Abstract
Atg8, a member of a novel ubiquitin-like protein family, is an essential component of the autophagic machinery in yeast. This protein undergoes reversible conjugation to phosphatidylethanolamine through a multistep process in which cleavage of Atg8 by a specific protease is followed by ubiquitin-like conjugation processes. Here, we identify two essential sites in Atg8, one of them involving residues Phe 77 and Phe 79 and the other, located on the opposite surface of Atg8, residues Tyr 49 and Leu 50. We show that these two sites are associated with different functions of Atg8: Phe 77 and Phe 79 seem to be part of the recognition site for Atg4, a cystein protease that acts also as a deubiquitination enzyme, whereas Tyr 49 and Leu 50 act downstream of the lipidation step. These two newly identified distinct sites that are essential for Atg8 activity provide an explanation for the many protein–protein interactions of this low-molecular-weight protein.
Keywords: Atg8, autophagy, ubiquitin-like
Introduction
The autophagic pathway mediates delivery of cytosolic proteins and organelles for degradation within the lysosomes/vacuole compartments. Recent studies linked autophagy with numerous physiological conditions, including liver diseases, muscular disorders, neurodegeneration, pathogen infections and cancer (Cuervo, 2004; Shintani & Klionsky, 2004a).
Genetic screens in the yeast Saccharomyces cerevisiae identified 16 autophagy-related genes (ATGs) that are essential for this process (Klionsky et al, 2003). This group of genes includes two unique ubiquitin-like (UBL) systems that take part in early stages of autophagosome biogenesis (Ohsumi & Mizushima, 2004). The first includes Atg12, a UBL molecule that is specifically conjugated to Atg5 to form a stable Atg12–Atg5 conjugate. This process is mediated by both Atg7, an E1-activating enzyme, and Atg10, an E2-conjugating enzyme. Together with Atg16, which forms a complex with the Atg12–Atg5 conjugate (Kuma et al, 2002), these molecules may act upstream of the second UBL system that conjugates Atg8 to phosphatidylethanolamine (PE; Ichimura et al, 2000). This lipidation process serves to associate Atg8 covalently with the isolation membrane and autophagosomes, thus rendering it a genuine marker for these membranes.
The mechanism of autophagosome biogenesis remains enigmatic, and the exact role of Atg8 in this process is not yet fully understood. This evolutionarily conserved molecule is part of a novel protein family with many homologues in mammals (Elazar et al, 2003) and plants (Yoshimoto et al, 2004), some of which are specifically associated with autophagosomes (Kabeya et al, 2004). Despite being a low-molecular-weight protein consisting of a single UBL fold decorated by two amino-terminal short helices, this protein is recognized specifically by several proteins, including the cysteine protease Atg4 (Kirisako et al, 2000), as well as by Atg7 and Atg3, which subsequently direct it to PE on the membrane (Ichimura et al, 2000). Here, we report the identification of two distinct functional sites that are required for the autophagic activity of Atg8.
Results and Discussion
Identification of essential residues on Atg8
Atg8 was initially identified as an essential protein for autophagy (Lang et al, 1998; Kirisako et al, 1999) and for cytosol-to-vacuole transport (Cvt; Huang et al, 2000) in yeast. This protein first becomes conjugated to PE, a process that starts with the removal of the carboxy-terminal arginine from the newly synthesized Atg8 by the Atg4 protease to expose a C-terminal glycine residue (Atg8G116; Kirisako et al, 2000). Atg8G116 is then activated by Atg7 (E1) and transferred to Atg3 (E2), which conjugates it to PE by an amide bond between the C-terminal glycine and the amino group of PE (Ichimura et al, 2000). The latter process is then terminated by the deubiquitination activity of Atg4. To clarify the physiological role of this conjugation process, it is imperative, first, to identify the functional domains on Atg8 and, second, to determine whether lipidation is the end point of Atg8 role in this process or whether it functions downstream of the conjugation step. To address these questions, we undertook a systematic mutagenesis approach aiming to identify the functional domains of Atg8.
