Xm/XpI-GFPn-I and Xm/XpII-GFPn-II females (Fig. 2D), where Xm is the maternal X chromosome and Xp is the paternal X chromosome. Thus, flanking insulators continue to block repression of the transgene at the Hprt locus on the Xa after the completion of development.
We conclude that, because cHS4 does not have canonical enhancer qualities of its own (8), it acts as a barrier to repression of the transgene at the Hprt locus on the Xa during in vitro differentiation and in adults and that this barrier function is more effective with two flanking copies than with one. Our experiments clearly demonstrate the dosage effect of cHS4’s barrier activity because we have targeted single copies of all of our reporter transgenes (with zero, one, or two flanking copies of cHS4) to the same location.
cHS4 DNA Insulators Do Not Prevent Random X Inactivation.
To analyze the effects of flanking insulators on the expression of the GFPn transgene during X inactivation, males carrying the different GFPn transgenes were bred to WT females. Embryos at 7.5 days postcoitum were then dissected for analysis. This scheme achieved three purposes. First, because only female progeny inherit the GFPn transgene, female embryos could be identified by their fluorescence. Second, because the female embryos also carry a WT X chromosome we could determine the effect of insulators on random X inactivation, which normally inactivates either Xm or Xp in embryonic lineages. Third, because the transgene is paternally inherited, embryos could also be used to examine imprinted X inactivation, which preferentially inactivates the Xp in extraembryonic lineages.
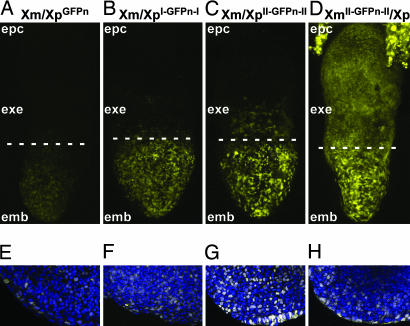
Fig. 3A–C shows green fluorescent images (pseudocolored yellow) of female embryonic day 7.5 embryos that are heterozygous for a paternally inherited GFPn transgene with zero (Xm/XpGFPn), one (Xm/XpI-GFPn-I), and two (Xm/XpII-GFPn-II) copies of the flanking insulator. Fig. 3 E–G shows images of the embryonic ectoderm (emb) that merge the GFP fluorescence with TO-PRO-3 fluorescence (pseudocolored blue) used to visualize nuclei. The GFP fluorescence seen in the emb is clearly greater when the transgene is flanked by insulators, confirming the observations in ES cells, WBCs, and splenocytes presented in Fig. 2.
Fig. 3.
Expression patterns of paternally and maternally inherited X-linked GFPn transgenes in embryonic day 7.5 embryos. (A–D) z projections of confocal images of the GFP fluorescence signal (pseudocolored yellow) from female embryos dissected at embryonic day 7.5. The dashed line marks the division between emb and extraembryonic (exe) regions of the embryo. (E–H) Single confocal sections of the embryo proper of embryonic day 7.5 females. Embryos were counterstained with TO-PRO-3 to visualize nuclei, which was pseudocolored blue. The merged images are shown, where the combination of yellow and blue produces white. In A–C and E–G, the female embryos are heterozygous for the GFPn, I-GFPn-I, or II-GFPn-II transgene, respectively, which is on their Xp. In D and H, the female embryo is heterozygous for the II-GFPn-II transgene, which is on the Xm. The ectoplacental cone (epc) shows GFP fluorescence only when the transgene is inherited from the mother (D). (Magnifications: A–D, ×20; E–H, ×40.)
Fig. 3 A and E shows a mosaic pattern of GFP fluorescence in the emb of an Xm/XpGFPn female, as expected for an X-linked transgene that undergoes random X inactivation in a heterozygous female (27, 28). The same mosaic pattern is seen when the transgene is flanked by one copy of the insulator (Fig. 3 B and F) or two copies (Fig. 3 C and G). The mosaic pattern persists in adults and is unaffected by the presence of the insulators or the parental origin of the transgene as judged by FACS analysis from heterozygous females in which approximately half the cells do not fluoresce (Fig. 2D and Tables 1 and 2, which are published as supporting information on the PNAS web site). This finding indicates that flanking insulators do not prevent random X inactivation from silencing of the GFPn transgene on the Xp or Xm.
