The biogenesis of translatable eukaryotic messenger RNA (mRNA) requires a series of post-transcriptional processing steps. Initially, the 5′-end of the nascent pre-mRNA acquires a cap structure. Then, the intron regions are successively spliced out from the growing RNA chain. Finally, the mRNA 3′-end is cleaved and a poly(A) tail is added. To coordinate efficient and faithful mRNA production, a complex network of dynamic physical and functional interactions are formed between the elongating RNA polymerase (pol) II and the different mRNA-processing machines that carry out each of these steps (Ares & Proudfoot, 2005). In vertebrates, the intron regions of protein-coding genes frequently encode small-nucleolar RNAs (snoRNAs). These snoRNAs are post-transcriptionally processed from the spliced out and debranched host intron by exonucleolytic activities. It has long been assumed that the only role of the RNA pol II transcription/splicing machine in intronic snoRNA expression is to provide the linear substrate RNA for the processing exonucleases. However, recent studies from several laboratories have revealed more intimate functional interplays between the machines that are devoted to snoRNA processing and mRNA biogenesis.
The intronic snoRNAs fall into two structurally and functionally well-defined classes. SnoRNAs carrying the conserved box C and D elements function mainly as guides in the site-specific 2′-O-methylation of ribosomal RNAs (rRNAs), whereas box H/ACA snoRNAs direct the pseudouridylation of rRNAs (Terns & Terns, 2002). To form functional small-nucleolar ribonucleoproteins (snoRNPs), the mature box C/D snoRNAs associate with fibrillarin, the 15.5 kDa protein, and nucleolar proteins Nop56 and Nop58, whereas the box H/ACA snoRNAs form complexes with dyskerin, Nop10, non-histone chromosome protein 2 (Nhp2) and glycine arginine rich protein 1 (Gar1).
The possibility that there is a synergy between intronic snoRNA processing and pre-mRNA splicing was first raised by members of the group of Joan Steitz. They noticed that human box C/D snoRNAs have a preferential intronic location with respect to the 3′ splice site (Hirose & Steitz, 2001). Subsequent in vivo and in vitro processing experiments confirmed that an optimal distance (about 50 nucleotides) between the snoRNA-coding region and the branch point is crucial for efficient snoRNA production (Hirose et al, 2003). This suggests that the splicing machine participates directly in box C/D snoRNP processing. By applying an in vitro system that couples pre-mRNA splicing and snoRNA processing, the Steitz group showed that the 15.5 kDa protein, which is the first to bind to box C/D snoRNAs and nucleate snoRNP assembly, is recruited to the intronic snoRNA through an active mechanism at the C1 splicing-complex stage (Fig 1A). The authors proposed that a putative box C/D snoRNP-assembly factor (AF) recruits and deposits the 15.5 kDa protein onto the intronic snoRNA. The hypothetical assembly factor that is responsible for connecting box C/D snoRNP assembly to pre-mRNA splicing probably interacts, directly or indirectly, with the U2 spliceosomal small-nuclear RNP (snRNP) or other splicing factors associated with the branch-point region in the C1 splicing complex.
Figure 1.
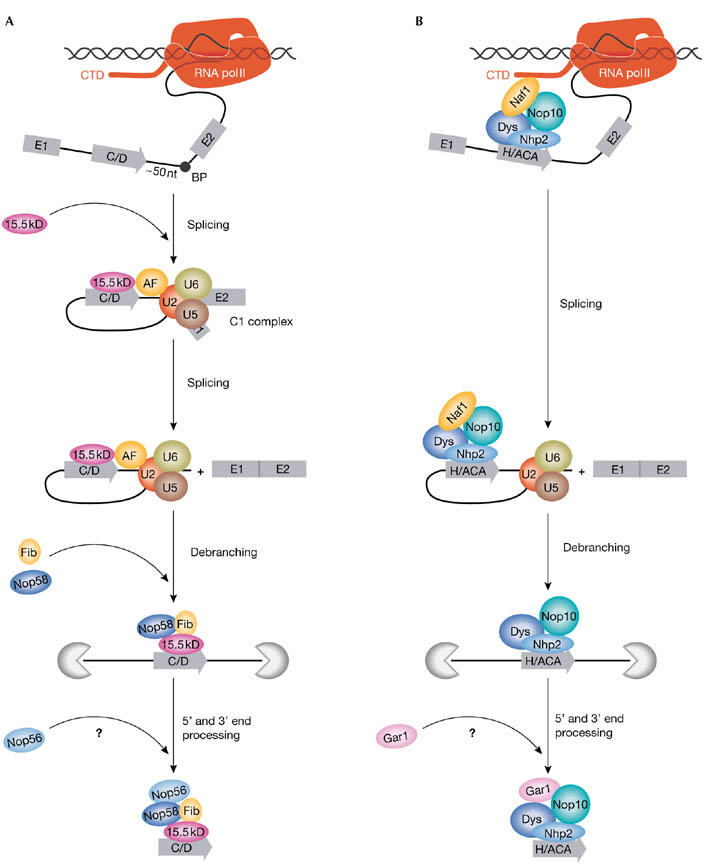
Models for coupling intronic small-nucleolar ribonucleoprotein assembly with pre-messenger RNA transcription and splicing. (A) Splicingdependent assembly of intronic box C/D small-nucleolar ribonucleoproteins (snoRNPs). The majority of mammalian intronic box C/D snoRNAs are located about 50 nucleotides (nt) upstream of the branch point (BP) and are processed in a splicing-dependent manner. At the C1-complex stage of splicing, the 15.5 kDa (kD) central core protein is actively recruited to the intronic snoRNA by a putative assembly factor (AF) that probably interacts with splicing factors associated with the BP region. Fibrillarin (Fib) and the nucleolar protein Nop58 are believed to bind directly after 15.5 kDa docking, but Nop56 might bind at a later stage of snoRNP biogenesis. (B) Transcription-dependent assembly of intronic box H/ACA snoRNPs.Assembly of H/ACA presnoRNPs occurs co-transcriptionally. Nuclear-assembly factor 1 (Naf1), which is an essential H/ACA assembly factor, might interact with the carboxyterminal domain (CTD) of RNA polymerase (pol) II, and could facilitate recruitment and binding of the H/ACA core proteins to the nascent snoRNA sequences. The correct timing of glycine arginine rich 1 (Gar1) binding is still unclear. It is important to note that assembly of a minority group of box C/D snoRNPs that have suboptimal intronic positions and carry external intronic stem structures might follow a transcription-dependent processing pathway similar to that described for H/ACA snoRNPs (Hirose et al, 2003). Dys, dyskerin; Nhp2, non-histone chromosome protein 2.
