Abstract
Cytokinesis in Saccharomyces cerevisiae occurs by the concerted action of the actomyosin system and septum formation. Here we report on the roles of HOF1, BNI1, and BNR1 in cytokinesis, focusing on Hof1p. Deletion of HOF1 causes a temperature-sensitive defect in septum formation. A Hof1p ring forms on the mother side of the bud neck in G2/M, followed by the formation of a daughter-side ring. Around telophase, Hof1p is phosphorylated and the double rings merge into a single ring that contracts slightly and may colocalize with the actomyosin structure. Upon septum formation, Hof1p splits into two rings, disappearing upon cell separation. Hof1p localization is dependent on septins but not Myo1p. Synthetic lethality suggests that Bni1p and Myo1p belong to one functional pathway, whereas Hof1p and Bnr1p belong to another. These results suggest that Hof1p may function as an adapter linking the primary septum synthesis machinery to the actomyosin system. The formation of the actomyosin ring is not affected by bni1Δ, hof1Δ, or bnr1Δ. However, Myo1p contraction is affected by bni1Δ but not by hof1Δ or bnr1Δ. In bni1Δ cells that lack the actomyosin contraction, septum formation is often slow and asymmetric, suggesting that actomyosin contraction may provide directionality for efficient septum formation.
INTRODUCTION
Cytokinesis in animal cells is thought to occur through the contraction of the actomyosin system (Satterwhite and Pollard, 1992; Fishkind and Wang, 1995; Rappaport, 1996), followed by the sealing and cleavage of a narrow intercellular bridge with a central midbody (Mullins and Biesele, 1973, 1977; Mullins and MacIntosh, 1982; Sanger et al., 1985). The contractile apparatus, which includes nonmuscle myosin II and F-actin, is assembled around anaphase and is located at the midpoint of the cell, making cytokinesis a temporally and spatially regulated event (Satterwhite and Pollard, 1992; Fishkind and Wang, 1995; Rappaport, 1996).
A similar contractile system also exists in yeasts, including Schizosaccharomyces pombe (Marks and Hyams, 1985; McCollum et al., 1995; Bezanilla et al., 1997; Kitayama et al., 1997) and Saccharomyces cerevisiae (Bi et al., 1998; Lippincott and Li, 1998b). There are at least three major differences among the systems. The first is in the timing and mechanism of site selection. In S. cerevisiae, the site for cell division is determined at the beginning of the cell cycle (Pringle et al., 1995; Drubin and Nelson, 1996). In S. pombe, the division site is thought to be chosen around the onset of mitosis, with the position of the premitotic nucleus appearing to play a role (Chang and Nurse, 1996; Chang et al., 1996). In animal cells, F-actin and myosin II are concentrated at the presumptive cleavage site around anaphase. Their position is thought to be determined by the position of the spindle (Satterwhite and Pollard, 1992; Rappaport, 1996). The second difference between the systems is that, in S. pombe and S. cerevisiae, cytokinesis involves septum formation, whereas animal cells have no cell wall. Septum formation is coordinated temporally and spatially with the action of the actomyosin system. Time-lapse analysis in S. cerevisiae has shown that cell division normally occurs through the contraction of the actomyosin system, followed closely by the centripetal growth of the septum and subsequent cell separation (Bi et al., 1998). The third difference is that the actomyosin contractile ring is not essential for cytokinesis in S. cerevisiae (Rodriguez and Paterson, 1990; Bi et al., 1998; Lippincott and Li, 1998a), as it is in S. pombe and mammalian cells. This represents a unique advantage for S. cerevisiae because the effects of gene deletions on actomyosin contraction can be monitored. Analyzing the similarities and differences among these systems will allow an accelerated and more complete understanding of the molecular mechanisms of cytokinesis.
Because cytokinesis in S. cerevisiae can occur independently of actomyosin ring contraction, there must be at least one other mechanism by which it can take place. It is possible that in the absence of the actomyosin system, septum formation can drive cell division. Consistent with this possibility, deletion of the two major chitin synthases, encoded by CHS2 and CHS3, causes lethality, with cells arrested in chains, indicating a defect in cytokinesis or cell separation (Shaw et al., 1991). Both mechanisms of cytokinesis must depend on septins because mutations in the septin-encoding genes completely block cytokinesis (Hartwell, 1971; Pringle and Hartwell, 1981; Longtine et al., 1996). Consistent with this view, the localization of Myo1p depends on septins (Bi et al., 1998; Lippincott and Li, 1998b), as does the neck localization of Chs3p and proteins involved in its activation or spatial regulation (DeMarini et al., 1997).
Elucidation of the molecular mechanisms of cytokinesis requires the identification and characterization of proteins involved in the actomyosin system, in septum formation, and in coordinating these two processes. Analysis of conditional mutants and subsequent molecular studies have identified proteins in S. pombe involved in the formation and/or contraction of the medial actin ring and in the formation of the septum (Gould and Simanis, 1997). Several of these proteins are conserved (Gould and Simanis, 1997). For example, cdc12p is a member of the formin family that includes the Drosophila protein Diaphanous and the S. cerevisiae proteins Bni1p and Bnr1p. All are involved in cytokinesis and other actin-dependent processes (Castrillon and Wasserman, 1994; Kohno et al., 1996; Chang et al., 1997; Evangelista et al., 1997; Fujiwara et al., 1998; Heil-Chapdelaine et al., 1999; Lee et al., 1999; Miller et al., 1999). The S. pombe cdc15p and Imp2p proteins are members of a family characterized by an N-terminal coiled-coil domain, followed by a PEST sequence and a C-terminal SH3 domain. Both proteins are involved in cytokinesis (Fankhauser et al., 1995; Demeter and Sazer, 1998). For instance, conditional mutations in cdc15 block septation, possibly by preventing the recruitment of actin patches to the division site (Nurse et al., 1976; Balasubramanian et al., 1998). A mammalian cdc15p family member, PSTPIP, is associated with the cleavage furrow (Spencer et al., 1997), although its role in cytokinesis remains to be determined.
In S. cerevisiae, there are two cdc15-related genes: HOF1 (Homologue Of Fifteen, also named CYK2) and HOF2. The role of Hof2p in cytokinesis is unclear (Chant, personal communication). We hope to elucidate the mechanisms for cytokinesis in S. cerevisiae and decided to characterize HOF1 because, based on the activities of the other cdc15-related proteins, it seemed quite likely that Hof1p might function in this process. If so, we wished to determine whether it was involved in the regulation of the actomyosin system or in septum formation or in the coordination of the two. This issue is not clearly addressed even for the cdc15 family members in S. pombe, which are the most well studied. Finally, we wanted to understand the relationship between HOF1 and other known genes involved in cytokinesis in S. cerevisiae. While our work was in progress, two papers on Hof1p were published (Kamei et al., 1998; Lippincott and Li, 1998a). Hof1p was hypothesized to regulate actomyosin ring dynamics and septin distribution (Lippincott and Li, 1998a). However, our results conflict with this and instead suggest that Myo1p and Bni1p are involved in actomyosin contraction, whereas Hof1p and Bnr1p may be involved in coupling septum formation to the actomyosin system. In addition, evidence presented in this report suggests that actomyosin contraction in S. cerevisiae may provide a guiding cue for efficient septum formation.
MATERIALS AND METHODS
Strains, Growth Conditions, and Genetic Methods
Yeast strains are listed in Table 1. Standard media and genetic methods were used (Guthrie and Fink, 1991). Where noted, YM-P, a buffered rich medium (Lillie and Pringle, 1980), was used for yeast growth in liquid culture. All yeast strains were grown at 23°C, unless indicated otherwise. In some cases, 1 mg/ml 5-FOA (Toronto Research Chemicals, North York, ON, Canada) was added to select for the loss of URA3-containing plasmids (Sikorski and Boeke, 1991). Escherichia coli strain DH12S (Life Technologies, Gaithersburg, MD) was used as a plasmid host.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| YEF473 | a/α his3/his3 leu2/leu2 lys2/lys2 trp1/trp1 ura3/ura3 | Bi and Pringle, 1996 |
| YEF473A | a his3 leu2 lys2 trp1 ura3 | Bi and Pringle, 1996 |
| YEF1929 | As YEF473 except HOF1/hof1Δ::Kan | Text |
| YEF1951 | a his3 leu2 lys2 trp1 ura3 hof1Δ::Kan | YEF1929 |
| YEF2082 | As YEF473 except hof1Δ::Kan/hof1Δ::Kan | This studya |
| YEF1976 | As YEF473 except HOF1/HOF1::HA::HIS3 | Text |
| YEF2016 | a his3 leu2 lys2 trp1 ura3 HOF1:HA::HIS3 | YEF1976 |
| YEF2019 | α his3 leu2 lys2 trp1 ura3 HOF1:HA::HIS3 | YEF1976 |
| YEF2062 | a his3 leu2 lys2 trp1 ura3 HOF1:HA::Kan | Text |
| YEF2063 | α his3 leu2 lys2 trp1 ura3 HOF1:HA::Kan | Text |
| 4078-14-3a | a can1 leu2 his7 ura3 cdc14 | L. Hartwell |
| YEF2100-2B | a his3 leu2 lys2 trp1 ura3 cdc14 HOF1:HA::Kan | YEF2063 × 4078-14-3a |
| J230-2D | a ade1 leu2 trp1 ura3 dbf2-1 dbf20Δ::TRP1 | L. Johnston |
| YEF2098-11C | a his3 leu2 lys2 trp1 ura3 dbf2-1 dbf20Δ::TRP1 HOF1:HA::Kan | YEF2063 × J230-2D |
| DLY1872 | α ade1 his2 leu2 trp1 ura3 cdc15-2 | D. Lew |
| YEF2102-10A | a ade1 his3 leu2 lys2 trp1 ura3 cdc15-2 HOF1:HA::Kan | YEF2062 × DLY1872 |
| YEF1930 | As YEF473 except HOF1/HOF1:GFP::Kan | Text |
| YEF1986 | As YEF473 except HOF1:GFP::Kan/HOF1:GFP::Kan | Cross of segregants of YEF1930 |
| YJZ426 | a his3 leu2 lys2 trp1 ura3 bnilΔ::HIS3 | J. Pringle |
| HH798 | As YEF473 except bnilΔ::HIS3/bnilΔ::HIS3 | J. Pringle |
| YEF1689 | As YEF473 except BNR1/bnr1Δ::HIS3 | Text |
| YEF1730 | a his3 leu2 lys2 trp1 ura3 bnr1Δ::HIS3 | YEF1689 |
| YEF1732 | α his3 leu2 lys2 trp1 ura3 bnr1Δ::HIS3 | YEF1689 |
| YEF1822 | As YEF473 except bnr1Δ::HIS3/bnr1Δ::HIS3 | YEF1730 × YEF1732 |
| YEF1681 | a his3 leu2 lys2 trp1 ura3 MYO1:GFP::Kan | Bi et al., 1998 |
| YEF1682 | α his3 leu2 lys2 trp1 ura3 MYO1:GFP::Kan | Bi et al., 1998 |
| YEF1698 | As YEF473 except MYO1:GFP::Kan/MYO1:GFP::Kan | Bi et al., 1998 |
| YEF1854 | As YEF473 except bnr1Δ::HIS3/bnr1Δ::HIS3 MYO1:GFP::Kan/MYO1:GFP::Kan | Cross of segregants of YEF1732 × YEF1681 |
| YEF1839 | As YEF473 except bnilΔ::HIS3/bnilΔ::HIS3 MYO1:GFP::Kan/MYO1:GFP::Kan | This studyb |
| YEF2124 | As YEF473 except hof1Δ::Kan/hof1Δ::Kan MYO1:GFP::Kan/MYO1:GFP::Kan | This studyc |
| YEF2026 | As YEF473 except HOF1:HA::HIS3/HOF1:HA::HIS3 | Cross of segregants of YEF1976 |
| M-238 | α his3 leu2 trp1 ura3 cdc12-6 | J. Pringle |
| YEF2086 | As YEF473 except HOF1:HA::HIS3/HOF1:HA::HIS3 cdc12-6/cdc12-6 | YEF2016 × M-238 |
| YEF1751 | As YEF473 except MYO1/myo1Δ::HIS3 | Bi et al., 1998 |
| YEF2125 | As YEF473 except myo1Δ::HIS3/myo1Δ::HIS3 HOF1:HA::HIS3/HOF1:HA::HIS3 | This studyd |
| YEF2126 | As YEF473 except bnilΔ::HIS3/bnilΔ::HIS3 HOF1:HA::HIS3/HOF1:HA::HIS3 | This studye |
| YEF2095 | α his3 leu2 lys2 trp1 ura3 bnr1Δ::HIS3 HOF1:HA::HIS3 | YEF1730 × YEF2019 |
Plasmids pRS315-HOF1 and pRS316-HOF1 were transformed into YEF1929. Transformants were sporulated to generate hof1Δ::Kan strains. Two that carried different plasmids were mated, and the plasmids were lost by continued growth under nonselective conditions to generate strain YEF2082.
