Abstract
INSM1/IA-1 (insulinoma-associated 1) is a developmentally regulated zinc-finger transcription factor, exclusively expressed in the foetal pancreas and nervous system, and in tumours of neuroendocrine origin. We have identified an INSM1 binding site in the neuroD/β2 promoter and demonstrated transcriptional repressor activity of INSM1 by transient transfection assay. A chromatin immunoprecipitation assay confirmed that in vivo INSM1 is situated on the promoter region of the neuroD/β2 gene. In an attempt to elucidate the molecular mechanism of transcriptional repression by the INSM1 gene, cyclin D1 was identified as an interacting protein by using a 45-day-old human foetal brain cDNA library and a yeast two-hybrid screen. The physical association between INSM1 and cyclin D1 was confirmed by in vitro and in vivo pull-down assay. Cyclin D1 co-operates with INSM1 and suppresses neuroD/β2 promoter activity. Co-immunoprecipitation of INSM1, cyclin D1 and HDACs (histone deacetylases) in mammalian cells revealed that INSM1 interacts with HDAC-1 and -3 and that this interaction is mediated through cyclin D1. Overexpression of cyclin D1 and HDAC-3 significantly enhanced the transcriptional repression activity of INSM1 on the neuroD/β2 promoter. A further chromatin immunoprecipitation assay confirmed that HDAC-3 occupies this same region of the neuroD/β2 promoter, by forming a transcription complex with INSM1. Thus we conclude that INSM1 recruits cyclin D1 and HDACs, which confer transcriptional repressor activity.
Keywords: cyclin D1, histone deacetylase (HDAC), insulinoma-associated 1 (INSM1/IA-1), neuroD/β2, repression complex, yeast two-hybrid
Abbreviations: Ad-INSM1, adenoviral-INSM1; CDK4, cyclin-dependent kinase 4; ChIP, chromatin immunoprecipitation assay; CMV, cytomegalovirus; Cpep, C-terminal peptide; CREB, cAMP-response-element-binding protein; EMSA, electrophoretic-mobility-shift assay; HDAC, histone deacetylase; HEK-293, human embryonic kidney-293; INSM1/IA-1, insulinoma-associated 1; Npep, N-terminal peptide; NP40, Nonidet P40; PPARγ, peroxisome-proliferator-activated receptor γ; Rb, retinoblastoma; STAT, signal transducer and activator of transcription
INTRODUCTION
The INSM1/IA-1 (insulinoma-associated 1) gene encodes a 58 kDa protein that contains five zinc-finger DNA binding motifs. It was originally isolated from a human insulinoma subtraction library [1]. Functional studies have revealed that the N-terminus of the INSM1 protein possesses repressor activity and that the zinc-finger motifs recognize the conserved target sequence, TG/TC/TT/AGGGGG/TCG/A, which is located in the promoter region of the neuroD/β2 and INSM1 genes [2]. One potential target gene, neuroD/β2, was chosen for further study of its regulation by INSM1. NeuroD/β2 is a basic helix-loop-helix transcription factor that has been shown to be associated with late neuronal differentiation in Xenopus laevis [3]. Gene-targeting experiments revealed that deletion of the neuroD/β2 gene resulted in defective pancreatic morphogenesis and abnormal enteroendocrine differentiation, which leads to the development of early diabetes [4]. Furthermore, neuroD/β2 is required for the differentiation of granule cells in the cerebellum and hippocampus [5,6]. The expression pattern of the INSM1 gene is highly restricted in the human foetal pancreas and embryonic tissues of the nervous system, as well as in tumours of neuroendocrine origin [7–9]. Since the INSM1 transcription factor is expressed transiently in embryonic stages of development, we hypothesized that its biological function could be to act as a differentiation antigen during pancreatic and nervous system development. In order to reveal the molecular mechanisms by which the INSM1 protein functions during neuroendocrine cell development, a yeast two-hybrid screen was employed to isolate cellular proteins that interact with the INSM1 protein in developing foetal brain tissue. We identified a cell cycle regulator, cyclin D1, that interacts with the INSM1 protein.
Cyclin D1 is regulated by mitogen stimulation and its promoter responds to several transcription factors such as STAT (signal transducer and activator of transcription) proteins, NF-κB (nuclear factor-κB), EGR-1 (early growth response gene product-1), CREB (cAMP-response-element-binding protein), PPARγ (peroxisome-proliferator-activated receptor γ) and nuclear receptors [10–17]. Inactivation of the cyclin D1 gene produces a smallmouse phenotype with defects in eye and mammary-gland development [18,19]. The primary function of cyclin D1 is to initiate cell cycle progression by association with CDK4 (cyclin-dependent kinase 4), which phosphorylates the Rb (retinoblastoma) protein and disrupts its association with the DNA-binding transcription factor, E2F, allowing transcriptional activation of S-phase genes. Cyclin D1 also plays an important role in malignant transformation, and is considered to be an oncogene [20,21]. Additionally, cyclin D1 displays CDK4-independent roles in transcriptional regulation [22]. Cyclin D1 inhibits the transcriptional activation of MyoD, PPARγ, DMP1 (dentin matrix protein 1), Sp-1, STAT3, v-Myb, neuroD/β2, thyroid-hormone receptor and androgen receptor [17,23–31]. By contrast, cyclin D1 also binds to the estrogen receptor and potentiates its activation [32–34]. These multi-functional features of cyclin D1 clearly support its major role in cell growth, gene regulation and tumorigenesis.
In the present study, we provide evidence to support the idea that INSM1 is a transcriptional repressor of the neuroD/β2 promoter, and have identified the recruitment of cyclin D1 and HDACs (histone deacetylases) as components for INSM1-mediated transcriptional repression. This interaction was confirmed by co-immunoprecipitation of cyclin D1, INSM1 and HDACs both in vivo and in vitro. The N-terminal region of the INSM1 protein, which was previously shown to possess transcriptional repressor activity [2] is required for cyclin D1 binding. We sought to determine whether cyclin D1 has any functional effect on the transcriptional repressor activity of INSM1, and found that cyclin D1 enhances its activity. The combination of INSM1, cyclin D1 and HDAC-3 has been demonstrated in the present study to greatly enhance the repressive activity of INSM1 on the neuroD/β2 promoter. Furthermore, INSM1 and HDAC-3 were shown to occupy the same region of the mouse neuroD/β2 gene promoter in vivo, by ChIP (chromatin immunoprecipitation) assay. Significantly, our studies highlight the major components of the INSM1 regulatory complex that confer transcriptional repressor activity.
MATERIALS AND METHODS
Cell culture and reagents
HEK (human embryonic kidney)-293, and medulloblastoma, D283Med cells were obtained from American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium with 10% foetal bovine serum, 50 μg/ml penicillin, 0.25 μg/ml streptomycin and 2 mM L-glutamine at 37 °C under 5% CO2. Mouse insulinoma, βTC-1 cells were kindly provided by Dr E. H. Leiter (Jackson Laboratory, Bar Harbor, ME, U.S.A.). The mouse and rabbit anti-cyclin D1, mouse anti-HA (haemagglutinin), mouse anti-Myc, mouse and rabbit anti-HDAC-3, and mouse and rabbit anti-FLAG antibodies were obtained from Biosource (Camarillo, CA, U.S.A.), Covance Research Product (Berkeley, CA), Clontech (Palo Alto, CA, U.S.A.) and Sigma (St. Louis, MO, U.S.A.) respectively.
Plasmid constructs
For the yeast two-hybrid screen, the INSM1 bait-plasmid was constructed by subcloning the full-length INSM1 cDNA [1] into the pGBKT7 vector (Clontech), which was designated as pGAL4-BD-INSM1. For mammalian cell expression, the HA-tagged INSM1, INSM1-Npep (N-terminal peptide; amino acids 1–263), INSM1-Cpep (C-terminal peptide; amino acids 257–510) fusion proteins and full-length cyclin D1 were cloned into a eukaryotic expression vector, pcDNA3 (Invitrogen). The cyclin D1-GH and -KE mutant constructs were kindly provided by Dr R. Weinberg (Department of Biology, Whitehead Institute for Biomedical Research, Cambridge, MA, U.S.A.) and Dr K. E. Knudsen (Department of Cell Biology, University of Cincinnati, Cincinnati, OH, U.S.A.). The FLAG-tagged HDAC-1, -2 and -3 expression vectors were provided by Dr E. Seto (Department of Medical Microbiology and Immunology, and Department of Biochemistry and Molecular Biology, University of South Florida, FL, U.S.A.).