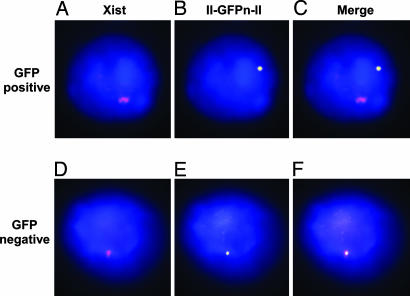
Although the mosaic pattern of GFP expression seen in the emb is consistent with silencing of the GFPn transgene on the Xi, it does not exclude the possibility that the expressed GFPn transgene is on the Xi and is protected from silencing by the insulators. To test for this possibility, we sorted splenocytes from females heterozygous for the II-GFPn-II transgene into GFP+ and GFP− populations and performed FISH for the II-GFPn-II transgene and Xist RNA. Fluorescent images in Fig. 4A–C are of a single nucleus from a GFP+ splenocyte. Fig. 4A shows the Xist signal (pseudocolored magenta) that marks the Xi, Fig. 4B shows the II-GFPn-II signal (pseudocolored yellow), and Fig. 4C is the merge. In all GFP+ splenocytes in which we have detected both the signal for the transgene and the signal for Xist the two signals are nonoverlapping (Fig. 4C), which demonstrates that the II-GFPn-II transgene is not on the Xi. Overlapping Xist and II-GFPn-II signals, indicating the II-GFPn-II transgene is on the Xi, were observed in the GFP− splenocytes (Fig. 4F). These results confirm that flanking cHS4 insulators do not prevent random X inactivation from silencing a transgene at the Hprt locus.
Fig. 4.
Fluorescence in situ hybridization for Xist RNA and the II-GFPn-II transgene. Fluorescent images are shown of a GFP+ splenocyte (A–C) and GFP− splenocyte (D–F) from a female heterozygous for the II-GFPn-II transgene. (A and D) RNA FISH for Xist is shown. The Xist signal (pseudocolored magenta) indicates the Xist RNA coating the Xi. (B and E) DNA FISH for the II-GFP-II transgene (pseudocolored yellow) is shown. The nucleus is stained with DAPI (blue). (C and F) The merge of the Xist and II-GFPn-II transgene images. Fifty of 50 GFP+ splenocytes had the localization pattern shown in C. Twenty-four of 25 GFP− splenocytes had the localization pattern shown in F; the 1 of 25 showing the pattern in C probably was caused by contamination with GFP+ cells during FAC sorting. (Magnification: ×100.)
cHS4 DNA Insulators Do Not Prevent Imprinted X Inactivation.
To test whether insulators prevent imprinted X inactivation, we compared the GFP fluorescence of female embryos that inherited the II-GFPn-II transgene from their father with that of female embryos that inherited the transgene from their mother. Fig. 3 C, D, G, and H show that the emb of the Xm/XpII-GFPn-II and XmII-GFPn-II/Xp embryos has essentially indistinguishable mosaic fluorescence, confirming that random X inactivation is not affected by the parent of origin of the transgene. However, in the extra-emb and the ectoplacental cone, fluorescence is only apparent when the transgene is inherited from the mother (XmII-GFPn-II/Xp). The paternally derived transgene (Xm/XpII-GFPn-II) is silent. (A few fluorescent cells above the dashed line in Fig. 3 A–C are most likely embryonic cells that have migrated from the epiblast. The difference in the number of visible fluorescent cells in Fig. 3C compared with Fig. 3 A and B probably reflects the greater level of fluorescence in the embryo with the transgene flanked by two copies of the insulator compared with the embryo with the uninsulated transgene.) Thus the insulators do not prevent imprinted inactivation from silencing a transgene on the Xp.
Some heterozygous female embryonic day 7.5 embryos receiving a transgene with or without flanking insulators from their fathers were examined with Reichert’s membrane intact. These embryos (Fig. 6, which is published as supporting information on the PNAS web site) showed that some trophoblast giant cells and trophoblasts attached to Reichert’s membrane are fluorescent. Although these cells are clearly extraembryonic, their escape from imprinted X inactivation has been reported (27) and was unaffected by the presence of flanking insulators, as demonstrated by our results.
DNA Methylation of the Transgene on Xa but Not Xi Is Affected by the Number of Insulators.