Atg8 shows significant amino-acid sequence homology to a novel UBL mammalian protein family (Fig 1A). The crystal structures of three members of this family, namely GATE-16, GAPARAP and LC3, are almost identical (Paz et al, 2000; Knight et al, 2002; Sugawara et al, 2004). On the basis of these structures and the sequence homology, we used SWISS MODEL to predict the structure of Atg8 with a root-mean-square deviation (r.m.s.d.) value of 0.48 Å (Fig 1B). We then systematically mutated the conserved amino-acid residues located on the predicted protein surface to alanine and tested the ability of these mutants to complement autophagy. Autophagic activity was monitored in Δatg8 cells using aminopeptidase I (API) maturation under nitrogen starvation. Altogether, we mutated 13 amino acids, including Arg 20, Arg 24, Arg 28, Lys 46, Tyr 49, Leu 50, Tyr 62, Lys 66, Phe 77, Phe 79 and Asp 100 (underlined in the Atg8 amino-acid sequence presented in Fig 1A). Nine of these mutations had no effect on the protein activity. However, because of limitations of the API assay, we cannot exclude the possibility that some of these residues are involved in other autophagic activities of Atg8. Mutations of residue Tyr 49 or Leu 50 blocked the Atg8 autophagic activity, whereas mutations of Phe 77 and Phe 79, when introduced in tandem, inhibited its activity (all four residues are highlighted in the structure presented in Fig 1C). On the basis of these results, we proposed that Atg8 contains two distinct sites, located on opposite sides of the protein, which are essential for its autophagic activity.
Figure 1.
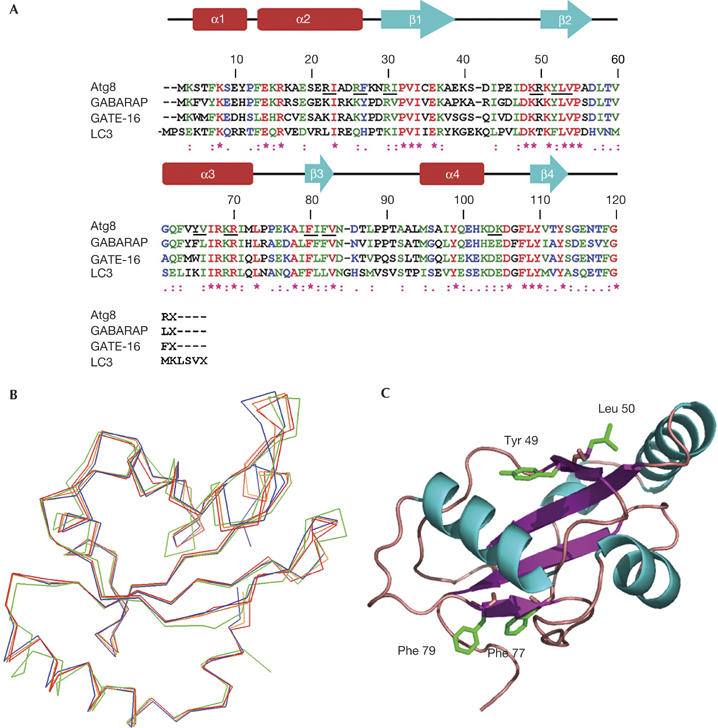
Modelling the three-dimensional structure of Atg8. (A) Multiple sequence alignment of Atg8 homologues. Amino-acid sequences of Atg8 and its mammalian homologues are aligned using the Clustal_W program. Underlined residues represent mutated amino acids in Atg8. (B) Alignment of Atg8 predicted three-dimensional structure with its mammalian homologues. SWISS MODEL was used to predict Atg8 structure and PyMol was used for presentation. Orange represents the predicted Atg8 structure, and red, blue and green represent GATE-16, GABARAP and LC3, respectively. (C) Three-dimensional model of Atg8. The predicted structure contains a ubiquitin fold decorated by two further amino-terminal helices (amino acids Tyr 49, Leu 50, Phe 77 and Phe 79 are marked in green).