Transient transfection and neuroD/β2 reporter assay
The mouse neuroD/β2 promoter sequence (−419/+59 bp) was subcloned into a luciferase reporter vector, pGL3-basic (Promega) [35]. Three copies of the INSM1 binding site derived from the neuroD/β2 promoter were subcloned into the GL3-basic vector, in front of the E1bTATA-basic promoter (ND3-E1bTATA) and the luciferase gene. The pcDNA3-HA–INSM1 and pcDNA3-cyclin D1 expression vectors were co-transfected into HEK-293 cells with the reporter vector, neuroD/p2-pGL3-basic, by using FuGENE™ 6 (Roche) to examine the effect of INSM1 or cyclin D1, or both on neuroD/β2 or ND3-E1bTATA promoter activity. CMV (cytomegalovirus)-β-galactosidase activity, and the total protein concentration in lysates were used as internal controls against which to normalize the transfection efficiency. An empty pcDNA3 vector was also included to ensure that each transfection had an equal amount of DNA. Post-transfection (48 h), cells were washed with PBS and collected for luciferase and β-galactosidase activity assay (Promega). Each experiment was performed at least four times, and the values are the means±S.E.M.
Electrophoretic mobility shift assay (EMSA)
An EMSA was performed using a double-stranded oligonucleotide derived from the neuroD/β2 promoter, A−185GTGGTGGTGGAAGGGGGCGGGAGGAAAGT −156. The double-stranded oligonucleotide was end-labelled using [γ-32P]ATP (3000 Ci/mmol; PerkinElmer Life Sciences) and T4 polynucleotide kinase (New England Biolabs). The INSM1 Cpep was transcribed and translated from the pGBKT7-Cpep vector in vitro by using a TNT® Quick-Coupled Rabbit Reticulocyte Lysate Kit (Promega). Production of Cpep was confirmed by Western blot analysis by using an anti-Cpep antibody. The EMSA binding reaction mixture contained 1 μl of in vitro-translated Cpep in a binding buffer composed of 10 mM Tris/HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5% (v/v) glycerol, 500 ng of poly(dI·dC) and 50000 c.pm. of the probe. For the supershift assay, 1 μl of the mouse anti-Cpep antibody or control mouse IgG was added first with the in vitro-translated Cpep at 4 °C for 25 min, and then the probe was added for an additional 25 min at 4 °C. For the competition assay, 50×, 100× and 200× of the unlabelled oligonucleotide was incubated with in vitro-translated lysate first, followed by the addition of the radiolabelled probe. The protein–DNA complexes were resolved on a 5% PAGE (40:1) gel in 0.25× Tris/borate/EDTA (1×TBE=45 mM Tris/borate/1 mM EDTA) buffer. The gels were dried and analysed by autoradiography.
ChIP assay
A ChIP assay was performed on the βTC-1 cells using the ChIP kit (Upstate) according to the manufacturer's instructions, with some modifications as described [36,37]. Briefly, chromatin in 80% confluent βTC-1 cells was cross-linked by adding formaldehyde directly to the tissue-culture medium to a final concentration of 1%. Fixation was monitored at 37 °C for 10 min, and cells were washed twice with cold PBS buffer containing protease inhibitors. The fixed cells were collected by centrifugation and re-suspended in lysis buffer (supplemented with protease inhibitors). After incubation for 30 min on ice, the cells were Dounce-homogenized to facilitate nuclei release. Nuclei were collected by centrifugation at 2700 g, and resuspended in sonication buffer [1% SDS, 10 mM EDTA, 50 mM Tris/HCl (pH 8.0), 0.5 mM PMSF, 100 ng of leupeptin and 100 ng of aprotinin]. Samples were sonicated on ice to an average length of 200–500 bp and then centrifuged at 20000 g at 4 °C for 12 min. The chromatin solution was pre-cleared with the addition of Protein G beads for 2 h at 4 °C. The pre-cleared chromatin was incubated with the following antibodies: 10 μg of a mouse anti-INSM1 antibody, 10 μg of normal mouse IgG serum (Southern Biotechnology Associates, Inc), or no antibody as a negative control. After overnight incubation with the antibodies described above, the antibody/chromatin mixtures were precipitated with Protein G beads, and the beads were sequentially washed with ChIP immunoprecipitation wash buffer. The complexes were eluted twice by using 250 μl of elution buffer for each ChIP reaction. Cross-linking was reversed by adding 4 μl of 5M NaCl and 1 μl of RNase, and samples were incubated at 65 °C overnight. Protein was removed from DNA by adding proteinase K at 42 °C for 2 h, and DNA was purified by phenol/chloroform extraction and ethanol precipitation. The PCR primers used to detect target sequences were as follows: neuroD/β2 (−291/−16 bp) 5′-AAATAGGCAGGTCACGTGGTT-3′; 5′-CGCTAGGGTTATATAGCCGAG-3′. For the anti-HDAC-3 ChIP assay, βTC-1 cells were transduced with Ad-INSM1 (adenoviral-INSM1) for two days and were subjected to the same protocol as described above.
Construction of recombinant adenovirus for gene transfer
An AdEasy XL adenoviral vector system (Stratagene) was used. This system includes BJ5183 cells pre-transformed with the pAdEasy-1 plasmid (BJ5183-AD-1 cells). This feature dramatically decreases background caused by the non-recombinant shuttle plasmid. Briefly, FLAG-tagged human INSM1 cDNA was subcloned into the multiple cloning sites of the pCMV-shuttle vector. The recombinant human INSM1 shuttle vector was electroporated into BJ5183-AD-1 cells and the smaller colonies present on the kanamycin plate (the small size of the colony indicates a recombinant clone) were selected. The clones produce either 4.5 kb and/or 3 kb bands after PacI enzyme digestion, indicating that the recombination either took place at the origins of replication or between the left arms. After selecting the correct recombinant clones, 10–20 μg of PacI-digested recombinant Ad-plasmid DNA was used to transfect AD-293 cells using an MBS Transfection Kit (Stratagene). The primary viral stock was amplified, measured for viral titre and verified for INSM1 gene expression. Large amounts of the recombinant adenovirus were purified from five large plates (150 cm2) using a repeated CsCl centrifugation method, to give a high viral titre. For determination of the adenoviral titre, a BD Adeno X Rapid Titre Kit was used to stain the virus-infected cells using an anti-hexon antibody (BD Biosciences Clontech).
Construction of a yeast two-hybrid library from 45-day-old foetal brain tissue
Total RNA was extracted from the brain of a 45-day-old human foetus (obtained from the Central Laboratory for Human Embryology, University of Washington, Seattle, WA, U.S.A.) with TriZOL® reagent (Invitrogen). Approx. 2 μg of total RNA was used to construct a MATCHMAKER cDNA library according to the manufacturer's instructions, by using the Clontech MATCHMAKER Library Construction & Screening Kit (PT3529-1). The total number of library transformants was approx. 3.5×106 colonies. The yeast plasmid DNA was isolated by using the MasterPure™ Yeast DNA Purification Kit (Epicentre).
Yeast transformation and two-hybrid library screening
Bait-INSM1 (pGal4-BD-INSM1) and a cDNA library (pGal4-AD-library) were used to transform AH109 yeast cells according to the manufacturer's (Clontech) protocol. Yeast DNA (containing both the bait-and target-plasmid) was prepared and used to transform the Escherichia coli strain, DH5α. The ampicillin resistant plasmid clones containing the target DNA were selected for further confirmation and sequence analysis.
Co-immunoprecipitation and Western blot analysis
For in vitro co-immunoprecipitation, pGAL4-BD-INSM1 (Myc-tagged) containing the entire open reading frame of human INSM1 and pGal4-AD-cyclin D1 (HA-tagged) containing the entire open reading frame of human cyclin D1 were translated in vitro in a TNT Quick-Coupled Rabbit Reticulocyte Lysate system (Promega) in the presence of [35S]methionine/cysteine (Amersham). An equal amount of radiolabelled INSM1 or cyclin D1 was mixed and incubated overnight at 4 °C with a monoclonal anti-Myc (Clontech) or anti-HA antibody (BAbCO, MMS-101R) in an immunoprecipitation buffer [20 mM Tris/HCl (pH 7.4), 150 mM NaCl, 1% NP40 (Nonidet P40), 0.1% SDS and 2 mM EDTA]. The next morning, 50 μl of recombinant Protein G–agarose (Invitrogen) was added for 1 h at 4 °C, the slurry was then washed with immunoprecipitation buffer 4 times, and analysed by SDS/10% PAGE. For transient transfection, HEK-293 cells were seeded in a 100 mm-diameter culture dish. FuGENE6™ (12 μl) (Roche):4 μg of test DNA (2 μg of pcDNA3-HA–INSM1 and 2 μg of pcDNA3-cyclin D1) 3:1, (v/v) was used to transfect 80% confluent HEK-293 cells. The cells were harvested at 48 h post-transfection for co-immunoprecipitation and Western blot analysis. Cell lysates were prepared using protein binding buffer [20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 0.5% NP40, 0.1% (v/v), Triton-X 100 and 1 mM PMSF]. For co-immunoprecipitation, 300 μg of the cell lysate was pre-cleared with recombinant Protein G–agarose and incubated for 2 h at 4 °C with an anti-HA antibody, followed by incubation overnight at 4 °C with recombinant Protein G–agarose beads. The beads were washed five times with binding buffer, separated by SDS/(10%) PAGE, and transferred on to a nitrocellulose membrane (Invitrogen, Carlsbad, CA, U.S.A.). HA–INSM1 or cyclin D1 expression was detected by Western blotting (1:1000) with either an anti-HA or anti-cyclin D1 antibody respectively. The horseradish peroxidase-conjugated anti-mouse secondary antibody (1:4000) and chemiluminescent substrate (Pierce) were used for detection of INSM1 or cyclin D1 protein.