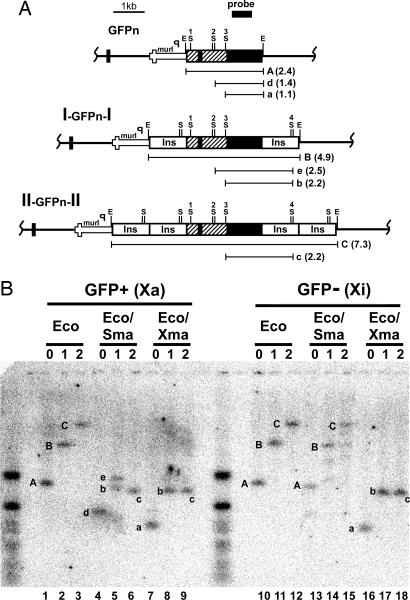
CpG methylation correlates with transcriptional silencing (29) and is characteristic of the promoters of silenced genes on the Xi (30, 31). To assess whether the cHS4 insulator affected the DNA methylation of the transgene, we determined its CpG methylation status by using the methylation-sensitive restriction enzyme SmaI. Splenocytes from female mice heterozygous for the three different transgenes were sorted by FACS into GFP+ and GFP− populations, corresponding to cells having the GFPn transgene on their Xa or Xi. Southern blot analyses were performed on DNA isolated from the sorted splenocytes after digestion with EcoRI, which is insensitive to methylation, EcoRI and SmaI, and EcoRI and XmaI, an isoschizomer of SmaI that is not affected by DNA methylation.
The Southern blot in Fig. 5B shows the results of one of four comparable tests made with digests of DNA from fluorescent splenocytes with the transgene on the Xa [GFP + (Xa)] and from nonfluorescent splenocytes with the transgene on the Xi [GFP − (Xi)]. After digestion with EcoRI, the GFPn transgene gives band A (2.4 kb), the I-GFPn-I transgene gives band B (4.8 kb), and the II-GFPn-II transgene gives band C (7.3 kb). When DNA from GFP− splenocytes is digested with SmaI in addition to EcoRI (Fig. 5B, lanes 13–15), bands A, B, and C are still the predominant bands, demonstrating that the SmaI sites in the β-actin promoter and cHS4 insulator are resistant to cleavage and are predominantly methylated when the transgene is on the Xi. The average darkness of these bands in the four tests was indistinguishable (A, 127 ± 52; B, 133 ± 48; C, 127 ± 47). Thus the insulators do not affect this DNA methylation. [The SmaI sites are intact because when the samples were digested with EcoRI and XmaI band A was completely converted to a (1.1 kb), B to b (2.2 kb), and C to c (2.2 kb).]
Fig. 5.
DNA methylation status of the GFPn transgenes on the Xa and Xi. (A) The structure of the transgenes integrated into the Hprt locus. The location of EcoRI (E) and SmaI (S) restriction sites in the transgenes and the sizes of the fragments detected after Southern blot analysis are shown. The small black rectangle represents the probe. (B) A Southern blot using DNA isolated from splenocytes of female mice heterozygous for the GFPn (zero copies, 0), I-GFPn-I (one copy, 1), or II-GFPn-II (two copies, 2) transgene. Lanes 1–9 used DNA isolated from GFP+ splenocytes. Lanes 10–19 used DNA isolated from GFP− splenocytes. In lanes 1–3 and 10–12, the DNA was digested with EcoRI (Eco). In lanes 4–6 and 13–15, the DNA was digested with EcoRI and SmaI (Sma). In lanes 7–9 and 16–18, the DNA was digested with EcoRI and XmaI (Xma). The letters (A–C and a–e) correspond to the restriction fragments illustrated in A.
Fig. 5B Left shows digests with DNA from GFP+ splenocytes with the transgene on the Xa [GFP + (Xa)]. When the transgene is flanked by two copies of the insulator, digestion with EcoRI and SmaI (Fig. 5B, lane 6) converts band C to c (2.2 kb) approximately as completely as do EcoRI and XmaI (Fig. 5B, lane 9). This hybridization pattern demonstrates that the SmaI sites in the insulator (S4) and the GFPn transgene (S1, S2, and S3) in II-GFPn-II are essentially free from methylation. When the transgene is flanked by one copy of the insulator, digestion with EcoRI and SmaI leads to the conversion of B to b in ≈50% of the cells and of B to e (2.5 kb) in the other 50%. This finding demonstrates that in approximately half of the cells with the transgene flanked by one copy of the insulator site S3 between the β-actin promoter and GFPn is methylated, and in the other half S3 is unmethylated. When the transgene is devoid of insulators, site S3 is fully methylated and site S2 is completely unmethylated, as judged by the complete absence of band a in Fig. 5B, lane 4, and its replacement by the longer band d (1.4 kb). We observed the same methylation pattern in DNA from splenocytes of male mice that have the transgene on the single Xa (Fig. 7, lanes 1–9, which is published as supporting information on the PNAS web site). However, the methylation pattern on the Xa occurs after differentiation because in undifferentiated ES cells the S3 site is almost completely unmethylated independent of the flanking insulators (Fig. 7, lanes 13–15).