Residues Phe 77 and Phe 79 of Atg8 recognize Atg4
Green fluorescent protein (GFP)–Atg8 has been previously shown to complement both Cvt and autophagy in Δatg8 cells (Suzuki et al, 2001). To further characterize the mutants described above, we have introduced GFP–Atg8 harbouring mutations at residues Phe 77 and Phe 79 into Δatg8 cells. We have made use of API, a vacuolar resident hydrolase that travels to the vacuole by means of the Cvt pathway under normal growth conditions and by autophagy under starvation conditions. Its N-terminal propeptide containing the vacuole targeting signal is cleaved off by Pep4 following delivery to the vacuole (Fig 2A). As depicted in Fig 2A, GFP–Atg8F77A or GFP–Atg8F79A complemented API maturation under the different growth conditions, whereas the double mutant GFP–Atg8F77/79A failed to do so. Notably, similar results were obtained when the different Atg8 mutants were expressed with their endogenous promoter (data not shown). These results indicate that Phe 77 and Phe 79 are indispensable for both the Cvt and autophagy pathways.
Figure 2.
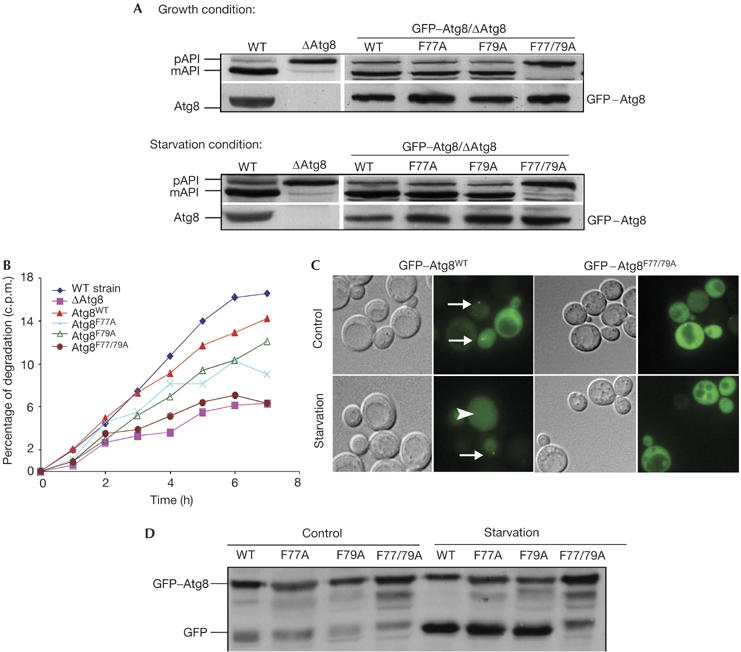
GFP–Atg8F77/79A double mutant shows a defect in the autophagic process. GFP–Atg8WT, GFP–Atg8F77A, GFP–Atg8F79A or GFP–Atg8F77/79A was transformed into Δatg8 cells. (A) Cells were grown to mid-exponential phase and then incubated in the presence of 3.5% galactose to induce expression of GFP–Atg8. Aminopeptidase I (API) maturation was tested in cells incubated in control or starvation medium by western blot analysis using anti-Atg8 (right bottom panel), anti-GFP (left bottom panel) or anti-API antibodies (upper panel). (B) Cells were pulse-labelled with [35S]methionine and chased on non-radioactive starvation medium. Aliquots were taken at the indicated times and acid-soluble small peptides generated by proteolysis were determined. (C) Cells were grown in control or in starvation medium and visualized by light microscopy. Arrows point to the pre-autophagosomal structure (PAS), and arrowheads point to GFP within the vacuoles. (D) Cell extracts were subjected to SDS–polyacrylamide gel electrophoresis followed by immunoblot analysis using anti-GFP antibodies. GFP, green fluorescent protein; mAPI, mature aminopeptidase I; pAPI, premature aminopeptidase I; WT, wild type.
To study further the role of these phenylalanine residues in Atg8 activity, we used different approaches to assess autophagy in living cells. First, we tested the ability of GFP–Atg8F77A, GFP–Atg8F79A and GFP–Atg8F77/79A to complement starvation-induced protein degradation in the Δatg8 cells. Consistent with their activity in the API maturation assay, both GFP–Atg8F77A and GFP–Atg8F79A partly complemented starvation-induced protein degradation, whereas the GFP–Atg8F77/79A double mutant strain did not (Fig 2B). These results lend support to the idea that these two adjacent phenylalanine residues are essential for autophagy.