INSM1 and cyclin D1 co-immunoprecipitate HDAC-1 and -3
The interaction between INSM1, cyclin D1 and HDAC-1, -2, or -3 was investigated in a co-immunoprecipitation experiment. In HA–INSM1, cyclin D1 and FLAG–HDAC-1, -2, or -3 transfected mammalian cells, an anti-HA antibody was used to precipitate INSM1 that was subsequently subjected to Western blotting with anti-HA, anti-cyclin D1 and anti-HDAC antibodies to reveal that INSM1 could pull down HDAC-1 and -3. An anti-HA antibody was used to confirm its ability to pull down itself and cyclin D1.
Statistical analysis
Statistical analysis was performed using the Student's t test for unpaired comparisons. Data are presented as means±S.E.M., P<0.05 was considered significant.
RESULTS
INSM1 is a transcriptional repressor of the neuroD/β2 gene
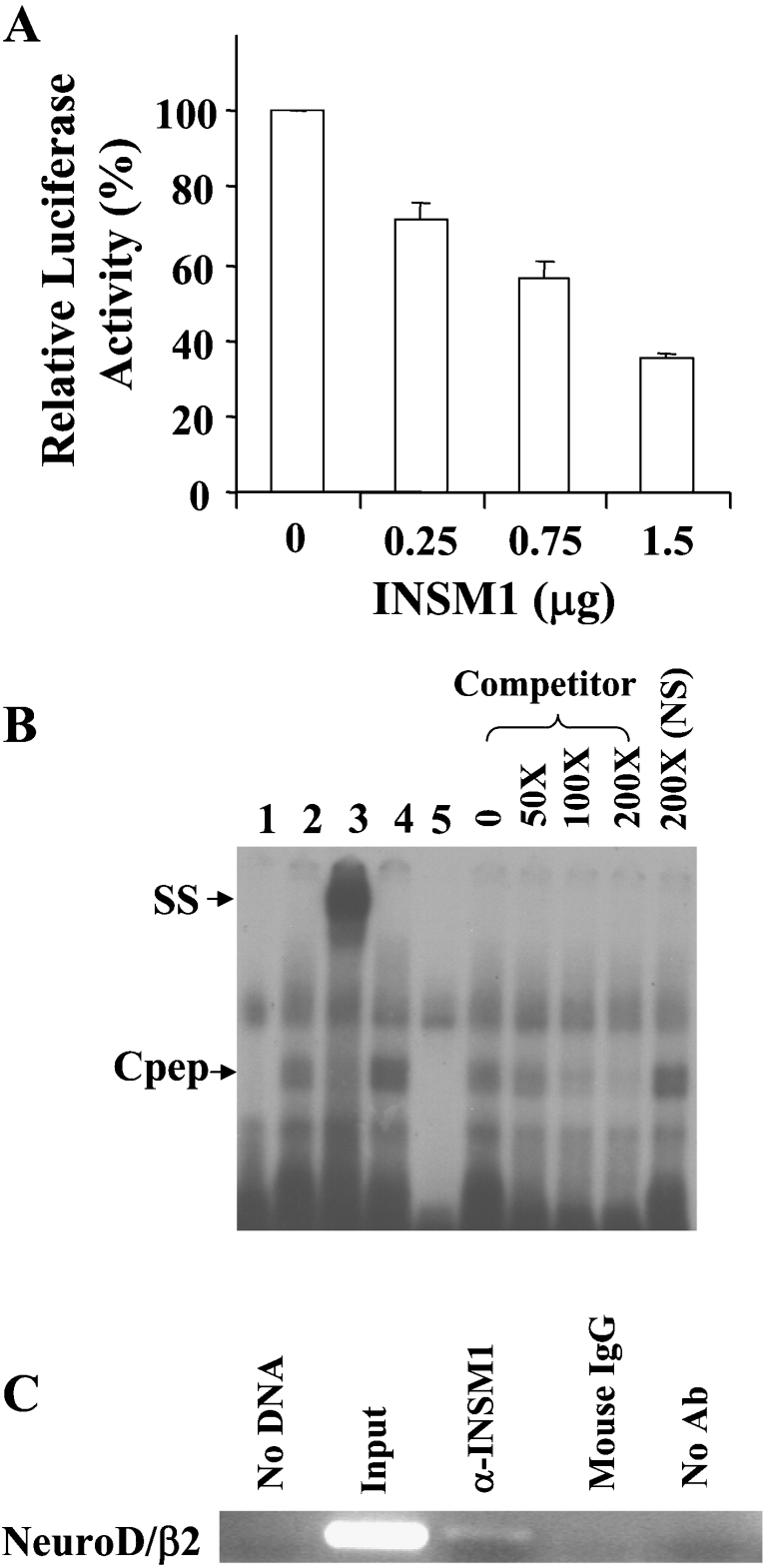
A high-affinity INSM1 binding site in the mouse neuroD/β2 promoter was previously identified but not extensively characterized [2]. Therefore, by using different amounts of the CMV-INSM1 expression vector in HEK-293 cells, we demonstrated transcriptional repression of the mouse neuroD/β2 promoter (−419/+59 bp) linked to a luciferase reporter gene (Figure 1A). INSM1 clearly exerted a dose-dependent suppressive effect on the 479 bp promoter-driven reporter. A 62% inhibition of neuroD/β2 promoter activity was observed at the highest concentration of the INSM1 expression vector. A search for INSM1 consensus binding sites in eukaryotic promoter sequence databases revealed that the mouse neuroD/β2 promoter (−177/−166 bp) contains a potential INSM1 binding site. Therefore an oligonucleotide spanning −185/−156 bp or a non-specific oligonucleotide was synthesized for the EMSA. As shown in Figure 1(B), a specific band-shift was observed with an in vitro-translated C-terminal segment (amino acids 257–510) of the INSM1 protein. A super-shifted band was observed upon the addition of a mouse monoclonal antibody against Cpep, but not when using the normal mouse IgG as a control. Various amounts (50–200×) of unlabelled competitor oligonucleotide demonstrated that Cpep binding to the INSM1 binding site is specific. We further confirmed the interaction of the endogenous INSM1 transcription factor with the neuroD/β2 promoter using an in vivo ChIP assay. A mouse βTC-1 cell line was chosen for the ChIP assay. The ChIP assay was carried out by using a mouse anti-INSM1 antibody or a control antibody. As shown in Figure 1(C), the anti-INSM1 antibody specifically pulled down the neuroD/β2 promoter sequence (−291/−16 bp). Therefore we concluded that INSM1 functions as a transcriptional repressor of the neuroD/β2 gene.
Figure 1. The neuroD/β2 promoter is a target gene suppressed by INSM1.
(A) Dose-dependent suppression of the neuroD/β2 promoter by INSM1 in HEK-293 cells. In HEK-293 cells, 1 μg of the mouse neuroD/β2 promoter (−419/+59 bp) linked to a luciferase reporter gene was co-transfected with increasing amounts (0.25–1.5 μg) of a CMV-INSM1 expression vector. Repressive effects on neuroD/β2 promoter activity using different amounts of CMV-INSM1 are shown as the percentage of suppression±S.E.M. The transfections were carried out and normalized in three sets of experiments. (B) EMSA. The double-stranded oligonucleotide spanning the −185 to −156 bp region of the mouse neuroD/β2 promoter, or a non-specific (NS) oligonucleotide was end-labelled as the probe. Probe alone (lane 1), with Cpep lysate (lane 2), Cpep lysate plus an anti-Cpep antibody (lane 3), Cpep lysate plus an anti-(mouse IgG) control antibody (lane 4), and the non-specific (NS) probe with the Cpep lysate (lane 5) are shown in the left of the panel. The right of the panel shows different concentrations (50–200×) of unlabelled competitor oligonucleotides. The presence of a specific band-shift (Cpep) and supershifted (SS) band indicated that the INSM1 C-terminal peptide (amino acids 257–510) is capable of binding to the INSM1 site in the neuroD/β2 promoter. (C) ChIP assay. Formaldehyde cross-linked chromatin from β-TC1 cells was incubated with a mouse antibody against INSM1 (lane 3). Immunoprecipitated genomic DNA was analysed by PCR using primers specific for the transcriptional regulatory sequences of the mouse neuroD/β2 (276 bp) promoter (see Materials and methods section). As a control, PCR reactions were carried out with no DNA (lane 1), input DNA (lane 2), DNA immunoprecipitated by normal IgG serum that was species-matched to the source of the test antibody (lane 4), and DNA that was immunoprecipitated in the absence of antiserum (lane 5). ChIP assays were repeated in two separate experiments. Ab, antibody.