YJZ426 and YEF1682 were crossed and sporulated to generate bnilΔ::HIS3 MYO1:GFP::Kan strains of opposite mating type. These were transformed with YEp13 or YEp352-BNI1 and mated. The diploid was grown nonselectively to lose both plasmids, resulting in YEF1839.
A MATα hof1Δ::Kan segregant containing plasmid pRS316-HOF1 from YEF1929 (pRS316-HOF1) was crossed to YEF1681. After plasmid loss, the diploid was sporulated to generate hof1Δ::Kan MYO1:GFP::Kan. Two segregants were then transformed with YEplac181 or pRS316 and then crossed. Loss of both plasmids generated YEF2124.
YEF1751 was transformed with YCp50-MYO1 and sporulated to generate a MATa myo1a::HIS3 strain carrying YCp50-MYO1. This strain was crossed to YEF2019, followed by plasmid loss under nonselective growth. The resulting diploid was sporulated to generate myo1Δ::HIS3 HOF1:HA::HIS3 segregants of opposite mating type. One was transformed with plasmid pRS316 and one with YEplac181, and they were then crossed to form a diploid. Loss of both plasmids generated YEF2125.
A strain from the cross of YJZ426 and YEF2019 was sporulated to generate two bnilΔ::HIS3 HOF1:HA::HIS3 segregants of opposite mating types. These were then transformed with YEplac181 or pRS316 and crossed. After losing both plasmids, YEF2126 was obtained.
Construction of Plasmids and Yeast Strains Carrying Deletions or Tagged Genes
Oligonucleotide primers (Table 2) were purchased from Integrated DNA Technologies (Coraville, IA). pRS315-HOF1 and pRS316-HOF1 were constructed by cloning an ∼2.7-kilobase BamHI-SalI PCR-amplified fragment into pRS315 (CEN LEU2) and pRS316 (CEN URA3) (Sikorski and Hieter, 1989). Chromosomal DNA from YEF473 was the template. Primer HOF1-forward-B1 corresponds to 446–422 nucleotides upstream of the HOF1 start codon, and primer HOF1-reverse-S1 (the SalI site was introduced) corresponds to 198–224 nucleotides downstream of the HOF1 stop codon. The PCR reaction was carried out with the use of Expand (Boehringer Mannheim, Indianapolis, IN). YEp13 (2μ, LEU2) (Rose and Broach, 1990), YEplac181 (2μ, LEU2) (Gietz and Sugino, 1988), YEp352 (2μ, URA3) (Hill et al., 1986), YEp352-BNI1 (carrying the full-length BNI1), and YCp50-MYO1 (CEN URA3; carrying the full-length MYO1; kindly supplied by Dr. Susan Brown, University of Michigan, Ann Arbor) were also used.
Table 2.
Primers used in this study
| Name | Sequence |
|---|---|
| PCR-mediated deletionsa | |
| HOF1-forward | TTGGAAAGTGTACTACTAATATTCAGAAAAAGGTGAAAGACGGATCCCCGGGTTAATTAA |
| HOF1-reverse | TCTTTTATCAGAAAACTAGTAAAATTGATATACATCGAGAGAATTCGAGCTCGTTTAAAC |
| BNR1-forward | GTGACACAAAAGCAGATAAAAAAATAGCACAATCATCAGCGGATTGTACTGAGAGTGCACC |
| BNR1-reverse | TTATATAAGCTCCACAACTACATAAAATACTAAGTCTTCACTGTGCGGTATTTCACACCG |
| Tagging with HA or GFPb | |
| HOF1-forward-2 | AATTCCCTATAATTTCATTCAGCTACTGCATCAAGGTCTTCGGATCCCCGGGTTAATTAA |
| Cloning of HOF1c | |
| HOF1-forward-B1 | GAACCTTTAGGATCCACTGAATTA |
| HOF1-reverse-S1 | CCTAACGTCGACAATTATCGTGATTATTGTGTC |
Underlined sequences are immediately upstream or downstream of the ORF to be deleted.
The underlined sequence is immediately upstream of the stop codon of HOF1.
BamHI and SalI sites are underlined.
A complete BNR1 deletion was constructed (Baudin et al., 1993). A pair of hybrid primers, BNR1-forward and BNR1-reverse, was used for PCR. The 5′ ends of the primers correspond to sequences immediately upstream or downstream of BNR1 ORF, whereas the 3′ ends of primers correspond to sequences flanking the HIS3 gene in pRS303 (Sikorski and Hieter, 1989). The amplified fragment was transformed into YEF473, and stable His+ colonies were selected to generate YEF1689. A deletion of HOF1 was constructed by following protocols described by Longtine et al. (1998) and Wach et al. (1997), except the primers were HOF1-forward and HOF1-reverse and the template was pFA6a-kanMX6 (Wach et al., 1997; Longtine et al., 1998). The amplified fragment was transformed into YEF473, and cells were plated onto YPD plates at 30°C. After 2 d, cells were replica plated onto YPD plates containing 200 μg/ml G418 (Life Technologies) twice to select stable Kanr colonies, generating YEF1929.
To tag HOF1 at its 3′ end with an in-frame HA or GFP epitope, primer HOF1-forward-2, consisting of sequences immediately upstream of the HOF1 stop codon at the 5′ end and the vector sequences at the 3′end, and the same reverse primer used for hof1 deletion were used to amplify the HA tag from plasmid pFA6a-3HA-HisMX6 or the GFP tag from plasmid pFA6a-GFP(S65T)-kanMX6 (Wach et al., 1997; Longtine et al., 1998). The amplified fragments were transformed into YEF473, and stable His+ or Kanr colonies were selected, resulting in YEF1976 and YEF1930. To construct strains carrying HOF1-HA marked with the Kanr gene, the same primers were used to amplify a fragment from pFA6a-3HA-kanMX6 (Wach et al., 1997; Longtine et al., 1998). The fragment was transformed into YEF2016 (a HOF1:HA::HIS3) and YEF2019 (α HOF1:HA::HIS3), and stable Kanr colonies were selected and screened for His− growth, resulting in YEF2062 (a HOF1:HA::Kan) and YEF2063 (α HOF1:HA::Kan). Deletions and tagging were verified by PCR. In each case, at least two independent transformants were dissected to ensure that segregants with the appropriate modifications had the same phenotypes.
Morphological Observations
All cells were fixed by adding formaldehyde directly into the medium to a final concentration of 3.7%. Cell morphologies were observed by differential interference contrast (DIC) microscopy. Chitin was visualized by staining with calcofluor (Sigma Chemical, St. Louis, MO). Indirect immunofluorescence was carried out by following protocols described by Pringle et al. (1991). Actin ring and actin patches were visualized by staining with rhodamine-conjugated phalloidin (Molecular Probes, Eugene, OR) at 2 U/ml for actin ring and 20 U/ml for actin patches for 5–30 min at 23°C. DNA was stained with 1 μg/ml bis-benzimide (Sigma Chemical). Mouse monoclonal anti-HA epitope (HA.11) (Berkeley Antibody, Richmond, CA) was used for immunofluorescence at 1:100. Monoclonal rat anti-tubulin antibody (YOL1/34; Accurate Chemical and Scientific, Westbury, NY) was used at 1:100 (Kilmartin et al., 1982; Kilmartin and Adams, 1984). Rabbit polyclonal antibodies against Cdc11p were used to visualize septins (Ford and Pringle, 1991). The secondary antibodies were Cy3-conjugated donkey anti-mouse immunoglobulin G (IgG) (used at 1:400), Cy2-conjugated donkey anti-rat IgG (used at 1:400), FITC-conjugated goat anti-rat IgG, and rhodamine-conjugated goat anti-rabbit IgG. All secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
Time-lapse Microscopy
To visualize HOF1-GFP or MYO1-GFP, exponentially growing cells in synthetic complete (SC) medium were spotted onto a thin layer of SC medium containing 25% gelatin (Yeh et al., 1995). A computer-controlled microscope (Eclipse E800, Nikon, Tokyo, Japan) with motorized focus and a cooled, high-resolution charge-coupled device camera (model C4742–95, Hammamatsu Photogenics, Bridgewater, NJ) were used. DIC images (exposure time, 0.6 s) and corresponding GFP images (exposure time, 2 s) were acquired and analyzed by a modified version of Image-Pro Plus imaging software (Media Cybernetics, Silver Spring, MD; now called Phase 3 Imaging Systems, Glen, PA). The diameter of the Hof1p-GFP or the Myo1p-GFP ring was determined by measuring the distance between the bright ends of the band (a side view of a ring). The contrast of the images was enhanced with the use of Phase 3 systems and/or Adobe PhotoShop version 4.0 (Adobe Systems, San Jose, CA) systems. GFP images in Figure 6A were processed with a sharpening filter.
Figure 6.
(A) Merging of Hof1p-GFP from two rings into a single ring. Cells of strain YEF1986 (a/α HOF1:GFP/HOF1:GFP) were observed by time-lapse photomicroscopy. DIC (top image, left panel) and GFP images were recorded at the indicated times (min). Bar, 1 μm. (B) Contraction of Hof1p-GFP ring relative to septum formation and cell separation. Cells of strain YEF1986 were observed by time-lapse photomicroscopy. DIC (left) and GFP (right) images were recorded at the indicated times (min). Bar, 1 μm.