GFP–Atg8 has been localized to the cytosol and to a dot-like structure, near the vacuole, termed the pre-autophagosomal structure (PAS; Suzuki et al, 2001). Moreover, after starvation, GFP–Atg8 is translocated from the cytosol to the vacuole in an autophagy-dependent manner (Mizushima et al, 2004). We therefore examined the subcellular localization of wild-type GFP–Atg8 and the mutant GFP–Atg8F77/79A in Δatg8 cells under different growth conditions. As depicted in Fig 2C, under control conditions, wild-type GFP–Atg8 is mainly localized to the cystosol and the PAS, whereas after starvation it accumulates within the vacuole. In contrast, GFP–Atg8F77/79A did not localize to the PAS and remained cytosolic even in starved cells. After delivery of GFP–Atg8 to the vacuole, the GFP moiety is proteolytically removed, and the free GFP can serve as a biochemical marker to monitor vesicle delivery (Shintani & Klionsky, 2004b). The failure of GFP–Atg8F77/79A to undergo translocation to the vacuole was further confirmed by western blot analysis using anti-GFP antibodies (Fig 2D). Notably, the vacuole in cells expressing GFP–Atg8F77/79A appeared slightly fragmented.
The inability of Atg8F77/79A to support autophagic activities may have resulted from defects in its processing machinery. To test whether the aforementioned mutations affected C-terminal cleavage by Atg4 protease, haemagglutinin (HA) tag was fused to the C terminus of the protein and expressed in the Δatg8 cells under normal and starvation conditions. As shown in Fig 3A, the C-terminal cleavage of the HA tag was not affected by the mutation at residue Phe 79, whereas mutation at residue Phe 77 significantly inhibited cleavage under normal growth conditions and, to a lesser extent, under starvation. Cleavage of Atg8F77/79A–HA, however, was completely inhibited under both growth conditions (Fig 3A). Similar results were obtained when a low-copy plasmid was used for the expression of Atg8 (data not shown). From these results, we concluded that both Phe 77 and Phe 79 residues of Atg8 are essential for cleavage by Atg4 activity. Notably, these residues are found at the centre of an interface surface predicted to participate in protein–protein interactions (Fig 3B; for details, see supplementary information online). To test whether Phe 77 and Phe 79 participate in Atg8–Atg4 interaction, recombinant Atg4 tagged with six histidine residues at its N terminus was coupled to Talon beads and used in a pull-down assay with cell extracts obtained from the different strains. As depicted in Fig 3C, mutations in either Phe 77 or Phe 79 significantly reduced the binding of Atg8 to Atg4, whereas the double mutant had a larger effect. Atg8 was not detected in eluates when the different extracts were incubated with Talon beads in the absence of Atg4 (data not shown). These results indicate that Phe 77 and Phe 79 are involved in Atg8–Atg4 interaction. Consistently, overexpression of Atg4 suppressed the autophagic phenotype of Atg8F77/79A (Fig 3D,E). To test whether residues Phe 77 and Phe 79 are also essential for the conjugation process, we expressed the different mutants with the C-terminal glycine exposed (Atg8F77/79AΔR), thus overcoming the need for the initial cleavage by the protease. As shown in Fig 4A, both Atg8F77A and Atg8F79A, but not Atg8F77/79A, undergo PE lipidation under starvation condition. Notably, Atg8F77/79A migrated in the urea SDS–polyacrylamide gel electrophoresis (SDS–PAGE) slightly faster with a trail of a higher-molecular-weight band. This phenomenon was previously reported for an uncleaved protein (Kirisako et al, 2000). Atg8F77/79AΔR, conversely, was lipidated (Fig 4A), indicating that these mutations specifically affect the protease but not the conjugation machinery. This was further confirmed by the ability of GFP–Atg8F77/79AΔR to be localized to the PAS (Fig 4B), a lipidation-dependent process. These mutants, however, failed to reconstitute autophagy in the Atg8-null strain (Fig 4C), consistent with the notion that deconjugation of Atg8 by Atg4 is essential for this process (Kirisako et al, 2000).
Figure 3.