Physical association of cyclin D1 and INSM1
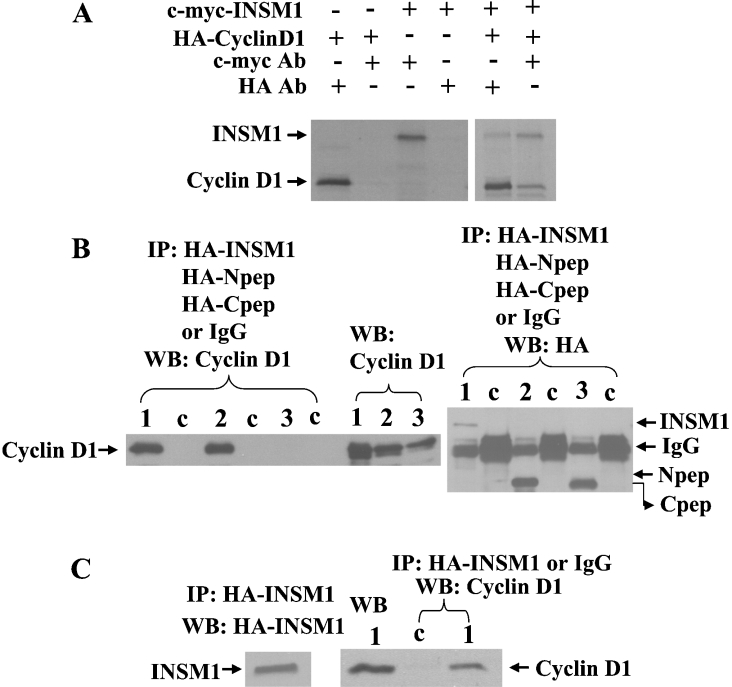
We identified cyclin D1 as an INSM1-interacting-protein in a yeast two-hybrid screen of a 45-day-old human foetal brain cDNA library. In order to confirm the physical interaction between INSM1 and cyclin D1, further pull-down experiments were performed both in vivo and in vitro. Radiolabelled full-length Myc–INSM1 and HA–cyclin D1 were translated in vitro in a rabbit reticulocyte lysate system.
Co-immunoprecipitation of INSM1 and cyclin D1 was carried out using the corresponding antibody to the epitope-tag. As shown in Figure 2(A), an anti-Myc antibody co-precipitated cyclin D1 and an anti-HA antibody co-precipitated INSM1 respectively. To further analyse the region of INSM1 to which cyclin D1 was bound in cultured cells, we performed an in vivo transfection study. The HA-tagged full-length INSM1, INSM1 N-terminal peptide (1–263 amino acids), or INSM1 C-terminal peptide (257–510 amino acids) expression vector was co-transfected with cyclin D1 into HEK-293 cells. Post-transfection, in order to demonstrate correct processing and expression of the cyclin D1 protein from transfected cDNA, lysates were subjected to Western blot analysis with a monoclonal anti-cyclin D1 antibody (Figure 2B, middle panel). To investigate whether the full-length INSM1 protein (lane1), N-(lane 2) or C-terminal peptide (lane 3) was bound to cyclin D1, cell lysates were first immunoprecipitated with the anti-HA antibody or normal IgG serum, and were subsequently subjected to Western blot analysis with an anti-cyclin D1 or anti-HA antibody. Normal mouse IgG serum was used as a control (lane c). Figure 2(B) indicates that cyclin D1 binds to both the full-length INSM1 and N-terminal portion of the INSM1 protein, which confers transcriptional repressor activity [2]. Furthermore, we demonstrated that INSM1 can pull down endogenous cyclin D1, which confirms their physical interaction in vivo (Figure 2C).
Figure 2. In vitro and in vivo co-immunoprecipitation.
(A) Myc epitope-tagged full-length INSM1 and HA-tagged cyclin D1 were translated in vitro in a rabbit reticulocyte lysate system with the addition of [35S]cysteine/methionine. An anti-Myc monoclonal antibody or an anti-HA monoclonal antibody precipitated the tagged protein and pulled down the interacting counterpart. (B) HEK-293 cells were co-transfected with a CMV- promoter-driven full-length cyclin D1 and full-length HA–INSM1 (amino acids 1–510, lane 1), HA–INSM1-Npep (amino acids 1–263, lane 2) or HA–INSM1-Cpep (amino acids 257–510, lane 3). Normal mouse IgG (lane c) was used as a control. Cellular lysates were separated by SDS/12.5% PAGE. The SDS/PAGE gel was transferred on to a nitrocellulose membrane and subject to Western blot analysis with the rabbit anti-cyclin D1 antibody or a mouse anti-HA antibody (1:1000). Both full-length and N-terminal INSM1 proteins co-precipitated cyclin D1 in vivo. The middle panel (lane 1–3) shows direct Western blotting of cell lysates with an anti-cyclin D1 antibody. (C) HA–INSM1 was transfected into HEK-293 cells. In this experiment, HA–INSM1 is capable of pulling down the endogenous cyclin D1 (lane 1) in experiments in which mouse IgG was used as a control (lane c). The left panel shows Western blotting (WB) of the immunoprecipitated HA–INSM1 and the endogenous cyclin D1.
Cyclin D1 co-operates with INSM1 to suppress neuroD/β2 promoter activity
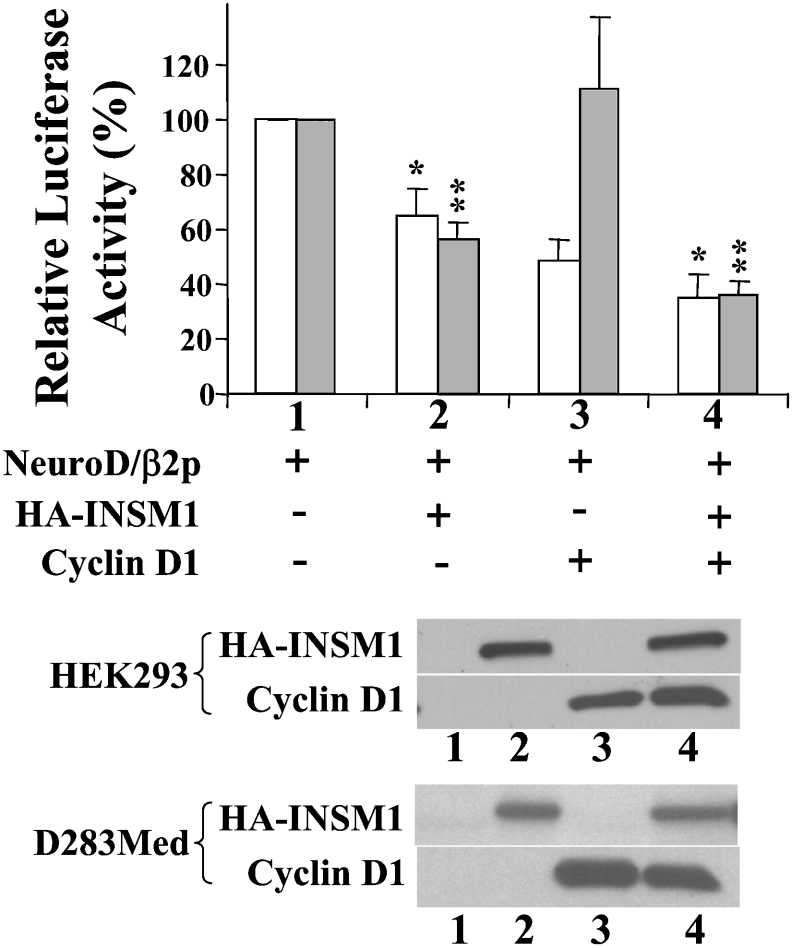
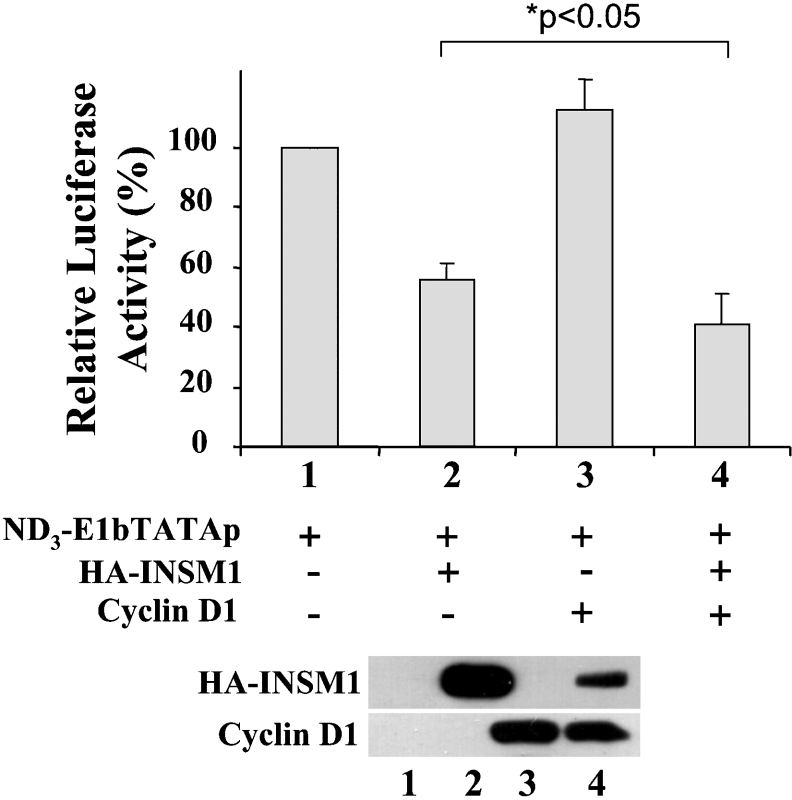
EMSA, the luciferase reporter transfection study, and the in vivo ChIP assay revealed the positive interaction between the INSM1 protein and the neuroD/β2 promoter sequence, supporting a role for INSM1 in the regulation of the neuroD/β2 gene. As described below, the suppressive effect of the INSM1 protein on the neuroD/β2 promoter was further tested in the presence of cyclin D1 in order to determine the functional significance of their physical association (Figure 3). Overexpression of the INSM1 protein suppresses 40–50% of the activity of the mouse neuroD/β2 promoter. The combination of INSM1 and cyclin D1 expression vectors suppressed the activity of the mouse neuroD/β2 promoter by up to 70%. Surprisingly, when the cyclin D1 expression vector and the neuroD/β2 promoter were co-transfected into D283Med cells, cyclin D1 alone induced a 50% decrease in neuroD/β2 activity. It is probable that this inhibitory effect is due to endogenous