Previous studies identified the yeast nuclear-assembly factor 1 (Naf1) protein as essential in box H/ACA snoRNP biogenesis (Dez et al, 2002; Fatica et al, 2002; Yang et al, 2002). Yeast Naf1 associates with actively transcribed H/ACA snoRNA genes, but not mature snoRNPs, indicating that it functions in an early step of H/ACA snoRNP synthesis (Yang et al, 2005; Ballarino et al, 2005). In a recent study, published in the May 2006 issue of RNA, the groups of Yves Henry and Michèle Ferrer have characterized the human orthologue of Naf1, and shown that it is essential for the accumulation of human intronic H/ACA snoRNPs (Hoareau-Aveilla et al, 2006). In another study, published in the 24 April 2006 issue of the Journal of Cell Biology, the group of Thomas Meier established a human cell line to visualize the transcription of intronic snoRNA genes, and showed that Naf1 and three H/ACA core proteins (dyskerin, Nhp2 and Nop10) associate specifically with transcriptionally active intronic H/ACA snoRNA genes (Darzacq et al, 2006). Using an in vitro translation/immunoprecipitation assay, the Meier group also showed that mammalian Naf1 interacts with the dyskerin/Nop10/Nhp2 H/ACA core complex. The Meier group propose that, at a later stage of snoRNP maturation, Naf1 is replaced by the Gar1 H/ACA protein, and the mature snoRNP is released from the transcription site and transported to the nucleolus (Fig 1B). The results of the Meier group, together with the previous observation that yeast Naf1 can interact with the carboxy-terminal domain (CTD) of the largest subunit of RNA pol II (Fatica et al, 2002), strongly suggest that Naf1 recruits box H/ACA snoRNP proteins to the newly synthesized H/ACA snoRNA through its interaction with the CTD of RNA pol II. Consistent with this view, in vivo processing experiments showed that the assembly of H/ACA snoRNPs depends on RNA pol II transcription, and confirmed that binding of H/ACA core proteins to intronic snoRNAs already occurs during pre-mRNA synthesis (Richard et al, 2006). Therefore, Naf1 might have a central role in coupling H/ACA snoRNP assembly with RNA pol II transcription.
Mammalian pre-mRNA introns that are removed by splicing are rapidly degraded. Therefore, active recruitment of snoRNP proteins during pre-mRNA synthesis or splicing seems to be crucial for efficient intronic snoRNP production. The demonstration that intronic snoRNP assembly is actively promoted by machines that are involved in pre-mRNA biogenesis indicates that the functional complexity of the mammalian ‘mRNA factory' exceeds our previous expectations (Ares & Proudfoot, 2005).


References
- Ares M Jr, Proudfoot NJ (2005) The Spanish connection: transcription and mRNA processing get even closer. Cell 120: 163–166 [DOI] [PubMed] [Google Scholar]
- Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I (2005) The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol 25: 5396–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT (2006) Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y (2002) Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol Cell Biol 22: 7053–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Dlakic M, Tollervey D (2002) Naf1p is a box H/ACA snoRNP assembly factor. RNA 8: 1502–1514 [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Shu MD, Steitz JA (2003) Splicing-dependent and -independent modes of assembly for intron-encoded box C/D snoRNPs in mammalian cells. Mol Cell 12: 113–123 [DOI] [PubMed] [Google Scholar]
- Hirose T, Steitz JA (2001) Position within the host intron is critical for efficient processing of box C/D snoRNAs in mammalian cells. Proc Natl Acad Sci USA 98: 12914–12919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M, Henry Y (2006) hNaf1 is required for accumulation of human H/ACA snoRNPs, scaRNPs and telomerase. RNA 12: 832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Kiss AM, Darzacq X, Kiss T (2006) Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 26: 2540–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Terns RM (2002) Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr 10: 17–39 [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G (2005) Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol 25: 3295–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G (2002) The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J Biol Chem 277: 45235–45242 [DOI] [PubMed] [Google Scholar]