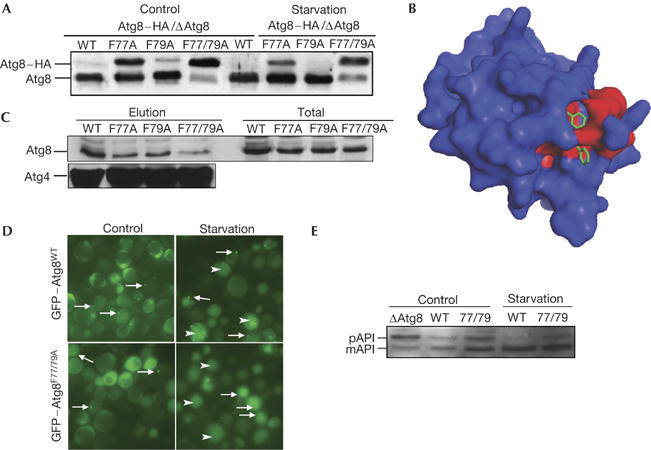
Atg8F77/79A does not undergo cleavage in the carboxyl terminus. (A) Atg8WT–HA, Atg8F77A–HA, Atg8F79A–HA or Atg8F77/79A–HA was transformed into Δatg8 cells. Cell extracts were prepared and subjected to 13.5% SDS–polyacrylamide gel electrophoresis (SDS–PAGE) containing 6 M urea, followed by immunoblot analysis with anti-Atg8 antibodies. (B) Prediction of protein–protein interaction sites in Atg8 (Neuvirth et al, 2004). A three-dimensional model of Atg8 is coloured according to ProMate prediction (see supplementary information online) with red representing the predicted protein–protein binding sites, and Phe 77 and Phe 79 are marked in green within the prediction site. (C) Pull-down assay of Atg8 mutants with Atg4. His–Atg4 was attached to cobalt beads (Talon metal-affinity resin), washed and incubated with total cell extracts of Atg8 mutants. Elution was subjected to SDS–PAGE followed by western blot analysis using anti-Atg8 and anti-Atg4 antibodies. (D) Localization of green fluorescent protein (GFP)–Atg8wt and GFP–Atg877/79 cells carrying Atg4 plasmid under normal (control) and starvation conditions. Arrows point to the pre-autophagosomal structure, and arrowheads point to GFP within the vacuoles. (E) Aminopeptidase I (API) maturation was tested in cells incubated in control or starvation medium by western blot analysis using anti-API antibodies. mAPI, mature aminopeptidase I; pAPI, premature aminopeptidase I; WT, wild type.
Figure 4.
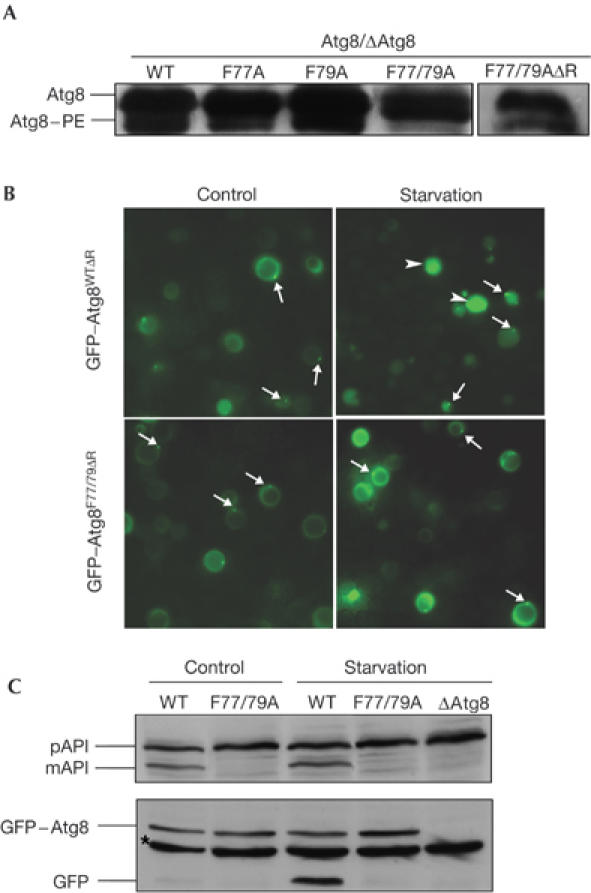
GFP–Atg8F77/79AΔR double mutant shows a defect in the autophagic process despite undergoing lipidation. GFP–Atg8WTΔR or GFP–Atg8F77/79AΔR was transformed into ΔAtg8 cells. (A) Cell extracts were subjected to 13.5% SDS–polyacrylamide gel electrophoresis containing 6 M urea, followed by immunoblot analysis using anti-Atg8-15N peptide antibodies. (B) Cells were grown in control or in starvation medium and visualized using fluorescence microscopy. Arrows point to the pre-autophagosomal structure, whereas arrowheads point to GFP within the vacuoles. (C) Aminopeptidase I (API) maturation and GFP cleavage were tested as described above. Asterisk represents nonspecific band. GFP, green fluorescent protein; mAPI, mature aminopeptidase I; pAPI, premature aminopeptidase I; WT, wild type.