In undifferentiated male ES cells the similar level of CpG hypomethylation among the insulated and uninsulated transgenes is associated with a similar level of GFP fluorescence. The increased GFPn expression seen in adult mice in the presence of cHS4 correlates with reduced CpG methylation. In agreement with observations by Pikaart et al. (11), we find that the insulators prevent loss of transcriptional activity rather than cause a gain of activity, and that there is an inverse relationship between the protective effects of the flanking insulators on the level of transgene expression and DNA methylation of the transgene promoter.
Although there is generally an inverse correlation between expression and CpG methylation, it is not one to one. The distribution of the expression of cells having the I-GFPn-I transgene is unimodal and approximately midway between that of the GFPn and II-GFPn-II transgenes, but the methylation pattern is bimodal with some of the cells having the S3 CpG site downstream of the β-actin promoter methylated, whereas other cells have the same site unmethylated. We infer that the insulator-induced changes in CpG methylation at this site do not determine expression of the transgene, but the insulator-induced changes in expression determine CpG methylation of the promoter site. This inference is compatible with a model proposed by Felsenfeld and his associates (12, 13) based on their analysis of chromatin modifications associated with a transgene flanked with two copies of cHS4. Our experiments show that two flanking copies of cHS4 are more potent than a single flanking copy in protecting the transgene from repression and its promoter from CpG methylation. We conclude that the more frequently the promoter is active, the more likely it is to escape methylation, but any given cell with intermediate expression can have its promoter methylated or unmethylated.
Conclusions
In summary, we report an investigation of the effects of the cHS4 DNA insulator at a predetermined site on the X chromosome. We have shown that when targeted adjacent to the X-linked Hprt locus flanking cHS4 insulators block transgene repression. This block in repression begins during development, persists in adults, and depends on the copy number of flanking cHS4 insulators. We have also shown that flanking cHS4 insulators, containing CTCF binding sites, are not sufficient to form a boundary that allows a transgene to escape X inactivation.
A recent report (24) has identified a sequence element with CTCF binding sites upstream of the X-linked Jarid1c gene, which escapes X inactivation but is immediately adjacent to a gene that is inactivated, suggesting that CTCF may play a role in establishing an escape domain boundary. However, our present results demonstrate that flanking cHS4 insulators with CTCF binding sites is not sufficient to establish an X inactivation escape domain at the Hprt locus, which implies that escape from X inactivation requires different or additional mechanisms that may be present at the Jarid1c escape domain. Given this result, it will be informative to test whether the region at the 5′ end of Jarid1c, with or without its CTCF binding sites, can cause a transgene to escape X inactivation when targeted to the Hprt locus.
Materials and Methods
Gene Targeting and Generation of Transgenic Mice.
The GFPn reporter transgene used a Renilla reniformis GFPn (Stratagene) and was expressed from a 1.3-kb genomic fragment containing the human β-actin promoter (25). A 1.2-kb fragment containing hypersensitive site 4 (cHS4) from the chicken β-globin locus (8), a gift from G. Felsenfeld (National Institutes of Health, Bethesda), was used as the DNA insulator. The reporter transgenes were cloned into the Hprt targeting vector, a modified version of pSKB1 (26). The targeting vectors were linearized with PmeI before electroporation. Cell culture, electroporation, selection, and microinjection of ES cells were as described by Bronson et al. (26). Southern blot analysis was performed with a random primer-labeled RsaI probe from intron 3 of the mouse Hprt gene. The mice were maintained in a facility accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International according to Institutional Animal Care and Use Committee-approved guidelines.
In Vitro Differentiation.