Synchronization Experiments
Haploid MATa cells containing Hof1p-HA (YEF2016 or YEF2018) were grown to an OD600 of 0.25 in YM-P. Cells were incubated with α-factor (4 μg/ml) for 90 min and then washed. Cells were resuspended in YM-P, and samples were removed every 12 min. Cultures were diluted during the time course to keep the cell number constant. For analysis of cdc14, cdc15, and dbf2ts dbf20Δ mutants, strains YEF2100-2B, YEF2102-10A, YEF2098-11C, and the wild-type strain YEF2016 were grown to log phase at 23°C. Half of each culture was added to an equal volume of YM-P prewarmed to 37°C. Cultures were incubated at 37°C for 3.5 h, by which time >90% of the mutant cells had large buds or projections.
For Western blotting, ∼7 OD600 units of cells were harvested by centrifugation. Cells were resuspended in cold 1× TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and transferred to a microfuge tube. Residual cells were fixed with 3.7% formaldehyde in 1× PBS, sonicated, and used to assay budding. Cell pellets were resuspended in 240 μl of 1.85 N NaOH, 1.06 M β-mercaptoethanol and incubated on ice for 10 min. An equal volume of 50% trichloroacetic acid was added, and samples were incubated on ice for an additional 10 min. Samples were microfuged for 10 min, and the resulting pellets were washed with ice-cold acetone and then respun. The acetone was aspirated, and pellets were resuspended in 100 μl of 0.5 M Tris, 5% SDS and sonicated. An equal volume of 75% glycerol, 0.12 M DTT, 0.05% bromophenol blue was added, and samples were heated for 10 min at 95°C before loading 10 μl onto an SDS-PAGE gel.
Proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P, Millipore, Bedford, MA) and detected with the use of anti-HA antibody at 1:1000 and peroxidase-conjugated goat anti-mouse antibody (Boehringer Mannheim) at 1:5000. Bound antibodies were detected with the use of ECL (Amersham Life Science, Buckinghamshire, UK).
For immunoprecipitation experiments, cells were removed 72 and 84 min after release from α-factor block. Cells were harvested by centrifugation and lysed under denaturing conditions in the presence of 1 mM PMSF, 1 μM pepstatin, 80 μM leupeptin, 1 mM NaVO4, 10 mM NaF, and 25 mM β-glycerophosphate (Kolodziej and Young, 1991). A fraction was saved as a total protein sample. Hof1p-HA protein was immunoprecipitated from the remainder with the use of 1 μl of HA.11 antibody per sample (Kolodziej and Young, 1991) and 30 μl of protein G–agarose (Life Technologies). After the immunoprecipitation, samples were washed three times with LSHNN (50 mM NaCl, 10 mM HEPES, pH 7.5, 10% glycerol, 0.1% NP-40) and once with phosphatase buffer (10 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM DTT). SDS sample buffer was then added to one-half of each sample. Phosphatase buffer with protease inhibitors (1 mM PMSF, 1 μM pepstatin, and 80 μM leupeptin) was added to the other half, and 1 U of calf intestinal alkaline phosphatase was added. Samples were incubated at 37°C for 30 min, at which time another 1 U of calf intestinal alkaline phosphatase was added, and incubation continued for 30 min. Samples were washed twice with LSHNN and once with LSHNN containing 250 mM NaCl. SDS sample buffer was added, and samples were heated for 30 min at 65°C and then for 10 min at 95°C before loading onto an SDS-PAGE gel. Proteins were transferred to a polyvinylidene difluoride membrane and detected as described above.
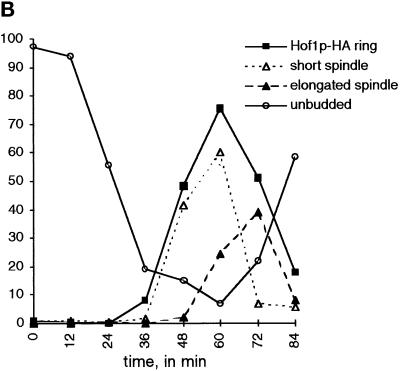
Between 200 and 400 cells were counted for each time point. Hof1p-HA ring staining refers to cells with a detectable ring or rings of HA-tagged protein; short spindles were bipolar spindles that may have been partially elongated, and elongated spindles stretched between the mother and daughter cells and were associated with separated nuclei, as determined by bis-benzimide staining.
RESULTS
Deletion of HOF1 Causes Temperature Sensitivity and a Defect in Cytokinesis
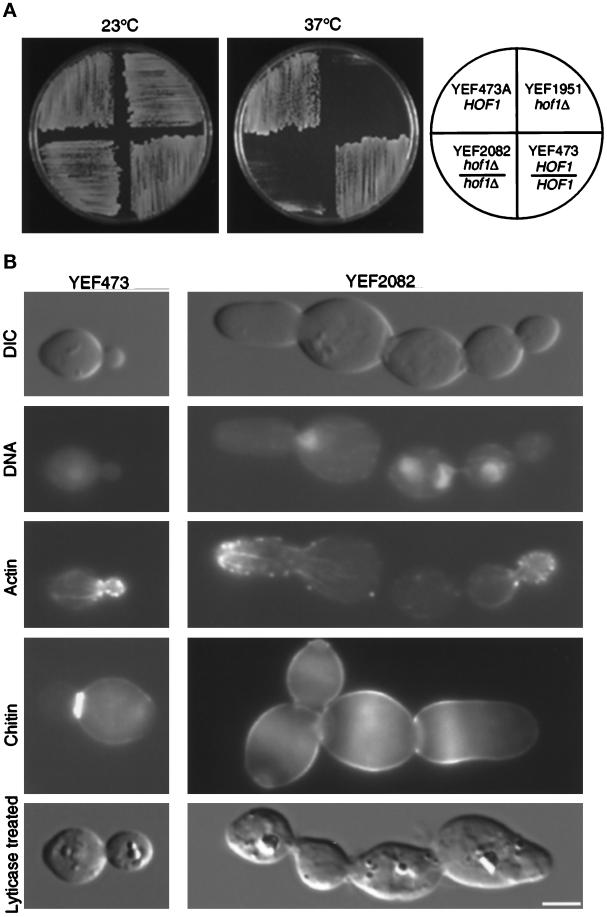
To determine whether HOF1 was involved in cytokinesis, we constructed and analyzed haploid and diploid strains deleted for HOF1. Both strains showed a temperature-sensitive growth phenotype (Figure 1A). Their growth rates in YM-P liquid medium at 23°C were indistinguishable from those of the corresponding wild-type strains, as measured by the increase in optical density. At 37°C, for the first 4 h, the growth rates of the hof1 deletion strains remained the same as those of their wild-type counterparts, and then they gradually decreased (data not shown). When shifted to 37°C for 5 h, the deletion strain contained 51% chained cells (n = 540; defined as three or more cell bodies connected together after mild sonication) (see Figure 1B), whereas the wild-type strain contained only 0.7% chained cells (n = 602). The deletion strain showed very little defect at 23°C (1.5% chained cells; n = 686). To determine whether the formation of cell chains was due to a defect in cytokinesis or cell separation, we treated fixed mutant cells with lytic enzyme to remove the cell wall (Pringle and Mor, 1975). The majority remained in chains, suggesting that deletion of HOF1 causes a cytokinesis defect at 37°C.
Figure 1.
(A) Deletion of HOF1 causes temperature-sensitive growth. Strains were streaked onto YPD plates and incubated for 2 d. (B) Deletion of HOF1 causes a cytokinesis defect. YEF473 and YEF2082 cells were shifted to 37°C for 4.5 h. Cells were fixed and stained for DNA, F-actin, and chitin. An aliquot was treated with lyticase to distinguish cytokinesis defects from cell separation defects (bottom row). The first three rows represent the same cell from strain YEF473 or strain YEF2082. Bar, 3 μm.
In addition to the cytokinesis defect, multinucleated cells were often observed with deletion strains. At 23°C, the mutant strain contained 10% multinucleated cells (n = 202). After shift to 37°C for 5 h, this proportion increased to 44% (n = 200). No wild-type cells (n = 203) were multinucleated at either temperature. To determine whether the formation of multinucleated cells was due to a defect in cell polarity, we monitored actin organization and chitin deposition. The actin cytoskeleton appeared polarized in a normal manner in hof1 cells (Figure 1B). In some cases, even large, multinucleated cells still had well-polarized actin cytoskeletons (data not shown). These results suggest that hof1 deletion does not alter gross cell polarity. Consistent with this finding, a majority of hof1 mutant cells were ovoid in shape, like the wild-type cells (Figure 1B). However, chitin was delocalized in hof1 mutant cells at 37°C (Figure 1B) but not at 23°C (data not shown). This suggests that at the nonpermissive temperature, either there is a mild polarity defect that is not manifest by the gross actin polarity or Hof1p is involved in restricting chitin to discrete locations.
Because multinucleate cells could arise from misoriented spindles (Li et al., 1993), we analyzed DNA and tubulin distribution in the wild-type strain and a deletion strain at 23 and 37°C. All cells, including those of the mutant at 37°C (n = 23), with DNA near the mother bud neck showed astral microtubules pointing to the bud, suggesting that spindle orientation was not affected by the hof1 deletion. Regardless of how multinucleation occurs, it is unlikely to be the cause of the cytokinesis defect, because chained cells with a single DNA mass in each cellular compartment were observed (data not shown).
Hof1p Localizes to the Bud Neck
All known cytokinesis proteins in S. cerevisiae, such as the septins (Longtine et al., 1996), Myo1p (Bi et al., 1998; Lippincott and Li, 1998b), and Iqg1p/Cyk1p (Epp and Chant, 1997; Lippincott and Li, 1998b), are localized to the bud neck during part or most of the cell cycle. To determine whether Hof1p functions during cytokinesis at the bud neck, we analyzed the localization of Hof1p after tagging the 3′ end of the chromosomal locus with HA or GFP. Neither construct caused temperature-sensitive growth or defects in cytokinesis, demonstrating that the fusion proteins were functional. We used two different tags for the following reasons. First, we wished to determine whether the patterns revealed by the HA- and GFP-tagged proteins were identical, which would eliminate any tag-specific effect on the localization. Second, each construct has unique advantages. The HOF1-HA construct can be used most easily for biochemical experiments, because there is a highly specific anti-HA antibody available commercially. Use of this antibody also allows correlation of protein levels and modifications with immunofluorescent images in cell synchrony experiments. The HOF1-GFP construct can be used for live-cell analysis. Finally, different reporters sometimes yield different strengths of signal for documentation.