Atg8 Tyr 49 and Leu 50 are essential for autophagy
As described in our initial screen, the two neighbouring residues Tyr 49 and Leu 50 seemed to be essential for autophagy. To further characterize the effect of these mutations on Atg8 autophagic activity, we expressed GFP–Atg8 fusion protein containing these mutations in Δatg8 cells. As shown in Fig 5A, neither GFP–Atg8Y49A nor GFP–Atg8L50A restored API maturation under normal or starvation conditions, implying that these mutants do not support the Cvt pathway or autophagy. To confirm the autophagic defect in these mutants, we tested their ability to complement starvation-induced protein degradation in Δatg8 cells. The control, GFP–Atg8WT, fully complemented starvation-induced protein degradation, whereas GFP–Atg8L50A or GFP–Atg8Y49A did not (Fig 5B). These results confirm the essential role of Tyr 49 and Leu 50 in Atg8 autophagic activity.
Figure 5.
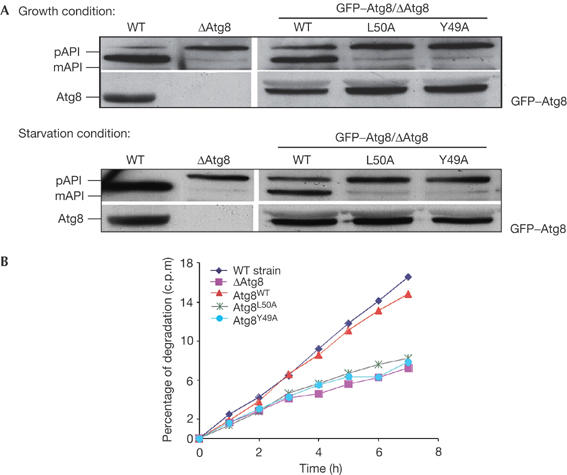
GFP–Atg8L50A or GFP–Atg8Y49A transformed into Δatg8 cells does not reconstitute. (A) Δatg8 cells transformed with GFP–Atg8WT, GFP–Atg8L50A or GFP–Atg8Y49A constructs were grown to mid-exponential phase and then incubated in the presence of 3.5% galactose to induce expression of GFP–Atg8. API maturation was tested using anti-Atg8 (right bottom panel), anti-GFP (left bottom panel) or anti-API antibodies. (B) Starvation-induced protein degradation of Δatg8 harbouring the different mutants was tested as described above. API, aminopeptidase I; GFP, green fluorescent protein; mAPI, mature aminopeptidase I; pAPI, premature aminopeptidase I; WT, wild type.
To monitor the localization of these Atg8 mutants, we examined the intracellular localization of GFP-tagged Atg8 in Δatg8 cells. As shown in Fig 6A, GFP–Atg8Y49A and GFP–Atg8L50A were found in the cytosol and in PAS-like structures under both control and starvation conditions. However, in response to starvation, GFP–Atg8WT was translocated to the vacuole, whereas GFP–Atg8Y49A did not, and only traces of GFP–Atg8L50A were detected in the vacuole. This was further confirmed by western blot analysis with anti-GFP antibodies (Fig 6B). Notably, the dot-like structures in these mutants were smaller than those of the wild-type protein. Nevertheless, the fact that mutant proteins are localized to the PAS under starvation conditions suggests that these mutations do not affect the overall folding of the protein. Moreover, as the localization of GFP–Atg8WT to the PAS apparently depends on the activity of the yeast conjugating machinery (Suzuki et al, 2001), we assume that the ability of the mutants to undergo conjugation to PE is unaffected.