INSM1 expression in D283Med cells. HEK-293 cells do not express INSM1, in contrast with D283Med cells, which express a high level of endogenous INSM1. Therefore, in HEK-293 cells overexpression of cyclin D1 alone does not demonstrate a suppressive effect on the neuroD/β2 promoter in the absence of INSM1. Both the INSM1-expressing cell line (D283Med) and non-INSM1-expressing cell line (HEK-293) exhibited a similar enhancement of the transcriptional suppressor activity of cyclin D1, which is statistically significant (D283Med, *P<0.01; HEK-293 **P<0.001). The enhancement of the suppressive effect could be due to contributions from both endogenous and overexpressed cyclin D1. In order to confirm that the INSM1 binding site is essential for transcriptional repression of the neuroD/β2 gene, we designed a reporter construct containing three copies of the INSM1 binding site sequence (the same sequence as in the neuroD/β2 promoter) in front of an E1bTATA-basic promoter and the luciferase gene. This transient transfection study revealed that INSM1 alone, or in combination with cyclin D1, exhibited 40–60% transcriptional repressor activity (*P<0.05), whereas cyclin D1 alone did not exhibit any transcriptional repressor activity in HEK-293 cells (Figure 4). This result suggests that the INSM1 transcription factor binds specifically to the INSM1-binding site in the neuroD/β2 promoter and recruits either endogenous and/or overexpressed cyclin D1 to enhance its acitivity as a transcriptional repression.
Figure 3. INSM1 co-operates with cyclin D1 to suppress neuroD/β2 promoter activity.
Mouse neuroD/β2 promoter activity (−419/+59 bp) was examined in the presence of the CMV-driven expression vector containing either INSM1, cyclin D1 or both. Two different cell lines, D283Med (open box) and HEK-293 (shaded box), were used as either INSM1-expressing (D283Med) or non-expressing (HEK-293) cells. INSM1 alone induces a 40–50% inhibition of neuroD/β2 promoter activity in both cell lines, whereas cyclin D1 alone induces a 50–60% inhibition of the neuroD/β2 promoter activity in D283Med, but not in HEK-293, cells. The combined expression of INSM1 and cyclin D1 enhances inhibition of neuroD/β2 promoter activity by up to 70% (D283Med, *P<0.01). A CMV-β-galactosidase construct was used to normalize the transfection efficiency. The graph shows the means for four separate experiments±S.E.M. A representative Western blot of the expressed proteins is shown below the histogram.
Figure 4. Transcriptional repressor activity is mediated through the INSM1 binding site.
A triple repeat of the INSM1 binding site was cloned into an E1bTATA-basic promoter driven luciferase reporter gene. Transfection experiments were performed in HEK-293 cells as described above, in order to identify the repressive activity of INSM1, cyclin D1 or both. INSM1 demonstrated a 50% repressive effect, whereas cyclin D1 failed to suppress the E1bTATA promoter. A combination of INSM1 and cyclin D1 enhanced suppressive activity by up to 60% (*P<0.05). This result revealed that the INSM1 binding site is crucial for cyclin D1 to co-operate with INSM1 for transcriptional repression. A CMV-β-galactosidase construct was used to normalize transfection efficiency. The histogram shows the means for four separate experiments±S.E.M. A representative Western blot of the expressed proteins is shown below the chart.
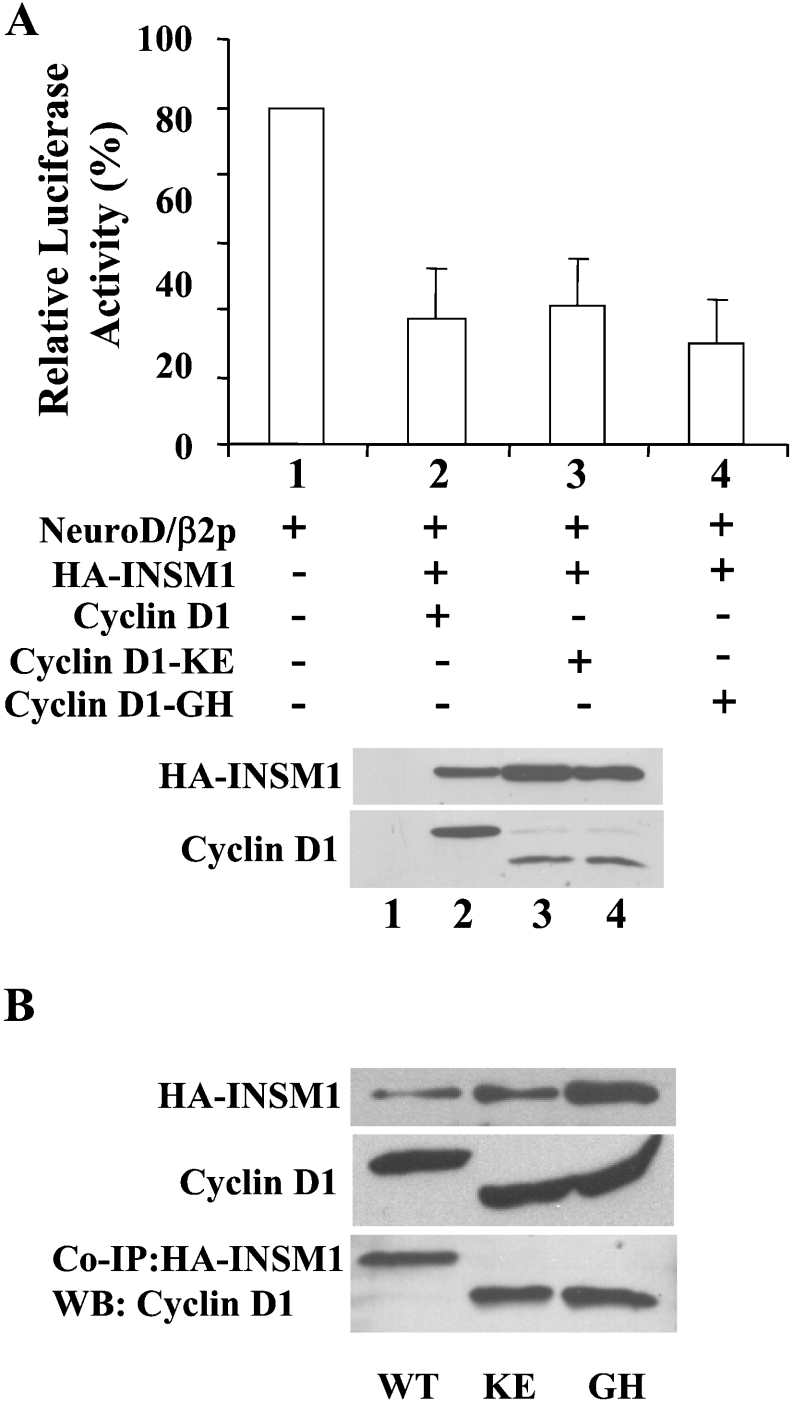
The transcriptional repression of neuroD/β2 promoter activity is independent of the cell cycle function of cyclin D1
The combined expression of INSM1 and cyclin D1 decreased the activity of the neuroD/β2 promoter by 60–70%. In order to elucidate whether transcriptional repressor activity is dependent upon the conventional cell cycle functions of cyclin D1, the cyclin D1-KE and cyclin D1-GH mutants were examined. These mutants had lost the ability to bind to CDK4 (cyclin D1-KE) or to Rb protein (cyclin D1-GH) [38]. Both mutants exhibited the same level of transcriptional repressor activity as wild-type cyclin D1, suggesting that transcriptional repression by cyclin D1 occurs independently of its cell cycle functions (Figure 5A). Co-immunoprecipitation of INSM1 and cyclin D1 mutants revealed a similar interaction as occurred with wild-type cyclin D1 (Figure 5B). The size discrepancy between the wild-type and the mutant cyclin D1 is due to the addition of the HA-tagged sequence present in the wild-type cyclin D1 expression vector.