Undifferentiated ES cells were cultured on γ-irradiated mouse embryonic feeder cells. In vitro differentiation of ES cells was performed as described (32). Cells were harvested for FACS analysis by treating them with 0.2% collagenase type IAS (Sigma) for 30 min at 37°C, followed by 0.0125% trypsin-EDTA for 10 min.
FACS Analysis.
Undifferentiated and differentiated ES cells and peripheral blood mononuclear cells were analyzed on a Becton Dickinson FACScan analytical flow cytometer. Single-cell suspensions of splenocytes were sorted with a DAKO Modular Flow cytometer. Histograms were processed with DAKO summit software, version 3.1.
Embryo Dissection and Confocal Microscopy.
Transgenic male mice were mated to WT C57BL/6 female mice. Embryos were dissected in PBS and 5% FBS at embryonic day 7.5, examined with a Leica MZLIII stereoscope equipped with a UV lamp and GFP filter, and fixed on ice in PBS and 4% paraformaldehyde for 15–30 min. Embryos were counterstained with TO-PRO-3 (Molecular Probes) as described (33). Images were acquired the same day with a Zeiss LSM5 Pascal confocal laser scanning microscope. The dynamic range was set by using the range palette function, and the photomultiplier gain settings were 980, 967, and 902 for embryos with the GFPn, I-GFPn-I, and II-GFPn-II transgenes, respectively. For each embryo, 35–60 optical sections at 2.2 μm were captured with a Zeiss 20/NA 0.75 objective. The stacks of optical sections were processed into a single z projection by using imagej software. Single optical sections were captured at ×40 with a Zeiss 40/NA 1.2 objective. The 8-bit grayscale images were pseudocolored with imagej software, adjusted with the same values, and merged in Adobe Systems (San Jose, CA) photoshop, version 7.0.1.
FISH.
Spleens from heterozygous female adult GFPn transgenic mice were dissociated into single-cell suspensions in RPMI medium 1640, 0.1% BSA (NEB, Beverly, MA), and 10 units/ml RNasin (Promega). Splenocytes were then sorted into GFP+ and GFP− populations with the DAKO Modular Flow cytometer. The cells were spun onto slides by using a Shandon (Pittsburgh) Cytospin at 112 × g for 5 min. DNA and RNA FISH was performed as described (34). The probes were generated by random priming by using the BioPrime DNA Labeling System (Invitrogen) and Cy3-dCTP (Amersham Pharmacia) for the 7-kb II-GFPn-II probe and FITC-dUTP (Roche) for the Xist exon 6 probe. Fluorescent images were captured with a Leica (Deerfield, IL) DML fluorescence microscope and spot rt software. The 8-bit grayscale images were pseudocolored with imagej software, adjusted, and merged in Adobe photoshop 7.0.1.
DNA Methylation Analysis.
Splenocytes from heterozygous female adult GFPn transgenic mice were sorted into GFP+ and GFP− populations by using the DAKO Modular Flow cytometer. DNA from ≈3 × 106 GFP+ and 3 × 106 GFP− cells from the three GFPn transgenic lines was digested with EcoRI (NEB). One-third of the EcoRI-digested DNA was digested with SmaI (NEB), and one-third was digested with XmaI (NEB). Digested DNA was analyzed by Southern blot with the GFPn coding sequence as a probe. Intensity of the hybridized bands was quantified by using imagej software.
Supplementary Material
Acknowledgments
We thank members of the University of North Carolina Animal Models Laboratory, the University of North Carolina Flow Cytometry Facility, and the Microscopy Services Laboratory for assistance; Gary Felsenfeld for the pJC-HS4 plasmid containing the cHS4 insulator; and Robert Bagnell, Colyn Crane-Robinson, Nobuyo Maeda, Nathan Montgomery, Kumar Pandya, and Barbara Panning for help and advice. This work was supported by National Institutes of Health Grant HL37001.