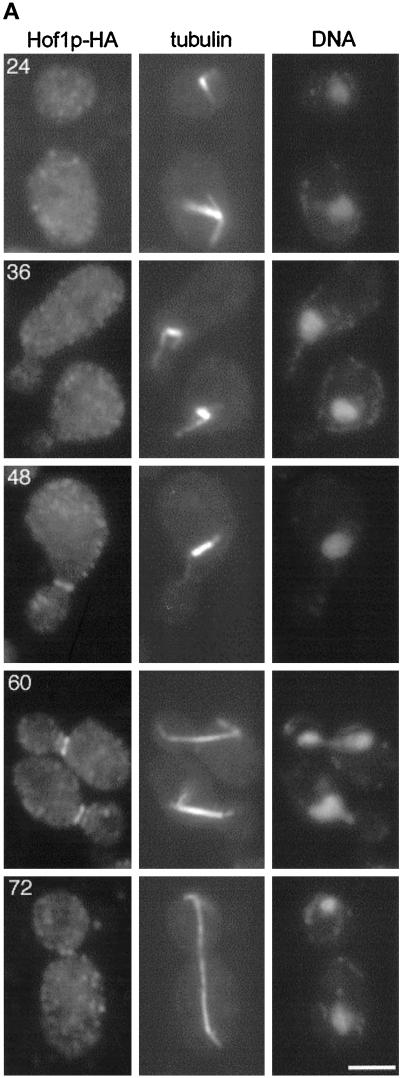
Analysis of the Hof1p-HA and Hof1p-GFP fusions showed that the proteins localized to the bud neck, although under the experimental conditions, the HA fusion consistently yielded a stronger signal than the GFP fusion (Figures 2 and 6). Double-labeling experiments with the use of anti-tubulin and anti-HA antibodies demonstrated that 92% of cells with short bipolar spindles had no detectable Hof1p-HA (n = 50). In contrast, 88% of the cells with partially elongated spindles (n = 50) and 98% of the cells with fully elongated or disassembled spindles (n = 100) had Hof1p-HA at the bud neck. We did not detect staining at the bud neck in untagged control strains at any cell cycle stage (data not shown).
Figure 2.
Epitope-tagged Hof1p localizes to the bud neck after bud emergence and formation of a bipolar spindle. (A) Hof1p-HA strain YEF2016 was synchronized with α-factor and then released from the block. Samples were fixed at 12-min intervals and stained for Hof1p-HA (left panels), tubulin (middle panels), and DNA (right panels). The numbers correspond to the minutes after release from arrest. Bar, 3 μm. Based on spindle morphology, it is likely that the cells at 24 min are in G1, the cells at 36 min are in S phase, the cell at 48 min is in G2, the cells at 60 min are in anaphase, and the cell at 72 min is in telophase. (B) Quantitation of cell staining and distribution after release from α-factor arrest. Short spindles are bipolar spindles, including those that may be partially elongated; elongated spindles stretch between the mother and daughter and are associated with divided chromatin.
To determine more precisely the timing of Hof1p localization, we examined the staining pattern of HA-tagged Hof1p in synchronously dividing cells. Cells were released from an α-factor block. There was no detectable Hof1p-HA staining in pheromone-arrested cells or in cells up to 24 min after release, although ∼50% of the cells had budded by this time (Figure 2). The number of cells with Hof1p-HA rings at the bud neck increased at later time points, peaking at 60 min after release, coincident with the peak of cells with short or partially elongated spindles. As cells completed cytokinesis and cell separation, the number of cells with Hof1p ring staining decreased, as shown by the increase in unbudded cells.
We observed cells with both single and double rings of Hof1p staining (Figure 2A). From the synchronization experiment, we determined that a single ring of Hof1p-HA staining in mother cells appeared first (Table 3) and peaked ∼48 min after release. Later, two rings of staining, one in the mother and one in the daughter, were evident; the staining in daughter cells was less bright at first and then became equal. Finally, late in the cell cycle, probably just before cytokinesis, we observed cells with only a single small ring located between the mother and daughter.
Table 3.
Hof1p localization in synchronously dividing cells
| Time (min) | No ring | M only | M>D | M=D | Single small ring |
|---|---|---|---|---|---|
| 0 | 99 | 1 | 0 | 0 | 0 |
| 12 | 99 | 0 | 1 | 0 | 0 |
| 24 | 100 | 0 | 0 | 0 | 0 |
| 36 | 92 | 7 | 1 | 1 | 0 |
| 48 | 51 | 34 | 13 | 2 | 0 |
| 60 | 24 | 20 | 54 | 2 | 0 |
| 72 | 49 | 14 | 16 | 9 | 12 |
| 84 | 82 | 7 | 7 | 1 | 2 |
The numbers are percentages of cells with a given staining pattern, and the time is in minutes after release from α-factor block. Between 200 and 400 cells were counted at each time. M and D refer to mother and daughter cells. “M only” refers to cells with a single ring of staining localized to the mother cell; “M>D” refers to cells with two rings of staining in which the intensity of staining in the mother cell is greater than that in the daughter cell; “M=D” refers to cells with two rings of staining in which the intensities appeared equal; “single small ring” refers to cells having a single small ring of staining localized between the mother and daughter. We never observed a ring in only a daughter or a ring in a daughter that was more intense than that in the mother.
Hof1p Localization Distinguishes among the Arrest of the Telophase Mutants cdc14, cdc15, and dbf2/dbf20
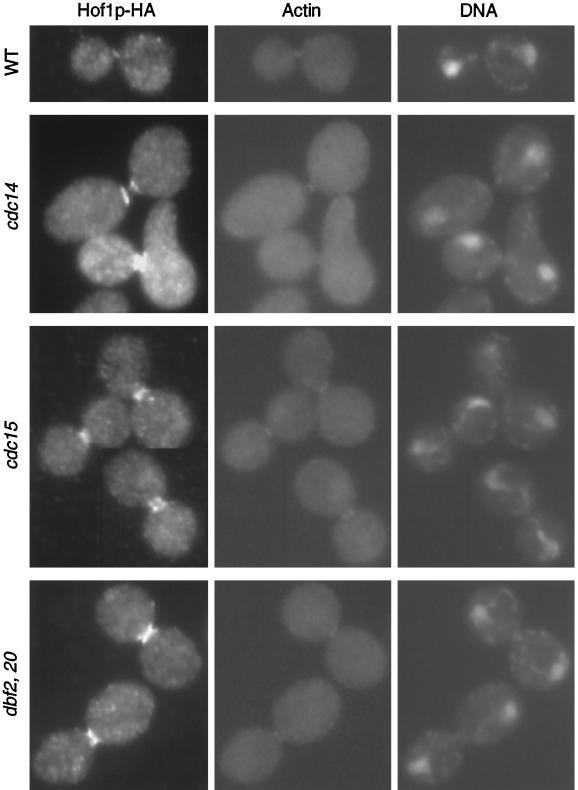
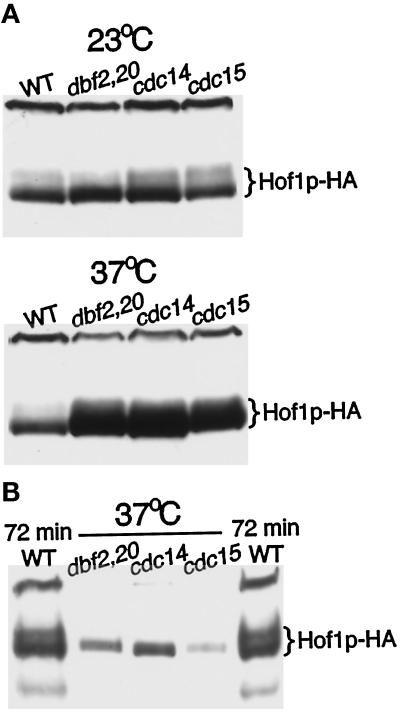
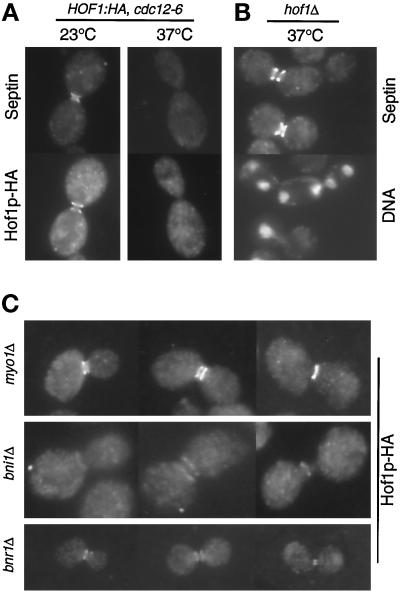
To correlate the changes in Hof1p localization more precisely with other cell cycle events, we analyzed its localization in mutants arrested at anaphase/telophase. Strains carrying HOF1-HA and cdc14, cdc15, or dbf2 dbf20 were grown at 37°C for 3 h. All of the mutant strains arrested predominantly with a typical anaphase spindle and an actin ring at the bud neck (Figure 3). Consistent with the time course, Hof1p was also present at the bud neck in all three mutant strains. However, the precise localization pattern differed among the three mutants (Figure 3). In the wild-type strain, 96% of the actin ring–containing cells had a Hof1p single ring that appeared to colocalize with the actin ring (n = 25). In contrast, in cdc14 cells, 100% of the actin ring–forming cells had Hof1p double rings of equal intensity (n = 51). In dbf2 dbf20 cells, 90% of the actin ring–containing cells showed what appears to be an intermediate-state localization pattern between the double rings and the single ring (n = 51; this pattern is referred to as intermediate ring). Hof1p localization in cdc15 cells was between that observed in cdc14 and dbf2 dbf20 cells. Among 50 actin ring–forming cells, 58% showed Hof1p double rings and 42% showed an intermediate ring–like structure.
Figure 3.
Localization of Hof1p in mutants arrested at telophase. Exponentially growing wild-type (YEF2016), cdc14 (YEF2100-2B), cdc15 (YEF2102-10A), and dbf2 dbf20 (YEF2098-11C) cells that contained HOF1-HA were shifted to 37°C for 3 h. Cells were fixed and stained for Hof1p-HA (left panels), F-actin (middle panels), and DNA (right panels). All cells are shown at the same magnification.
The Abundance and Phosphorylation State of Hof1p Is Cell Cycle Regulated
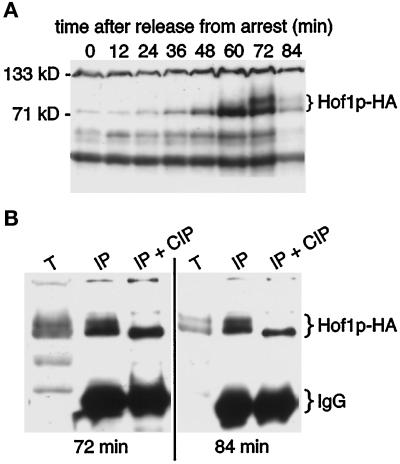
The experiments described above demonstrate that bud neck localization of Hof1p-HA is cell cycle dependent. HOF1 transcript levels fluctuate in the cell cycle, peaking during M phase (Spellman et al., 1998). To determine whether Hof1p levels were cell cycle regulated and whether this might explain the protein localization pattern, we examined HA-tagged Hof1p in synchronously dividing cells by Western blotting. Levels of Hof1p peaked ∼60 min after release from pheromone, just before or during anaphase (Figure 4A), coincident with the peak of Hof1p-HA neck ring staining (Figure 2B). In contrast, levels of an unrelated HA-tagged construct did not fluctuate during the cell cycle, nor did any background bands that were similar in size to Hof1p-HA (data not shown), demonstrating that the observed pattern is due specifically to Hof1p-HA.
Figure 4.
Hof1p is cell cycle regulated and phosphorylated. (A) Regulation of Hof1p levels. Strain YEF2016 (HOF1-HA) was synchronized with α-factor and then released from the block. Samples were removed at 12-min intervals. (B) Hof1p-HA mobility changes upon treatment with phosphatase. Protein was isolated from strain YEF2016 at 72 and 84 min after release from α-factor arrest. T, total protein; IP, immunoprecipitated Hof1p-HA protein; IP + CIP, immunoprecipitated Hof1p-HA protein treated with calf intestinal alkaline phosphatase.