Figure 5. Transcriptional repression of neuroD/β2 promoter activity is independent of the cell cycle function of cyclin D1.
The combined expression of INSM1 and cyclin D1 demonstrated an approx. 65% inhibition of neuroD/β2 promoter activity in HEK-293 cells. (A) To test the hypothesis as to whether the transcriptional repression is dependent upon the cell cycle function of cyclin D1, cyclin D1 mutants were used that had either lost the ability to bind to CDK4 (mutant CycD-KE), or Rb (mutant CycD-GH). Both mutants revealed the same repressor activity as did wild-type cyclin D1, suggesting that the repressor activity is independent of the cell cycle function of cyclin D1. A CMV-β-gal construct was used to normalize the transfection efficiency. The graph shows the means for four separate experiments±S.E.M. A representative Western blot of the expressed proteins is shown below the histogram. (B) Co-immunoprecipitation of INSM1 and wild-type cyclin D1 or mutant cyclin D1 was demonstrated by a pull-down experiment. Mutant cyclin D1-KE and -GH showed similar binding efficiency to INSM1, as did wild-type cyclin D1. The size discrepancy between the wild-type and the mutant cyclin D1 was due to the addition of the HA-tagged sequence on to the wild-type cyclin D1 expression vector. WB, Western blot.
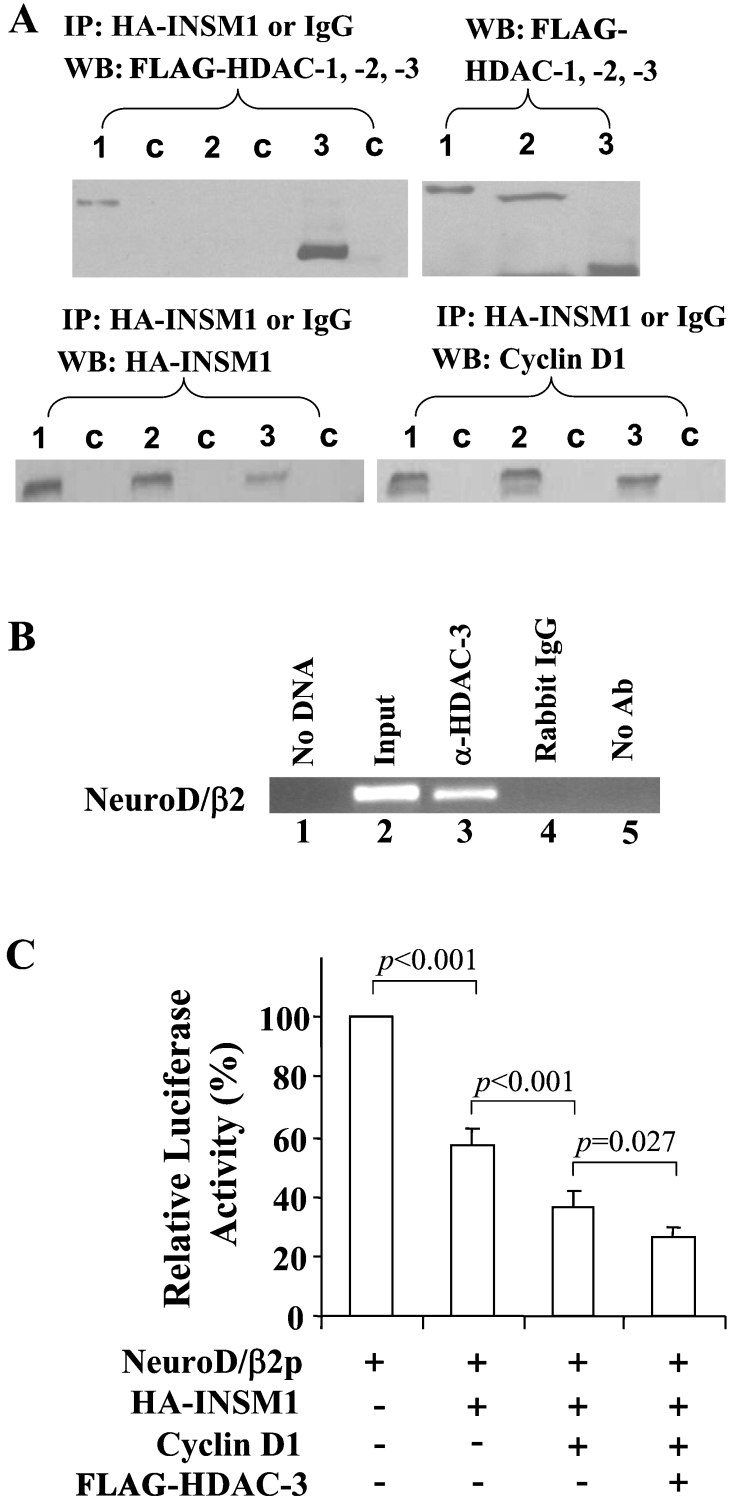
Recruitment of HDACs contributes to INSM1 transcriptional repressor activity
We examined whether INSM1 transcriptional repressor activity is associated with the recruitment of HDACs. Cyclin D1 has been shown to recruit HDAC-3 by forming a complex with the thyroid-hormone receptor [29]. HDAC-1, -2 and -3 were each transfected, along with HA–INSM1 and cyclin D1, into HEK-293 cells (Figure 6A). A co-immunoprecipitation study was carried out by using an anti-HA antibody or control mouse IgG, immunoprecipitates were then subjected to Western blot analysis with anti-HA–INSM1, anti-cyclin D1 or anti-FLAG–HDAC-1, -2 and -3 antibodies. Both cyclin D1 and HDAC-1 and -3 were precipitated by HA–INSM1 in HEK-293 cells (Figure 6A). The interaction, if any, between INSM1 and HDAC-2 is minimal, in contrast with a strong signal in the HDAC-3 lane and a moderate signal in the HDAC-1 lane. These results confirm that cyclin D1 is strongly associated with HDAC-3 and moderately with HDAC-1. Since we were able to pull down endogenous cyclin D1 using an anti-HA–INSM1 antibody, as shown in Figure 2(C), this suggested that the interaction of INSM1 with HDAC-1 and -3 is mediated through binding of endogenous or overexpressed cyclin D1. In order to show that cyclin D1 and HDAC-3 do indeed occupy the neuroD/β2 promoter region in vivo, we performed a ChIP assay using anti-cyclin D1 or anti-HDAC-3 antibodies. As shown in Figure 6(B), an anti-HDAC-3 antibody could efficiently pull down the same neuroD/β2 promoter sequence as an anti-INSM1 antibody. However, the anti-cyclin D1 antibody failed to pull down the same neuroD/β2 promoter sequence (results not shown). It is possible that cyclin D1 is located in between INSM1 and the HDACs, and is thus inaccessible to the antiserum used in the ChIP assay.
Figure 6. HDAC-3 is situated on the promoter region of the neuroD/β2 gene and contributed to the transcriptional suppressor activity.