Figure 6.
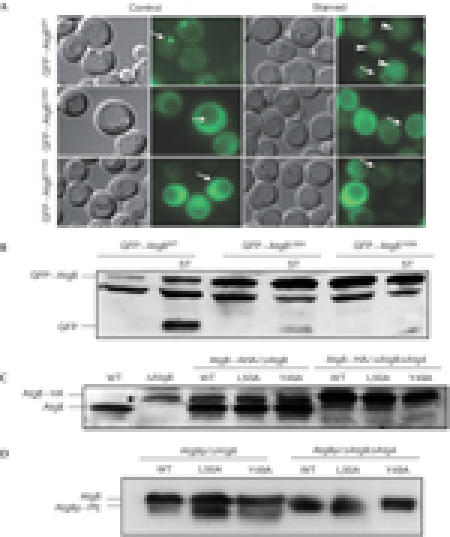
GFP–Atg8 mutants were tested for their carboxy-terminus processing, phosphatidylethanolamine conjugation and autophagic activity. Cells expressing GFP–Atg8WT or GFP–Atg8 mutants were grown in control or starvation medium. (A) GFP–Atg8 localization was determined using fluorescence microscopy (arrows depict pre-autophagosomal structure, and arrowheads point to GFP within vacuoles). Cells were also tested for the appearance of autophagic bodies by using light microscopy. (B) Immunoblot analysis of different cell extracts was carried out using anti-GFP antibodies (asterisk represents nonspecific band). (C) Atg8WT–HA, Atg8L50A–HA or Atg8Y49A–HA was transformed into Δatg8 and Δatg8Δatg4 double knockout strains and cell extracts were subjected to SDS–polyacrylamide gel electrophoresis (SDS–PAGE) followed by immunoblot analysis using anti-HA antibodies (lower panel; asterisk represents nonspecific band). (D) Atg8WT, Atg8L50A or Atg8Y49A was transformed into Δatg8 and Δatg8Δatg4 double knockout strains and grown under control or starvation conditions for 4 h. Cell extracts were subjected to 13.5% SDS–PAGE containing 6 M urea, followed by immunoblot analysis using anti-Atg8-N15 antibodies. GFP, green fluorescent protein; WT, wild type.
To verify whether these mutants undergo cleavage in the C terminus by Atg4, Δatg8 and Δatg8Δatg4 cells were transformed with Atg8WT–HA, Atg8L50A–HA or Atg8Y49A–HA fusion constructs in which the HA tag was introduced at the protein C terminus. Protein extracts were prepared from each strain, subjected to SDS–PAGE and analysed by immunoblot with anti-Atg8 (Fig 6C) and anti-HA antibodies (data not shown). Both Atg8Y49A–HA and Atg8L50A–HA were cleaved at their C termini. By using the Δatg8Δatg4 double knockout strain, we confirmed the involvement of Atg4 in this cleavage (Fig 6C). On the basis of these results, we conclude that the mutations at residues Tyr 49 and Leu 50 do not affect the C-terminal cleavage of the protein by Atg4, an essential step for the subsequent conjugation reaction. We next tested whether these mutants undergo PE lipidation. For this purpose, Atg8WT, Atg8L50A and Atg8Y49A were expressed in Δatg8 and Δatg8Δatg4 cells. Cells were grown under starvation conditions, and protein extracts were prepared and subjected to 13.5% SDS–PAGE containing 6 M urea. Both mutants acquired the lipidated form, which migrated faster only in the Δatg8 but not in the Δatg8Δatg4 cells (Fig 6D). Moreover, it has been reported that nascent Atg8 (cleaved by the Atg4 protease) migrates more slowly on SDS–PAGE urea gels (Kirisako et al, 2000). Consistently, it seems that on cleavage by Atg4 protease, the unlipidated form of the wild-type and mutant proteins of Atg8 migrated more slowly in these gels (Fig 6D).