A more slowly migrating form of Hof1p-HA was observed beginning at 72 min after pheromone release, during either the exit from mitosis or cytokinesis/cell separation. Again, a change in migration was not observed with an unrelated HA-tagged construct or background bands (data not shown). To determine whether the change in Hof1p-HA migration was due to a change in phosphorylation, we treated Hof1p-HA immunoprecipitates with calf intestinal phosphatase followed by Western blotting. Phosphatase treatment resulted in the formation of a single band, suggesting that the change in mobility was due to hyperphosphorylation of Hof1p (Figure 4B). The protein observed after phosphatase treatment appears to have a slightly faster mobility than that seen in cycling cells. It may be that Hof1p-HA is phosphorylated at one or a few sites at all times and becomes hyperphosphorylated late in the cell cycle.
To define further the timing of Hof1p-HA accumulation and modification, we analyzed Hof1p-HA from cells arrested during anaphase/telophase by temperature-sensitive mutations in cdc14, cdc15, and dbf2 dbf20 (Pringle and Hartwell, 1981; Toyn and Johnston, 1994). Levels of Hof1p were high in all three mutants at the nonpermissive temperature (Figure 5A), demonstrating that the protein accumulated at this stage of the cell cycle. The protein that accumulated was not hyperphosphorylated (Figure 5B), suggesting that hyperphosphorylation occurs after late anaphase/telophase, most likely during cytokinesis or cell separation.
Figure 5.
Increased levels of Hof1p-HA accumulate in a nonhyperphosphorylated form in cells arrested at telophase. (A) Exponentially growing wild-type (YEF2016), cdc14 (YEF2100-2B), cdc15 (YEF2102-10A), and dbf2 dbf20 (YEF2098-11C) cells containing Hof1p-HA were shifted to 37°C for 3.5 h. Protein was isolated from an equal number of cells and analyzed by Western blotting. (B) Protein isolated from mutant cells at 37°C was diluted 15-fold before analysis by SDS-PAGE and Western blotting. Samples flanking the mutant lysates were isolated from wild-type cells containing Hof1p-HA 72 min after release from an α-factor block.
Together, the data suggest that hyperphosphorylation of Hof1p is required either for its complete transition to a single ring structure or for its loss from the bud neck. Mutations in cdc14, cdc15, and dbf2/dbf20 may arrest cells at distinct points of the cell cycle that are before this event; alternatively, these proteins may function sequentially to modify Hof1p.
Hof1p Ring Does Not Contract Like the Actomyosin System
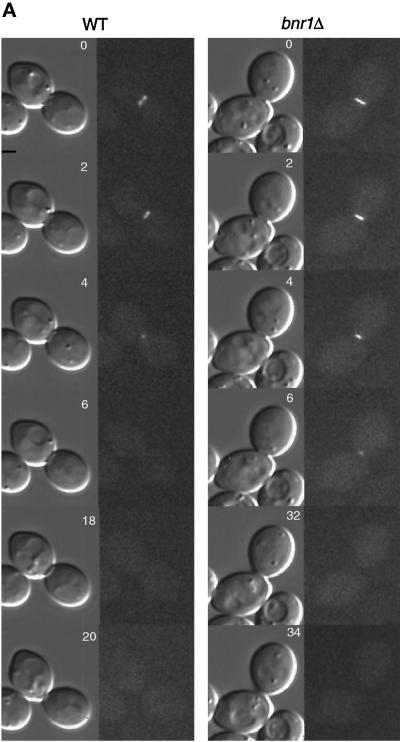
Because Hof1p is involved in cytokinesis and appears to colocalize with the actin ring at the bud neck late in the cell cycle, we asked whether the Hof1p ring contracts. A strain homozygous for HOF1-GFP was used for time-lapse experiments at 23°C. DIC and GFP images were taken for each individual cell, allowing us to follow the Hof1p localization pattern with respect to septum formation and cell separation (Figure 6, A and B). As expected from the Hof1p-HA analysis, only cells with a large bud have a visible Hof1p-GFP ring(s). Cells usually started with asymmetric double rings, with the brighter ring having a diameter of 1.27 ± 0.06 μm (n = 7) (Figure 6A). The double rings briefly became equal in intensity (not observed for all cells) and then fused into a single ring. When a clear septum was formed, the diameter of the ring reached the minimal size of 0.87 ± 0.08 μm, including its fuzzy edge (n = 7) (Figure 6B). At this time, the single ring split into faint and fuzzy double rings, which remained approximately the same size until cell separation. Hof1p signal disappeared when cell separation occurred. Such behavior is in sharp contrast to that of the myosin ring, which contracts to a single dot before septum formation is completed (Bi et al., 1998; Lippincott and Li, 1998b). These results indicate that Hof1p is not part of the actomyosin contractile system.
Hof1p and Myo1p May Define Alternative Pathways in Cytokinesis
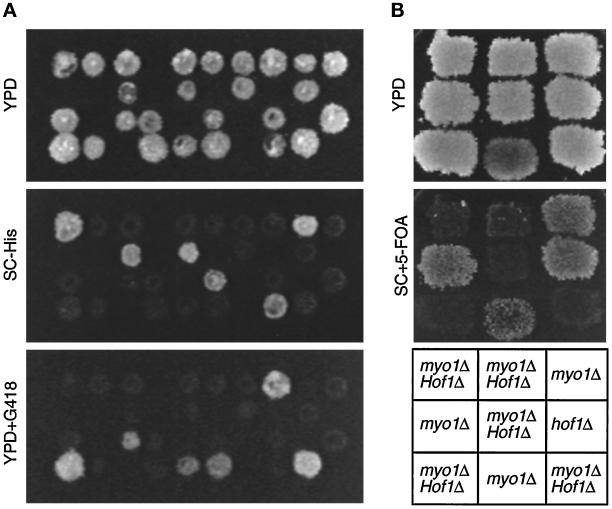
Myo1p function is not essential for cell viability or cytokinesis (Bi et al., 1998; Rodriguez and Paterson, 1990), suggesting that there is an alternative mechanism for cytokinesis in budding yeast. The fact that Hof1p is involved in cytokinesis yet does not behave like Myo1p suggested that Hof1p might act in this alternative pathway. If so, deletion of both genes would result in a more severe cytokinesis defect. To avoid complications caused by multinucleate cells in myo1 and hof1 deletion strains, we first dissected a diploid heterozygous for myo1 and carrying plasmid YCp50-MYO1 to produce strain YEF2056 [a myo1Δ::HIS3 (YCp50-MYO1)]. We used the same strategy to construct strain YEF2059 [α hof1Δ::Kan (pRS315-HOF1)]. These strains were then crossed, and both plasmids were cured, to generate strain YEF2083 (MYO1/myo1Δ::HIS3 HOF1/hof1Δ::Kan). This prevented the expression of myo1 or hof1 mutant phenotypes before analysis, including production of multinucleated cells. Among 20 tetrads dissected from YEF2083, 18 of 19 predicted double mutants (His+ G418 resistant) were inviable at 23°C, whereas only 5 of 20 myo1 mutants (His+) and 0 of 20 hof1 mutants (G418 resistant) were inviable (Figure 7A). The sole survivor of the double mutants may have acquired a suppressor mutation or an extra copy of the MYO1- or HOF1-containing chromosome. Thus, myo1 and hof1 deletions are synthetically lethal at 23°C.
Figure 7.
Synthetic lethality between myo1Δ and hof1Δ. (A) Tetrads from strain YEF2083 (a/α MYO1/myo1Δ::HIS3 HOF1/hof1Δ::Kan) were dissected onto YPD. Colonies were replica plated onto YPD, SC-His, and YPD plus G418 plates and incubated for 3–4 d. (B) Tetrads from YEF2083 carrying YCp50-MYO1 (CEN URA3) were dissected onto YPD. Nine spore colonies containing the plasmid and a single or double deletion were analyzed for their ability to lose the MYO1 plasmid by assaying their ability to grow on 5-FOA plates. Plates were photographed after 3–4 d.
The synthetic lethality could result from a germination defect. To examine this, we dissected tetrads from a diploid that was heterozygous for both myo1 and hof1 and carried the URA3-marked plasmid YCp50-MYO1. Resulting single and double mutants harboring YCp50-MYO1 were replica plated onto a YPD plate, which allowed some cells to lose the plasmid. After incubation at 23°C for 2 d, the YPD plate was replica plated onto a plate containing 5-FOA, which selects against Ura+ cells. Both myo1 and hof1 single mutants could easily lose the plasmid and grow on 5-FOA, whereas the double mutants could not (Figure 7B). We conclude that the myo1 and hof1 deletions are synthetically lethal during vegetative growth, probably as a result of defective cytokinesis. Thus, Myo1p and Hof1p likely define parallel pathways in cytokinesis.
Within the framework defined above, we wanted to determine how other genes are placed in these two cytokinesis pathways, because well-defined relationships among genes may provide valuable insight into their functions. Bni1p has been suggested to play a role in cytokinesis (Kohno et al., 1996). To determine whether it functions in the Myo1p or Hof1p pathway, we crossed YJZ426 (a bni1Δ::HIS3) to YEF2061 [α hof1Δ::Kan (pRS316-HOF1)] and then cured the plasmid, to generate YEF2068. Among 19 tetrads analyzed, all 18 predicted double mutants were inviable, whereas only 4 of 19 hof1 and 1 of 20 bni1 single mutants were inviable, indicating that bni1 and hof1 deletions were synthetically lethal. Moreover, most of the double mutants arrested with morphology consistent with a cytokinesis defect (three connected cellular compartments). These data suggest that Bni1p functions in parallel to Hof1p, most likely in the same pathway as Myo1p. This conclusion is supported by the observation that bni1 and myo1 are not synthetically lethal (data not shown).
One of us previously observed that complete deletions of bni1 and bnr1 are synthetically lethal at 23°C (Bi and Pringle, unpublished result). Among 20 tetrads analyzed from a cross between JF23 (a bni1Δ::LEU2) and YEF1845 (α bnr1Δ::HIS3), all 19 predicted double mutants were inviable at 23°C. All but one bni1 and bnr1 single mutants were viable. This result contradicts a previous report that used incomplete deletions of both genes (Imamura et al., 1997). Because bni1 was synthetically lethal with either hof1 or bnr1, we asked whether hof1 and bnr1 deletions were synthetically lethal at 23°C. Tetrad analysis showed that they were not (data not shown). This confirms a recent report (Kamei et al., 1998) and supports the conclusion that Bnr1p may function in the same pathway as Hof1p. Together, the data suggest that there are two pathways for cytokinesis in S. cerevisiae, one involving Myo1p and Bni1p and the other involving Hof1p and Bnr1p.