(A) INSM1 co-immunoprecipitation of HDAC-1 and -3. HEK-293 cells were transfected with HA–INSM1, cyclin D1, and FLAG-tagged HDAC-1, -2 or -3. Anti-HA antiserum co-precipitated HDAC-1 and -3, and HDAC-3 displayed the strongest interaction. The HA precipitates were also subjected to Western blotting (WB) for cyclin D1 and HA–INSM1. Numbers indicate HDAC-1, -2 and -3, and lane c shows normal mouse IgG used as control. (B) HDAC-3 is associated with the neuroD/β2 gene promoter sequence in vivo as revealed by ChIP assay. Formaldehyde cross-linked chromatin isolated from Ad-INSM1 transduced βTC-1 cells was incubated with a rabbit anti-HDAC-3 antibody (lane 3). Immunoprecipitated DNA was analysed by PCR using primers specific for the transcriptional regulatory sequence of the mouse neuroD/β2 (the same primers used in Figure 1). As a control, PCR reactions were carried out with no DNA (lane 1), input DNA (lane 2), DNA immunoprecipitated by normal immune sera that were species-matched to the source of the test antibody (lane 4), and DNA that was immunoprecipitated in the absence of antiserum (lane 5). ChIP assays were performed in two separate experiments. Ab, antibody. (C) Co-transfection of INSM1, cyclin D1 and HDAC-3 with the mouse neuroD/β2 promoter. Overexpression of each component significantly enhanced the repressive effect on neuroD/β2 promoter activity.
In addition, we performed a co-transfection reporter assay by adding each expression vector (INSM1, cyclin D1 and HDAC-3) to the neuroD/β2 promoter construct. NeuroD/β2 promoter activity was gradually decreased with the addition of each component. The combination of all three components induced an approx. 75% suppressive effect on neuroD/β2 promoter activity (Figure 6C). Therefore, it can be concluded that the association of INSM1, cyclin D1 and HDACs confers the repression of transcriptional activity on the neuroD/β2 promoter.
DISCUSSION
The results presented in this study define INSM1 as a transcriptional repressor of the neuroD/β2 gene. The molecular mechanism of INSM1 transcriptional repression is attributed to the recruitment of cyclin D1 and HDAC-1 and -3. Human foetal brain (45-day-old) was selected, from which to construct a cDNA library because it expresses INSM1 mRNA abundantly (H. W. Wang, M. B. Breslin and M. S. Lan, unpublished work). The N-terminal portion of the protein is required for binding to cyclin D1. In a previous study, it was shown that the repressor activity of INSM1 is located at the N-terminal portion of the protein [2]. Hence, it is logical to speculate that the binding of cyclin D1 would enhance the transcriptional repressor activity.
Transcriptional regulation is dependent upon chromatin structure and its remodelling, which includes transcription factor modification, acetylation, methylation, phosphorylation and ubiquitination [39]. The exact mechanism that accounts for INSM1 transcriptional repressor activity is still unclear. The recruitment of cyclin D1 to the INSM1 binding site suggests several possible mechanisms of controlling INSM1-mediated transcriptional repression. The co-repressor activity of cyclin D1 in regulating Sp1-mediated transcription was previously shown to be associated with TAFII250 [25]. Cyclin D1 has also been shown to have distinct mechanisms of action in other co-repressor activities. Cyclin D1 decreases the nuclear level of STAT3 in HepG2 cells, and downregulates STAT3 activity [26]. It has also been shown to regulate MyoD function by cyclin D1-dependent nuclear targeting of CDK4 [40]. The co-repressor activity of cyclin D1 has been demonstrated by us and others to be closely associated with the recruitment of HDACs [17,29]. Moreover, multiple nuclear receptor co-repressor complexes contain distinct HDACs [41]. By contrast, cyclin D1 has been demonstrated to potentiate the activation of the oestrogen receptor through the formation of a complex with the histone acetyltransferase, P/CAF (p300/CREB-binding protein) [33]. Although INSM1 alone exhibits significant transcriptional repression activity on the neuroD/β2 and ND3-E1bTATA promoters, it is probable that first INSM1 recruits endogenous cyclin D1, as we have shown in Figure 2(C), and then HDAC-1 and -3 are recruited to form a transcriptional complex. This hypothesis was further substantiated by examining the interaction of INSM1 with HDAC-1, -2 and -3 using a yeast two-hybrid screen. The colonies selected on Leu−/Trp− plates cannot grow on Ade−/His−/Leu−/Trp− quadruple selection plates, suggesting that INSM1 does not interact directly with HDACs (results not shown). This specific association of the HDACs complexes for binding of the INSM1 protein, mediated through cyclin D1 and resulting in transcriptional repression, has been demonstrated in our study and in an increasing number of other laboratories that have used different transcription factors [17,29]. Therefore, in the present paper we report for the first time that cyclin D1 serves as a co-repressor with the INSM1 transcriptional repressor to promote suppressive activity through the recruitment of HDAC-1 and -3.
In the present study, we demonstrate that cyclin D1 not only physically interacts with the INSM1 transcription factor, but also exhibits transcriptional repressor activity alone on the neuroD/β2 promoter in a medulloblastoma cell line, D283Med, but not in the HEK-293 cell line. This cell-type-dependent repressor activity is probably due to the specific nuclear factors present in this cell line. The neuroD/β2 promoter contains multiple regulatory elements in addition to the INSM1 binding site, such as Sp-1 and E-box elements. It is possible that the repressor activity of cyclin D1 alone on the neuroD/β2 promoter is mediated through these Sp-1 or E-box elements. The association of cyclin D1 with the TBP (TATA-binding protein)-associated factor, TAFII250, on an Sp-1 site has been reported to suppress Sp-1 transcription [25]. A previous study has also demonstrated that cyclin D1 represses the neuroD/β2 transcription factor through binding to its E-box element [28,35]. Other unknown regulatory elements present in the neuroD/β2 promoter may also contribute to the observed repression of its activity. Both HEK-293 and D283Med cells express cyclin D1 and HDAC-3, but only D283Med cells express INSM1 and neuroD/β2. INSM1 is a self-regulated transcription factor and has been shown to be activated by the neuroD/β2 transcription factor [8]. The co-expression of INSM1 and neuroD/β2 in D283Med tumour cells could attenuate the suppressive effect of INSM1 on the human neuroD/β2 promoter. Thus it is difficult to predict whether further overexpression of INSM1 will interrupt endogenous neuroD/β2 gene expression.
Previous reports have revealed that, when cyclin D1 is involved in modulating transcriptional activity, this usually occurs independently of its cell cycle functions. This possibility was further examined in our study using cyclin D1-KE and cyclin D1-GH mutants that have lost their ability to bind to either CDK4, or Rb protein [38]. Since both cyclin D1 mutants bound to the INSM1 protein equally well and exhibited the same level of repression of activity as did wild-type cyclin D1, it was further confirmed that the co-repressor activity of cyclin D1 is cell cycle independent. However, it cannot be ruled out that INSM1 is not involved in normal cell cycle functions. Intriguingly, the INSM1 gene is highly expressed in the developing pancreas and nervous system, as well as in tumours of neuroendocrine origin. Cell cycle progression is activated by the binding of D-type cyclins to CDK4 and CDK6 that induce Rb protein phosphorylation and initiate entrance into the cell cycle [20]. Our previous observations have shown that overexpression of INSM1 could induce hypophosphorylation of the Rb protein (W. D. Liu, M. B. Breslin and M. S. Lan, unpublished work). When INSM1 serves as a transcriptional repressor that binds to cyclin D1, it is possible that the binding of INSM1 to cyclin D1 not only mediates the recruitment of HDACs, but also interrupts normal cell cycle progression and promotes cellular differentiation. Interestingly, a recent report has described HDAC-inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells [42]. NeuroD/β2 was identified among the HDAC-inhibitor-up-regulated neuronal specific genes. Since neuroD/β2 is the target gene of INSM1, HDAC inhibition could diminish the repressive effects of INSM1 and up-regulate neuroD/β2 gene expression involved in neuronal differentiation. Furthermore, the cyclin D1 gene is frequently overexpressed in a broad range of human tumours [43]. A recent report using tumour gene expression databases has revealed that the oncogenic effect of cyclin D1 involves the transcription factor, CCAAT/enhancer-binding protein β [44]. The physical association of INSM1 and cyclin D1, particularly in neuroendocrine tumours, implies a direct role for INSM1 in cell transformation. Whether the significance of the INSM1 and cyclin D1 interaction is limited to modulating the repressor activity of INSM1, or whether there is also involvement in cell cycle functions and oncogenic events, remains to be determined.
Acknowledgments
We thank Dr S. H. Pincus, Dr P. Wang and Dr D. M. Silverstein for critical reading of this manuscript before submission. This work was supported by funds from the Research Institute for Children, Children's Hospital, and a grant from NIDDK, National Institutes of Health (DK 61436, to M.S.L.).