Taken together, these results indicate that residues Tyr 49 and Leu 50 are located in a functional domain of Atg8, which is essential for its autophagic activity. Interestingly, Ichimura et al (2004) have recently showed that after conjugation to PE, Atg8 changes its conformation at the N-terminal region. We therefore suggested that, after lipidation, the protein changes its conformation, thus exposing a hydrophobic pocket (containing residues Tyr 49 and Leu 50) that serves for its post-lipidation activity. Furthermore, residues Phe 77 and Phe 79, located on the other side of the molecule, are involved in Atg8-specific recognition by Atg4.
Methods
Site-directed mutagenesis. All mutations in the Atg8 open reading frame were created with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) based on the construct pYES2/ATG8 under the β-Gal promoter, pRS426 with PGK promoter or pRS316 with Atg8 endogenous promoter.
Western blotting. Whole-cell lysates were prepared by disrupting cells with glass beads in lysis buffer. SDS–PAGE and immunoblotting were performed as described (Legesse-Miller et al, 2000).
Protein turnover. The procedures for the protein degradation assay were performed as described (Straub et al, 1997).
Fluorescence microscopy. Fluorescence microscopy was performed using a confocal microscope (Olympus FV500+IX70). Cells were grown in the indicated medium to mid-exponential phase, induced with 3.5% galactose and shifted to SD-N starvation medium. To observe GFP-tagged proteins under a fluorescence microscope, we employed a band filter of 505–525 nm.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank G. Schreiber and D. Reichmann for fruitful advice in modelling Atg8 structure. Z.E. is incumbent of the Sholimo and Michla Tomarin Career Development Chair of Membrane Physiology. This work was supported in part by the Israel Science Foundation and by the Minerva center.
References
- Cuervo AM (2004) Autophagy: in sickness and in health. Trends Cell Biol 14: 70–77 [DOI] [PubMed] [Google Scholar]
- Elazar Z, Scherz-Shouval R, Shorer H (2003) Involvement of LMA1 and GATE-16 family members in intracellular membrane dynamics. Biochim Biophys Acta 1641: 145–156 [DOI] [PubMed] [Google Scholar]
- Huang WP, Scott SV, Kim J, Klionsky DJ (2000) The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 275: 5845–5851 [DOI] [PubMed] [Google Scholar]
- Ichimura Y et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492 [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Imamura Y, Emoto K, Umeda M, Noda T, Ohsumi Y (2004) In vivo and in vitro reconstitution of atg8 conjugation essential for autophagy. J Biol Chem 279: 40584–40592 [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T (2004) LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 117: 2805–2812 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, Noda T, Ohsumi Y (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 147: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151: 263–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ et al. (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5: 539–545 [DOI] [PubMed] [Google Scholar]
- Knight D, Harris R, McAlister MS, Phelan JP, Geddes S, Moss SJ, Driscoll PC, Keep NH (2002) The X-ray crystal structure and putative ligand-derived peptide binding properties of γ-aminobutyric acid receptor type A receptor-associated protein. J Biol Chem 277: 5556–5561 [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y (2002) Formation of the approximately 350-kDa Apg12–Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 277: 18619–18625 [DOI] [PubMed] [Google Scholar]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M (1998) Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J 17: 3597–3607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legesse-Miller A, Sagiv Y, Glozman R, Elazar Z (2000) Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J Biol Chem 275: 32966–32973 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuvirth H, Raz R, Schreiber G (2004) ProMate: a structure based prediction program to identify the location of protein–protein binding sites. J Mol Biol 338: 181–199 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Mizushima N (2004) Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol 15: 231–236 [DOI] [PubMed] [Google Scholar]
- Paz Y, Elazar Z, Fass D (2000) Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J Biol Chem 275: 25445–25450 [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ (2004a) Autophagy in health and disease: a double-edged sword. Science 306: 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ (2004b) Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 279: 29889–29894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub M, Bredschneider M, Thumm M (1997) AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol 179: 3875–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara K, Suzuki NN, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F (2004) The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 9: 611–618 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20: 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