Deletion of BNI1, BNR1, or HOF1 Does Not Abolish Formation of the Actin Ring or Patches
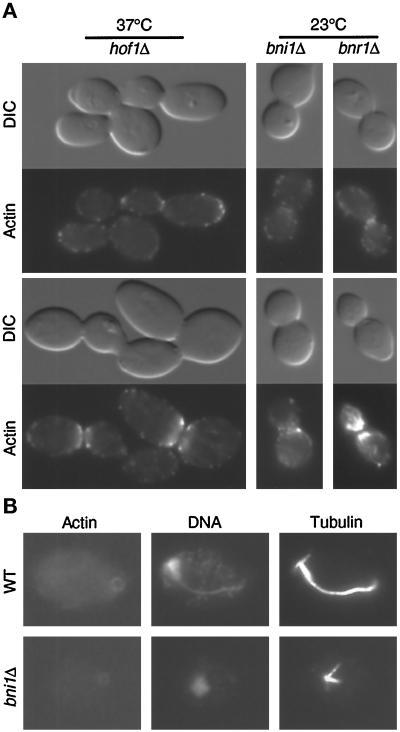
If there were two mechanisms for cytokinesis, as suggested above, the formation or contraction of the actomyosin ring might be affected by bni1 but not by bnr1 or hof1. When cells were grown at 23°C and subsequently fixed and stained for F-actin, 8.2% of the wild-type cells (n = 526) had an actin ring and 4.2% had actin patches at the neck. An actin ring and actin patches were also readily observed in YEF1822 (bnr1Δ::HIS3/bnr1Δ::HIS3) at 23°C (Figure 8A, right column) and in YEF2082 (hof1Δ::Kan/hof1Δ::Kan) at both 23°C (data not shown) and 37°C (Figure 8A, left column). Quantitative analysis demonstrated that the number of mutant cells with rings and patches was similar to that of the wild-type cells. In the bnr1 deletion strain, 5.7% of the cells (n = 508) had an actin ring and 2% had actin patches at the neck. In the hof1 deletion strain, 5.5% of the cells (n = 530) had an actin ring and 5.8% had actin patches at the neck. It was very difficult to get a meaningful count for hof1 cells that were grown at 37°C, because the cells were in chains. An actin ring and actin patches at the neck were also observed in strain YEF1628 (bni1Δ::HIS3/bni1Δ::HIS3; Figure 8A, middle column), although the actin ring was generally fainter and larger than that in wild-type cells. Actin patches in bni1 cells congregated at the bud neck, but they were generally less organized (Figure 8A, bottom cell in middle column). This disorganization was not simply due to an enlarged bud neck, because some hof1 cells with a larger neck appeared to have normally organized actin patches (Figure 8A, bottom cell in left column). Again, because of chain formation, quantitation of the percentage of bni1 cells with actin ring or patches was not attempted. Together, these results indicate that deletion of BNI1, BNR1, or HOF1 does not abolish the formation of actin ring or patches at the bud neck.
Figure 8.
(A) The formation of actin ring and actin patches at the bud neck is not affected by hof1Δ, bni1Δ, or bnr1Δ. Strains YEF2082 (a/α hof1Δ::Kan/hof1Δ::Kan; left panels), HH798 (a/α bni1Δ::HIS3/ bni1Δ::HIS3; middle panels), and YEF1822 (a/α bnr1Δ::HIS3/bnr1Δ:: HIS3; right panels) growing exponentially in YM-P were processed for DIC microscopy and stained for F-actin. YEF2082 was shifted to 37°C for 4.5 h before processing. Top two rows and bottom two rows represent different images of the same cell. (B) Contraction of actin ring is affected by bni1Δ. Strains YEF473 (wild type) and HH798 growing exponentially in YM-P were stained for F-actin, DNA, and tubulin. All cells are shown at the same magnification.
Actomyosin Ring Contraction Is Altered in bni1 Cells
Although the formation of the actin ring was not affected by bni1, if our hypothesis that there are two pathways for cytokinesis is correct, then the contraction of the actin ring might be affected by bni1. In processing cells for immunofluorescence, we noticed that the repeated washes and centrifugation caused some cells to break at late anaphase or telophase, appearing as “unbudded” cells still having an actin ring. In wild-type cells, 29 of 29 such cells had DNA located opposite to the actin ring with a half spindle connecting them (Figure 8B, top row). Strikingly, in a bni1 deletion strain, all seven unbudded cells with an actin ring had a G1-like spindle, and their DNA was often not opposite to the actin ring, suggesting that the actin ring had hardly contracted even when the cell had entered the next cell cycle.
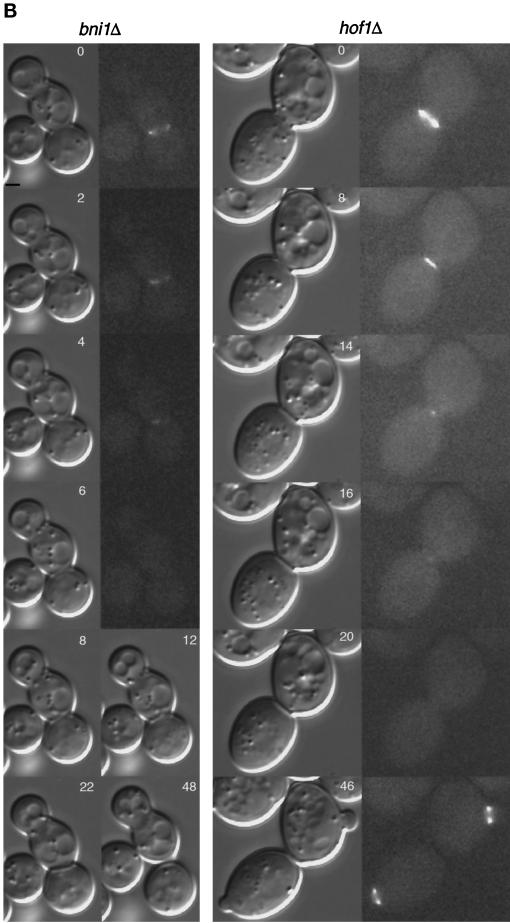
To investigate this more directly, we constructed a diploid strain homozygous for a bni1 deletion and containing MYO1-GFP. Similar bnr1 and hof1 strains were also made. A Myo1p ring formed in all three deletion strains, confirming that Bni1p, Bnr1p, and Hof1p are not required for the formation of the actomyosin ring. In wild-type cells, the ring diameter was 1.1 ± 0.1 μm (n = 9) and took 6–8 min to contract (Figure 9A, left column) (Bi et al., 1998). Septum formation followed contraction immediately or within 2–4 min. Cell separation, scored as rotation of cells from the original mother–daughter axis, occurred 10–14 min later. In contrast, among 18 bni1 cells, only 5 had relatively normal Myo1p contraction (forming a single dot before disappearing), 1 contracted ∼75%, 5 contracted 17–45%, and 7 never contracted. For example, a Myo1p ring of 2.3 μm contracted only ∼24% before its disappearance (Figure 9B, left column). Thus, Bni1p affects the myosin contraction process. The average diameter of the ring in bni1 cells was 1.8 ± 0.4 μm (n = 18), and the size varied significantly, ranging from 1.1 to 2.4 μm. Myosin rings that were larger than 1.6 μm never finished contraction. However, there was no obvious correlation between ring size and contractility when the ring was smaller than 1.6 μm.
Figure 9.
Effects of bnr1Δ, bni1Δ, and hof1Δ on the contraction of Myo1p ring. Time-lapse analysis of strains YEF1698 (a/α MYO1::GFP/ MYO1: GFP) (A, left panels), YEF1854 (a/α MYO1::GFP/MYO1::GFP bnr1Δ::HIS3/bnr1Δ::HIS3) (A, right panels), YEF1839 (a/α MYO1::GFP/ MYO1::GFP bni1Δ::HIS3/bni1Δ::HIS3) (B, left panels), and YEF2124 (a/α MYO1::GFP/MYO1::GFP hof1Δ::Kan/hof1Δ::Kan) (B, right panels) was carried out as described in Figure 6. GFP and DIC images were recorded for each strain. Bar, 1 μm.
In addition to the contraction defect in the bni1 mutant, there was a highly variable delay of several minutes to more than an hour in septum formation and cell separation. Interestingly, in cells with little or no contraction, septum formation was often asymmetric and/or misaligned (Figure 9B, time points 8–48, left column), initiating from one side of the bud neck, and often deviating from the neck axis. Septum in wild-type cells grew symmetrically and centripetally until the precise fusion of the growing ends from both sides (Figure 9A, left column). These data suggest that myosin contraction is correlated with the directionality of the septum growth.
In contrast to bni1, bnr1 and hof1 deletions had little effect on the rate of myosin contraction at 23°C. The size of the Myo1p ring in bnr1 cells was uniform, averaging 1.0 ± 0.1 μm (n = 8), and contracted to a single dot within 6–8 min (Figure 9A, right column). The Myo1p ring in hof1 cells was heterogeneous in size, ranging from 1.1 to 2.3 μm, with an average of 1.6 ± 0.4 μm (n = 6). Contraction to a single dot took 8–16 min, depending on the initial size of the ring. For example, a 2.3-μm ring (Figure 9B, right column) took 16 min to contract. The fact that a similarly sized Myo1p ring contracted in the hof1 cell but not in the bni1 cell suggests that there is a qualitative difference in the contractile ring between these mutants.
In both bnr1 and hof1 deletion strains, after Myo1p contracted to a single dot, it usually remained detectable for 2–4 min instead of disappearing quickly, as in wild-type cells. In addition, both mutants showed a variable delay in cell separation, ranging from several minutes to nearly an hour (Figure 9A, right column, and Figure 9B, right column, compared with Figure 9A, left column). These data suggest that Bnr1p and Hof1p play a role in the maturation of the septum and/or cell separation.
Hof1p Localization to the Neck Depends on Septins but Not Myo1p, Bni1p, and Bnr1p
Localization of the actomyosin system to the bud neck requires the septins (Bi et al., 1998; Lippincott and Li, 1998b). However, the septins, unlike MYO1, are essential for cytokinesis, suggesting that they might function in both cytokinesis pathways. To determine whether localization of Hof1p, a protein functioning in parallel to Myo1p, required septins, we constructed strain YEF2086, which was homozygous for HOF1-HA and cdc12-6. At 23°C, 82% of these cells (n = 50) with well-segregated DNA stained for both septins and Hof1p, whereas 100% of such cells (n = 50) stained for both proteins in the wild-type strain (YEF2026; data not shown). Although both the septins and Hof1p-HA localized to the neck region, they did not overlap precisely when photographed on the same focal plane (Figure 10A, left column). Similar results were obtained for the wild-type cells (data not shown). When strain YEF2086 was grown at 37°C for 1 h, 100% of the cells lost the neck signal of septins and Hof1p-HA (Figure 10A, right column), whereas in wild-type cells at the appropriate stage, 100% still had both proteins at the neck. Thus, the neck localization of Hof1p depends on the septin structure.
Figure 10.
(A) Localization of Hof1p to the bud neck depends on the septins. Strain YEF2086 (a/α HOF1:HA/HOF1:HA cdc12-6/cdc12-6) growing exponentially was shifted to 37°C for 1 h and then stained for a septin (Cdc11p) and Hof1p-HA. (B) Deletion of HOF1 does not affect septin organization. Exponentially growing YEF2082 (a/α hof1Δ::Kan/hof1Δ::Kan) was shifted to 37°C for 4.5 h. Cells were stained for a septin (Cdc11p) and DNA. (C) Localization of Hof1p to the bud neck does not depend on Myo1p, Bni1p, and Bnr1p. Strains YEF2125 (a/α myo1Δ::HIS3/myo1Δ::HIS3 HOF1:HA/HOF1:HA), YEF2126 (a/α bni1Δ::HIS3/bni1Δ::HIS3 HOF1:HA/HOF1:HA), and YEF2095 (α bnr1Δ::HIS3 HOF1:HA) were grown to exponential phase and stained for Hof1p-HA. All cells are shown at the same magnification.