References
- 1.Goto Y., DeSilva M. G., Toscani A., Prabhakar B. S., Notkins A. L., Lan M. S. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with zinc-finger DNA-binding motifs. J. Biol. Chem. 1992;267:15252–15257. [PubMed] [Google Scholar]
- 2.Breslin M. B., Zhu M., Notkins A. L., Lan M. S. Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 2002;30:1038–1045. doi: 10.1093/nar/30.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J. E., Hollenberg S. M., Snider L., Turner D. L., Lipnick N., Weintraub H. Conversion of Xenopus ectodrm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 4.Naya F. J., Huang H. P., Qiu Y., Mutoh H., DeMayo F. J., Leiter A. B., Tsai M. J. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in β2/NeuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyata T., Maeda T., Lee J. E. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu M., Pleasure S. J., Collins A. E., Noebels J. L., Naya F. J., Tsai M. J., Lowenstein D. H. Loss of β2/neuroD leads to malformation of the dentate gyrus and epilepsy. Proc. Natl. Acad. Sci. U.S.A. 2000;97:865–870. doi: 10.1073/pnas.97.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu M., Breslin M. B., Lan M. S. Expression of a novel zinc-finger cDNA, IA-1, is associated with rat AR42J cells differentiation into insulin-positive cells. Pancreas. 2002;24:139–145. doi: 10.1097/00006676-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Breslin M. B., Zhu M., Lan M. S. NeuroD1/E47 regulates the E-box element of a novel zinc-finger transcription factor, IA-1, in developing nervous system. J. Biol. Chem. 2003;278:38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie J. P., Cai T., Zhang H., Lan M. S., Notkins A. L. The zinc-finger transcription factor INSM1 is expressed during embryo development and interacts with the Cbl-associated protein. Genomics. 2002;80:54–61. doi: 10.1006/geno.2002.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushime H., Roussel M. F., Ashmun R. A., Sherr C. J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991;65:701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg J., Wrzeszczynska M., Devgan G., Zhao Y., Pestell R., Albanese C., Darnell J. Stat 3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 12.Matsumura I., Kitamura T., Wakao H., Tanaka H., Hashimoto K., Albanese C., Downward J., Pestell R. G., Kanakura Y. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J. 1999;18:1367–1377. doi: 10.1093/emboj/18.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttridge D. C., Albanese C., Reuther J. Y., Pestell R. G., Baldwin A. S., Jr NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan Y. X., Nakagawa H., Lee M. H., Rustgi A. K. Transforming growth factor-α enhances cyclin D1 transcription through the binding of early growth response protein to a cis-regulatory element in the cyclin D1 promoter. J. Biol. Chem. 1997;272:33181–33190. doi: 10.1074/jbc.272.52.33181. [DOI] [PubMed] [Google Scholar]
- 15.Lee R. J., Albanese C., Stenger R. J., Watanabe G., Inghirami G., Haines G. K. I. I. I., Webster M., Muller W. J., Brugge J. S., Davis R. J., Pestell R. G. pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J. Biol. Chem. 1999;274:7341–7350. doi: 10.1074/jbc.274.11.7341. [DOI] [PubMed] [Google Scholar]
- 16.Sabbah M., Courileau D., Mester J., Redeuilh G. Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11217–11222. doi: 10.1073/pnas.96.20.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu M., Rao M., Bouras T., Wang C., Wu K., Zhang X., Li Z., Yao T. P., Pestell R. G. Cyclin D1 inhibits PPARr-meidated adipogenesis through HDAC recruitment. J. Biol. Chem. 2005;280:16934–16941. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 18.Sicinski P., Donaher J. L., Parker S. B., Li T., Fazeli A., Gardner H., Haslam S. Z., Bronson R. T., Elledge S. J., Weinberg R. A. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 19.Fantl V., Stamp G., Andrews A., Rosewell I., Dickson C. Mice lacking cyclin D1 are small and show defects in eye and mammary gland development. Genes Dev. 1995;9:2364–2372. doi: 10.1101/gad.9.19.2364. [DOI] [PubMed] [Google Scholar]
- 20.Sherr C. J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 21.Donnellan R., Chetty R. Cyclin D1 and human neoplasia. J. Clin. Pathol. Mol. Pathol. 1998;51:1–7. doi: 10.1136/mp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coqueret O. Linking cyclins to transcriptional control. Gene. 2002;299:35–55. doi: 10.1016/s0378-1119(02)01055-7. [DOI] [PubMed] [Google Scholar]
- 23.Skaper S. X., Rhee J., Kim P. S., Novitch B. G., Lassar A. B. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperhohsphorylation. Mol. Cell. Biol. 1996;16:7043–7053. doi: 10.1128/mcb.16.12.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue K., Sherr C. J. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent kinase indenpendent mechanism. Mol. Cell. Biol. 1998;18:1590–1600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adnane J., Shao Z., Robbins P. D. Cyclin D1 associates with the TBP-associated factor TAFII250 to regulate Sp1-mediated transcription. Oncogene. 1999;18:239–247. doi: 10.1038/sj.onc.1202297. [DOI] [PubMed] [Google Scholar]
- 26.Bienvenu F., Gasean H., Coqueret O. Cyclin D1 represses STAT3 activation through a CDK4-independent mechanism. J. Biol. Chem. 2001;276:16840–16847. doi: 10.1074/jbc.M100795200. [DOI] [PubMed] [Google Scholar]
- 27.Ganter B., Fu S., Lipsick J. S. D-type cyclins repress transcriptional activation by the v-Myb but not the c-Myb DNA-binding domain. EMBO J. 1998;17:255–268. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratineau C., Petry M. W., Mutoh H., Leiter A. B. Cyclin D1 represses the basic helix-loop-helix transcription factor, β2/NeuroD. J. Biol. Chem. 2002;277:8847–8853. doi: 10.1074/jbc.M110747200. [DOI] [PubMed] [Google Scholar]
- 29.Lin H. M., Zhao L., Cheng S. Y. Cyclin D1 is a ligand-indenpendent co-repressor for thyroid hormone receptors. J. Biol. Chem. 2002;277:28733–28741. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 30.Knudsen K. E., Cavence W. K., Arden K. C. D-type cyclins complex with the androgen receptor and inhibit its transcriptional transactivation ability. Cancer Res. 1999;59:2297–2301. [PubMed] [Google Scholar]
- 31.Petre C. E., Wetherill Y. B., Danielsen M., Knudsen K. E. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J. Biol. Chem. 2002;277:2207–2215. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 32.Neuman E., Ladha M. H., Lin N., Upton T. M., Miller S. J., DiRenzo J., Pestell R. G., Hinds P. W., Dowdy S. F., Brown M., Ewen M. E. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol. Cell. Biol. 1997;17:5338–5347. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMahon C., Suthiphongchai T., DiRenzo J., Ewen M. E. P.CAF associates with cyclin D1 and potentiates its activation of the estrogen receptor. Proc. Natl. Acad. Aci. U.S.A. 1999;96:5382–5387. doi: 10.1073/pnas.96.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwijsen R. M. L., Wientjens E., Klompmaker R., van der Sman J., Bernards R., Michalides R. J. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–415. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 35.Huang H. P., Liu M., El-hodiri H. M., Chu K., Jamrich M., Tsai M. J. Regulation of the pancreatic islet-specific gene beta2 (neruoD) by neurogenin 3. Mol. Cell. Biol. 2000;20:3292–3307. doi: 10.1128/mcb.20.9.3292-3307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyd K. E., Wells J., Gutman J., Bartley S. M., Farnham P. J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl. Acad. Sci. U.S.A. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boyd K. E., Farnham P. J. Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol. Biol. Cell. 1999;19:8393–8399. doi: 10.1128/mcb.19.12.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dowdy S. F., Hinds P. W., Louie K., Reed S. I., Arnold A., Weinberg R. A. Physical interaction of the retinoblastoma protein with human D cyclins. Cell. 1993;73:499–511. doi: 10.1016/0092-8674(93)90137-f. [DOI] [PubMed] [Google Scholar]
- 39.Latchman D. S. Regulation of transcription factor activity. In: Latchman D. S., editor. Eukaryotic Transcription Factors. San Diego, CA, U.S.A.: Elsevier Academic Press; 2004. pp. 245–285. [Google Scholar]
- 40.Zhang J. M., Wei Q., Zhao X., Paterson B. M. Coupling of the cell cycle and myogenesis through the cyclin D1-dependent interaction of MyoD with CDK4. EMBO J. 1999;18:926–933. doi: 10.1093/emboj/18.4.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones P. L., Sachs L. M., Rouse N., Wade P. A., Shi Y. B. Multiple N-CoR complexes contain distinct histone deacetylases. J. Biol. Chem. 2001;276:8807–8811. doi: 10.1074/jbc.C000879200. [DOI] [PubMed] [Google Scholar]
- 42.Hsieh J., Nakashima K., Kuwabara T., Mejia E., Gage F. H. Histone deacetylase inhibition-mediated neuronal differentiatioin of multipotent adult neural progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall M., Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and cdk inhibitors in human cancer. Adv. Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 44.Lamb J., Ramaswamy S., Ford H. L., Contreras B., Martinez R. V., Kittrell F. S., Zahnow C. A., Patterson N., Golub T. R., Ewen M. E. A mechanism of cyclin D1 action encoded in the patterns of gene expression in human cancer. Cell. 2003;114:323–334. doi: 10.1016/s0092-8674(03)00570-1. [DOI] [PubMed] [Google Scholar]