Abbreviations
- cHS4
chicken hypersensitive site 4
- CTCF
CCCTC binding factor
- Xi
inactive X chromosome
- Xa
active X chromosome
- Xp
paternal X chromosome
- Xm
maternal X chromosome
- GFPn
GFP gene with a nuclear localization signal
- emb
embryonic ectoderm.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Weiler K. S., Wakimoto B. T. Annu. Rev. Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 2.Muller H. J. Genet. 1930;22:299–335. [Google Scholar]
- 3.Kellum R., Schedl P. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 4.Burgess-Beusse B., Farrell C., Gaszner M., Litt M., Mutskov V., Recillas-Targa F., Simpson M., West A., Felsenfeld G. Proc. Natl. Acad. Sci. USA. 2002;99:16433–16437. doi: 10.1073/pnas.162342499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West A. G., Gaszner M., Felsenfeld G. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 6.Saitoh N., Bell A. C., Recillas-Targa F., West A. G., Simpson M., Pikaart M., Felsenfeld G. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hebbes T. R., Clayton A. L., Thorne A. W., Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung J. H., Whiteley M., Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 9.Bell A. C., West A. G., Felsenfeld G. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 10.Recillas-Targa F., Pikaart M. J., Burgess-Beusse B., Bell A. C., Litt M. D., West A. G., Gaszner M., Felsenfeld G. Proc. Natl. Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pikaart M. J., Recillas-Targa F., Felsenfeld G. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West A. G., Huang S., Gaszner M., Litt M. D., Felsenfeld G. Mol. Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Mutskov V. J., Farrell C. M., Wade P. A., Wolffe A. P., Felsenfeld G. Genes Dev. 2002;16:1540–1554. doi: 10.1101/gad.988502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heard E. Curr. Opin. Genet. Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Lyon M. F. Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 16.Cohen D. E., Lee J. T. Curr. Opin. Genet. Dev. 2002;12:219–224. doi: 10.1016/s0959-437x(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 17.Carrel L., Cottle A. A., Goglin K. C., Willard H. F. Proc. Natl. Acad. Sci. USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown C. J., Greally J. M. Trends Genet. 2003;19:432–438. doi: 10.1016/S0168-9525(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 19.Disteche C. M. Trends Genet. 1995;11:17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- 20.Tsuchiya K. D., Willard H. F. Mamm. Genome. 2000;11:849–854. doi: 10.1007/s003350010175. [DOI] [PubMed] [Google Scholar]
- 21.Goldman M. A., Stokes K. R., Idzerda R. L., McKnight G. S., Hammer R. E., Brinster R. L., Gartler S. M. Science. 1987;236:593–595. doi: 10.1126/science.2437652. [DOI] [PubMed] [Google Scholar]
- 22.Whyatt D., Lindeboom F., Karis A., Ferreira R., Milot E., Hendriks R., de Bruijn M., Langeveld A., Gribnau J., Grosveld F., Philipsen S. Nature. 2000;406:519–524. doi: 10.1038/35020086. [DOI] [PubMed] [Google Scholar]
- 23.Chong S., Kontaraki J., Bonifer C., Riggs A. D. Mol. Cell. Biol. 2002;22:4667–4676. doi: 10.1128/MCB.22.13.4667-4676.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Filippova G. N., Cheng M. K., Moore J. M., Truong J. P., Hu Y. J., Nguyen D. K., Tsuchiya K. D., Disteche C. M. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama H., Niwa H., Makino K., Kakunaga T. Gene. 1988;65:135–139. doi: 10.1016/0378-1119(88)90426-x. [DOI] [PubMed] [Google Scholar]
- 26.Bronson S. K., Plaehn E. G., Kluckman K. D., Hagaman J. R., Maeda N., Smithies O. Proc. Natl. Acad. Sci. USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hadjantonakis A. K., Cox L. L., Tam P. P., Nagy A. Genesis. 2001;29:133–140. doi: 10.1002/gene.1016. [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Mager J., Chen Y., Schneider E., Cross J. C., Nagy A., Magnuson T. Nat. Genet. 2001;28:371–375. doi: 10.1038/ng574. [DOI] [PubMed] [Google Scholar]
- 29.Bird A. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 30.Migeon B. R. Trends Genet. 1994;10:230–235. doi: 10.1016/0168-9525(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 31.Weber M., Davies J. J., Wittig D., Oakeley E. J., Haase M., Lam W. L., Schubeler D. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 32.Wobus A. M., Wallukat G., Hescheler J. Differentiation. 1991;48:173–182. doi: 10.1111/j.1432-0436.1991.tb00255.x. [DOI] [PubMed] [Google Scholar]
- 33.Schejter E. D., Wieschaus E. Cell. 1993;75:373–385. doi: 10.1016/0092-8674(93)80078-s. [DOI] [PubMed] [Google Scholar]
- 34.Panning B., Dausman J., Jaenisch R. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.