In contrast, septin structure appeared normal in a hof1 strain grown at 23 or 37°C (data not shown and Figure 10B), suggesting that Hof1p function is not required for the localization or the organization of the septins. Interestingly, for cell chains, one bud neck always had strong septin signal, whereas others in the same chain had little or no signal (Figure 10B). It is not clear how this occurs in a chain with a continuous cytoplasm.
To map more completely the functional position of Hof1p, we tested whether Hof1p localization was affected by the deletion of MYO1, BNI1, or BNR1. None of the deletions abolished Hof1p localization (Figure 10C). In some of the myo1 and bni1 cells, the morphology of the Hof1p rings was altered significantly (data not shown and Figure 10C), presumably because the bud-neck structures in these cells were grossly different from those in wild-type cells. Particularly noteworthy, 63% (n = 27) of myo1 deletion cells with DNA masses at the opposite poles of the mother–daughter axis had the normal Hof1p single ring. Thus, the Hof1p single ring can form at the right stage of the cell cycle independently of the actomyosin system. These data also indicate that Bnr1p is not required for the localization of Hof1p, even though both proteins may function in the same pathway.
DISCUSSION
A Role for Hof1p in Septum Formation
Three sequential events at the terminal phase of the cell cycle are required for efficient cell division in S. cerevisiae: actomyosin contraction (cleavage of the cytoplasm), septum formation (cross-neck cell wall synthesis, including primary and secondary cell wall synthesis), and cell separation (removal of the primary septum). When the contraction system is eliminated by the deletion of myosin II, septum formation and cell separation can eventually occur (Rodriguez and Paterson, 1990; Bi et al., 1998), indicating that septum formation can drive cytoplasmic cleavage. Thus, if cytokinesis is the cleavage of one cytoplasm into two, there are apparently two ways that this process can take place: actomyosin contraction and septum formation.
As we and others have shown (Kamei et al., 1998; Lippincott and Li, 1998a; Chant, personal communication), deletion of HOF1 causes a specific defect in cytokinesis. However, it does not seem to alter either the formation or the contraction of the actomyosin ring, demonstrating that Hof1p does not affect cytokinesis by modulating this system. The hof1 cells are delayed before cell separation. This could reflect a defect in the maturation of the primary and/or the secondary septum or in cell separation. We favor a role for Hof1p in septum formation for three reasons. First, at the nonpermissive temperature, chitin deposition in hof1 null cells is delocalized (Kamei et al., 1998; Lippincott and Li, 1998a; this study), although actin polarity and the ovoid cell shape suggest that polarized cell growth occurs normally. Second, the only known proteins that clearly localize to the mother-cell side of the bud neck, as does Hof1p, are those that are either components of chitin synthase III (CSIII), such as Chs3p and Chs4p (Chuang and Schekman, 1996; DeMarini et al., 1997), or proteins involved in recruiting CSIII to the bud neck, such as Bni4p (DeMarini et al., 1997). Finally, if the hof1 deletion caused only a defect in cell separation, digestion of the cell wall would be sufficient to disrupt the cell chains, which is not what we observed. Together, these data suggest that Hof1p may be involved in septum formation, perhaps by restricting the septal and the bud-scar chitin to the neck region. Because the hof1 deletion is temperature sensitive, there may be another Hof1p-like activity that is sensitive to high temperature. It is unlikely that this activity is encoded by HOF2, because deletion of HOF2 did not produce any obvious effect on its own, and it also did not enhance the hof1 phenotype (Chant, personal communication).
This model for Hof1p function is quite different from that proposed previously (Lippincott and Li, 1998a). In addition to the data discussed above, three major pieces of novel evidence led us to these conclusions. First, we analyzed the localization of Hof1p in the cell cycle. Second, we found that myo1 deletion and hof1 deletion were synthetically lethal. Third, we visualized Myo1p-GFP signal and septum formation simultaneously in different mutants, which allowed us to analyze the roles of different genes in regulating actomyosin contraction and septum formation. We examine the basis for the differences and similarities between our work and the work of others below.
We observed that Hof1p was not localized to the bud neck until G2/M, which is supported by its expression profile at both the transcriptional and translational levels (Spellman et al., 1998; this study). In addition, we have observed that the Hof1p localization pattern at the bud neck changes in the cell cycle, which correlates with the phosphorylation state of Hof1p. Previously, Hof1p was reported by two different groups to localize to the bud neck throughout the cell cycle (Kamei et al., 1998; Lippincott and Li, 1998a). The discrepancy between our results and the previous results are likely attributable to the different constructs used in the localization experiments. In the paper by Kamei et al. (1998), an overexpressing GAL1–HOF1 fusion was used, which completely masked the cell cycle regulation and the dynamic pattern that we observed with the endogenous promoter. In the paper by Lippincott and Li (1998a), a plasmid carrying HOF1 with a 265-base pair 5′ sequence was used in the localization experiments in the presence, in some cases, of a chromosomal copy of HOF1. Whether lack of promoter sequences or interference from wild-type Hof1p is the cause of the observed differences is not clear. In our studies, HA or GFP tag was simply fused to the 3′ end of HOF1 at its physiological locus, thus producing minimal effect on its regulation.
The second piece of evidence leading us to suggest a function for Hof1p different from those suggested previously is the relationships we observed between Myo1p and Hof1p. A previous publication (Lippincott and Li, 1998a) suggested that Hof1p may regulate the dynamics of actomyosin contraction. The major evidence for this conclusion was that the rate of Myo1p contraction was increased twofold in hof1 cells versus wild-type cells. In contrast, we found that the rate of Myo1p contraction was approximately the same in both strains. There is no obvious way to explain the discrepancy between our results and the previous results except that the MYO1–GFP constructs and the background history of the strains used in the studies may differ. In our case, GFP was simply tagged to the 3′ end of the MYO1 at its physiological locus, whereas in the previous study, the construction process was not clearly described. Although one could argue that our measurements were not sensitive enough to detect a twofold difference, if Hof1p functions in cytokinesis by regulating actomyosin contraction, it is extremely difficult to reconcile the differences in myo1 and hof1 null phenotypes. hof1 deletion is temperature sensitive, whereas an isogenic myo1 deletion is not. In addition, we have shown in this study that myo1 deletion and hof1 deletion are synthetically lethal, suggesting that they have parallel roles in cytokinesis, which is inconsistent with the hypothesis that Hof1p regulates actomyosin contraction.
In addition to these points, another difference between our work and that of Lippincott and Li (1998a) is that we think it unlikely that Hof1p regulates the organization of septins. Lippincott and Li (1998a) proposed this mechanism for Hof1p function based on two observations. First, deletion of HOF1 was reported to stabilize the septins. Second, overexpression of Hof1p caused delocalization of septins. However, in our studies, we failed to observe multiple septin structures in hof1 null cells (Figure 10B, top cell). One possible difference is that we monitored septin organization with the use of antibodies against the native proteins, whereas they used a GFP-tagged septin gene carried on a plasmid. We made a GFP-tagged CDC3 some time ago and found that when it replaced the chromosomal copy of CDC3, most cells had normal morphology but some had septin mutant–like morphology, particularly at 37°C, suggesting that some GFP-tagged septin genes may not behave exactly like wild-type genes. The effect of Hof1p overexpression on septin organization is certainly consistent with an alternative explanation. Because Hof1p localization depends on the septins, overexpression of Hof1p may sequester septins into some nonfunctional complexes. In addition, we have demonstrated here that Hof1p does not localize to the bud neck until well after the septins are localized there. Although it is possible that septin localization becomes dependent on proper localization of Hof1p late in the cell cycle, it is clear that some septin localization is Hof1p independent. Thus, Hof1p may be one of a growing family of proteins that performs its function by anchoring to the septins, but it does not actively regulate septin organization.
The Actomyosin System May Be a Guide for Septum Formation
By dissecting cytokinesis into distinct processes and examining the interactions between genes involved in each process, a molecular framework for cytokinesis can be established. For example, the synthetic lethality between myo1 and hof1 cells suggests that Hof1p functions in a process parallel to Myo1p, such as septum formation. Using similar logic, we placed Bni1p and Myo1p in one functional group and Hof1p and Bnr1p in another. Other evidence also supports this classification. First, bni1 cells are defective in myosin contraction, whereas bnr1 and hof1 cells are not. Second, Bni1p localizes to the bud neck as a single ring at late anaphase or telophase and appears to colocalize with the actin ring (Longtine and Pringle, personal communication). Bnr1p localizes to the mother side of the bud neck throughout the cell cycle (Longtine and Pringle, personal communication), just as we show here for Hof1p in the early part of the mitotic cycle. Third, Hof1p physically interacts with Bnr1p but not with Bni1p (Kamei et al., 1998). Finally, both hof1 and bnr1 cells are delayed in the disappearance of the myosin dot formed at the terminal phase of the actomyosin contraction and in septum maturation and/or cell separation. Despite their similarities, Hof1p and Bnr1p must have at least one distinct function, because hof1 strains have a temperature-sensitive growth phenotype and no obvious defect with chitin deposition at 23°C, whereas bnr1 strains grow well at both 23 and 37°C. Unlike hof1, bnr1 is not synthetically lethal with myo1 (Vallen and Bi, unpublished results). This is not very surprising, because bnr1 cells have a much milder defect on septum formation than hof1 cells (discussed further below). The synthetic lethality observed between bnr1 and bni1 may reflect multiple functions shared by these two proteins.
It is likely that there is some interaction between the actomyosin system and septum formation, because myo1 null cells form cell chains with separated cytoplasm. This suggests that the actomyosin system plays a role in septum formation and/or cell separation (Bi et al., 1998; Rodriguez-Medina et al., 1998). The detailed analysis of Myo1p and septum behavior in bni1 cells described here provides some clues on the role of the actomyosin system in septum formation. In wild-type cells, immediately after myosin contraction, septum formation initiates symmetrically around the bud neck. The septum then moves progressively toward the center of the neck until it becomes a disk separating the mother and daughter cells. However, in bni1 cells that failed to finish myosin contraction, septum growth often initiated asymmetrically or unevenly and then moved away from the bud-neck axis. This asymmetric septum could change direction as it grew until it touched the opposite side of the bud neck. This suggests that, in wild-type cells, myosin contraction may provide the septum with directionality. For instance, perhaps the primary septum formation machinery is linked to the actomyosin ring uniformly along its circumference. Septum synthetic enzymes could then follow the contraction trail of the actomyosin system to synthesize the primary septum. This hypothesis explains why abnormally thick or branched septa were formed in cells when the actomyosin system was deleted or compromised in S. cerevisiae (Rodriguez and Paterson, 1990) and S. pombe (McCollum et al., 1995; Bezanilla et al., 1997; Kitayama et al., 1997).
Accumulating evidence suggests that the actomyosin system and the septum formation machinery target to the septins independently, which may explain why septins are essential for cytokinesis. In the absence of the actomyosin ring, septum formation can drive cytokinesis, albeit less efficiently, presumably as a result of the loss of directionality in septal growth. However, when the septum formation machinery is compromised to some degree, cytokinesis cannot occur without actomyosin contraction, presumably because septum formation is very inefficient in this case. For example, the ability to form a septum may be compromised more severely in hof1 cells, even at 23°C, than in bnr1 cells, which may explain why hof1, but not bnr1, is synthetically lethal with myo1.
A Molecular Model of Hof1p Function
Hof1p could function in septum formation either as part of the septum synthesis machinery or as an adaptor molecule that recruits or maintains the septum synthetic activities at the neck region. We think it is the latter, based on its localization pattern and the fact that hof1 cells form normal septa at 23°C, and synthesize plenty of chitin at 37°C. Hof1p is expressed at the G2/M transition and localizes initially to the mother side of the bud neck. This type of localization is shared by a limited number of proteins, i.e., those that are components of CSIII or that are involved in targeting CSIII to the mother side of the bud neck (Chuang and Schekman, 1996; DeMarini et al., 1997; Longtine and Pringle, personal communication). CSIII accounts for ∼90% of the total cellular chitin (Bulawa, 1993; Orlean, 1996) and is responsible for the chitin at the base of the bud (Shaw et al., 1991). Chs3p, a component of CSIII, is expressed at a constant level throughout the cell cycle and localizes to the neck region only before the G2/M transition and around the telophase (Chuang and Schekman, 1996). The localization pattern of Hof1p at G2/M to late anaphase, along with the other Hof1p phenotypes, is consistent with a role for Hof1p in restricting CSIII activity to the mother side of the bud neck. This could, for example, prevent continued chitin synthesis in the neck itself, which might interfere with the assembly and/or execution of the contractile system.
As the cell progresses toward late anaphase, Hof1p is phosphorylated. The hyperphosphorylation of Hof1p requires CDC14, CDC15, and DBF2/DBF20. Temperature-sensitive mutations in these genes cause cells to arrest at late anaphase or telophase. Because the hyperphosphorylated form of Hof1p did not accumulate in any of the mutants, it is likely that the phosphorylation event occurs very late in the cell cycle, possibly at telophase or later. In wild-type cells containing actin rings, Hof1p localizes as a single ring that appears coincident with the actin ring. However, although the cdc14-, cdc15-, and dbf2/dbf20-arrested cells contained actin rings, Hof1p remained localized as a double ring or what appears to be an intermediate stage between double and single ring structures. One possibility, given that Cdc15p, Dbf2p, and Dbf20p are all protein kinases (Schweitzer and Philippsen, 1991; Toyn and Johnston, 1994), is that hyperphosphorylation of Hof1p by one or more of these is required for the transition from double rings to a single ring. This could coordinate septum formation with the nuclear cycle.
Interestingly, the septum-specific chitin synthase gene, CHS2, is expressed only at late anaphase or telophase, in time for primary septum formation (Chuang and Schekman, 1996). It is tempting to suggest that the role of the hyperphosphorylated Hof1p single ring is to couple Chs2p to the actomyosin system to coordinate membrane contraction and primary septum formation.
The behavior of Hof1p, as observed in time-lapse analysis by us and others (Lippincott and Li, 1998a), is also consistent with a role for the protein in primary septum formation by interacting with the actomyosin system. These studies show that Hof1p-GFP contracted to some degree at approximately the same time as Myo1p, although the contraction patterns were quite different. The Myo1p ring contracted to a single dot and then quickly disappeared before septum formation was complete, whereas Hof1p-GFP remained bright in the center with fuzzy edges at both ends until cell separation. Thus, we hypothesize that the early function of Hof1p is to restrict CSIII activity to the mother side of the bud neck, and the late function is to couple CSII activity to the actomyosin system. This temporal and spatial change is most likely mediated by changes in the phosphorylation of Hof1p. Regulation by phosphorylation may be a common theme among cdc15 family members, because cdc15 and PSTPIP are both phosphoproteins (Fankhauser et al., 1995; Spencer et al., 1997) and cdc15, like Hof1p, is hyperphosphorylated during late anaphase or telophase of the cell cycle (Fankhauser et al., 1995).
Although the specific conclusions of our work cannot be extrapolated to mammalian cells because they do not have septa, it may be that cdc15-like proteins function generally either to restrict the localization of proteins to specific subcellular regions or to couple the actin cytoskeleton to other processes, as we have observed here. The localization of mammalian PSTPIP (Spencer et al., 1997) and its effects when overexpressed in S. pombe are consistent with this hypothesis (Spencer et al., 1997; Demeter and Sazer, 1998). In S. pombe, cdc15 appears to affect the assembly of the actin ring and/or the recruitment of actin patches to the septation site (Fankhauser et al., 1995; Balasubramanian et al., 1998), whereas imp2 appears to affect the medial ring stability in septating cells (Demeter and Sazer, 1998). In addition, imp2 displays genetic interactions with known septation genes (Demeter and Sazer, 1998), suggesting that it could function similarly to Hof1p in coordinating the function of the actin ring and septum formation.
ACKNOWLEDGMENTS
We thank L. Hartwell (University of Washington, Seattle), L. Johnston (National Institute for Medical Research, London, United Kingdom), D. Lew (Duke University, Durham, NC), S. Brown (University of Michigan Medical School), and M. Longtine and J.R. Pringle (University of North Carolina, Chapel Hill) for their generous gifts of strains, plasmids, and antibodies; M. Longtine and J. Chant for communicating unpublished results; J.M. Sanger for help with image analysis; M. Chou for phosphatase inhibitors; and S. DiNardo, S. Zigmond, and J.W. Sanger (University of Pennsylvania) for their encouragement. This work was supported by The University of Pennsylvania Research Foundation (E.B.), Swarthmore College (E.A.V.), and National Institutes of Health grant GM54300-01 (E.A.V.).
REFERENCES
- Balasubramanian MK, McCollum D, Chang L, Wong KCY, Naqvi NI, He X, Sazer S, Gould K. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla M, Forsburg SL, Pollard TD. Identification of a second myosin-II in Schizosaccharomyces pombe: myp2p is conditionally required for cytokinesis. Mol Biol Cell. 1997;8:2693–2705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, Pringle JR. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–1312. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Pringle JR. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Nurse P. How fission yeast fission in the middle. Cell. 1996;84:191–194. doi: 10.1016/s0092-8674(00)80973-3. [DOI] [PubMed] [Google Scholar]
- Chang F, Woolard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chuang J, Schekman R. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AEM, Fares H, DeVirgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeter J, Sazer S. imp2, a new component of the actin ring in the fission yeast Schizosaccharomyces pombe. J Cell Biol. 1998;143:415–427. doi: 10.1083/jcb.143.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell. 1995;82:435–444. doi: 10.1016/0092-8674(95)90432-8. [DOI] [PubMed] [Google Scholar]
- Fishkind DJ, Wang Y-L. New horizons for cytokinesis. Curr Opin Cell Biol. 1995;7:23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- Ford SK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC11 gene product and the timing of events at the budding site. Dev Genet. 1991;12:281–292. doi: 10.1002/dvg.1020120405. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gould KL, Simanis V. The control of septum formation in fission yeast. Genes Dev. 1997;11:2939–2951. doi: 10.1101/gad.11.22.2939. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. In: Abelson JN, Simon MI, editors. Methods in Enzymology. Vol. 194. San Diego, CA: Academic Press; 1991. pp. 1–933. [PubMed] [Google Scholar]
- Hartwell LH. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp Cell Res. 1971;69:265–276. doi: 10.1016/0014-4827(71)90223-0. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Adams NR, Cooper JA. Formin' the connection between microtubules and the cell cortex. J Cell Biol. 1999;144:809–811. doi: 10.1083/jcb.144.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src Homology 3 domain-containing Hof1p: implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces cerevisiae. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Wright B, Milstein C. Rat monoclonal antitubulin antibodies derived by using a new nonsectoring rat cell line. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, et al. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kolodziej PA, Young RA. Epitope tagging and protein surveillance. In: Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. Vol. 194. San Diego, CA: Academic Press; 1991. pp. 508–519. [DOI] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-Y, Yeh E, Hays TS, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: response to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998a;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998b;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, DeMarini DJ, Valencik ML, Al-Awar OS, Fares H, De Virgilio C, Pringle JR. The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol. 1996;8:106–119. doi: 10.1016/s0955-0674(96)80054-8. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, DeMarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marks J, Hyams JS. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- McCollum D, Balasubramanian MK, Pelcher LE, Hemmingsen SM, Gould KL. Schizosaccharomyces pombe cdc4+ gene encodes a novel EF-hand protein essential for cytokinesis. J Cell Biol. 1995;130:651–660. doi: 10.1083/jcb.130.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Matheos D, Rose MD. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Cytokinetic activities in a human cell line: the midbody and the intercellular bridge. Tissue Cell. 1973;5:47–61. doi: 10.1016/s0040-8166(73)80005-9. [DOI] [PubMed] [Google Scholar]
- Mullins JM, Biesele JJ. Terminal phase of cytokinesis in D-985 cells. J Cell Biol. 1977;73:762–684. doi: 10.1083/jcb.73.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins JM, MacIntosh JR. Isolation and initial characterization of the mammalian midbody. J Cell Biol. 1982;94:654–661. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Orlean P. Biogenesis of yeast wall and surface components. In: Pringle JR, Broach JR, Jones EW, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. pp. 229–362. [Google Scholar]
- Pringle JR, Adams AE, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harb Symp Quant Biol. 1995;60:729–743. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Saccharomyces cerevisiae cell cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 97–142. [Google Scholar]
- Pringle JR, Mor J-R. Methods for monitoring the growth of yeast cultures and for dealing with the clumping problem. Methods Cell Biol. 1975;11:131–168. doi: 10.1016/s0091-679x(08)60320-9. [DOI] [PubMed] [Google Scholar]
- Rappaport R. Cytokinesis in animal cells. In: Barlow PW, Bard JB, Green PB, Kirk DL, editors. Developmental and Cell Biology Series. Cambridge, UK: Cambridge University Press; 1996. pp. 1–386. [Google Scholar]
- Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskeleton. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Medina JR, Cruz JA, Robbins PW, Bi E, Pringle JR. Elevated expression of chitinase 1 and chitin synthesis in myosin II-deficient Saccharomyces cerevisiae. Cell Mol Biol. 1998;44:919–925. [PubMed] [Google Scholar]
- Rose AB, Broach JR. Propagation and expression of cloned genes in yeast: 2-μm circle based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- Sanger JM, Pochapin MB, Sanger JW. Midbody sealing after cytokinesis in embryos of the sea urchin Arbacia punctulata. Cell Tissue Res. 1985;240:287–292. doi: 10.1007/BF00222337. [DOI] [PubMed] [Google Scholar]
- Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- Schweitzer B, Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991;7:265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, Cabib E. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1991;114:111–123. doi: 10.1083/jcb.114.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Dowbenko D, Cheng J, Li W, Brush J, Utzig S, Simanis V, Lasky LA. PSTPIP: a tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J Cell Biol. 1997;138:845–860. doi: 10.1083/jcb.138.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Bracchat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]