Abstract
The heterodimeric transcription factor HIF (hypoxia-inducible factor) is central to the regulation of gene expression by oxygen. Three oxygen-dependent prolyl hydroxylase enzymes [PHD1 (prolyl hydroxylase domain 1), PHD2 and PHD3] control the abundance of HIF. In the presence of oxygen, they hydroxylate specific proline residues in HIF-α, allowing recognition by pVHL (von Hippel-Lindau protein) and subsequent ubiquitylation and proteasomal destruction. The precise roles and regulation of these enzymes are therefore of particular importance in understanding the physiological and pathological responses to hypoxia. In the present study, we define the existence of two species of PHD1 and provide evidence that they are generated by alternative translational initiation. We demonstrate that these alternative forms are both biologically active with similar HIF prolyl hydroxylase activity but that they differ in their responses to oestrogen, cell confluence and proteasomal inhibition. We show that the two PHD1 species are subject to proteolytic regulation but differ markedly in their protein stability. Though each isoform has the potential to interact with members of the Siah (seven in absentia homologue) ubiquitin ligase family, genetic studies indicated that other proteolytic mechanisms are responsible for control of stability under the conditions examined. The data define the existence of a further level of control in the pathway that regulates cellular responses to hypoxia.
Keywords: alternative initiation, hypoxia inducible factor (HIF), oxygen, prolyl hydroxylase domain 1 (PHD1), proteasomal inhibition, protein stability, Siah
Abbreviations: CODD, C-terminal oxygen-dependent degradation domain; DTT, dithiothreitol; EGLN2, Egl Nine like 2; HEK-293T cell, human embryonic kidney 293T cell; HIF, hypoxia-inducible factor; HUVEC, human umbilical-vein endothelial cells; IVTT, in vitro transcription/translation; mAb, monoclonal antibody; MEF, mouse embryonic fibroblast; NODD, N-terminal oxygen-dependent degradation domain; PHD1, prolyl hydroxylase domain 1; RING, really interesting new gene; Siah, seven in absentia homologue; siRNA, small interfering RNA; VHL, von Hippel-Lindau; pVHL, VHL protein
INTRODUCTION
The sensing of oxygen by cells and the resultant alterations in gene expression are critical for the maintenance of oxygen homoeostasis. The heterodimeric transcription factor HIF (hypoxia-inducible factor) is central to the regulation of gene expression by oxygen [1]. It consists of a constitutive β-subunit and a regulated α-subunit [2]. Oxygen-dependent prolyl hydroxylase enzymes control the abundance of the HIF-α subunits by hydroxylating two specific proline residues, generating hydroxyproline which permits the recognition and destruction by the product of the VHL (von Hippel-Lindau) gene and ubiquitin-mediated proteolysis [3–5]. Three 2-oxoglutarate-dependent dioxygenases have been identified that can catalyse this hydroxylation [6,7]. Named PHD1 (prolyl hydroxylase domain 1), PHD2 and PHD3 [6] {or alternatively EGLN2 (Egl Nine like 2), EGLN1 and EGLN3 [8]}, they differ in their tissue distribution and their relative contributions to cellular capacity for HIF-α hydroxylation [9–15]. These oxygen-sensing enzymes also show differences in regulation of their mRNA expression by hypoxia itself [6,10] and in their protein stability [16]. Although they share considerable homology in their catalytic domain, there are substantial differences in their N-terminal sequences but the function of these differing sequences is unclear. Differential expression and regulation may enable fine-tuning of the response to hypoxia under different tissues and physiological conditions and understanding how these processes operate has become an important focus in the analysis of the cellular responses to hypoxia.
In building an understanding of the likely roles and regulation of these oxygen-sensing enzymes, we have characterized their relative abundance and actions in a variety of cell lines [10]. During this analysis of the PHD enzymes, we observed that PHD1 commonly migrates as two species during electrophoresis [10]. Specific siRNA (small interfering RNA) oligonucleotides suppressed the expression of both forms, indicating that they were both the products of PHD1 mRNA [10]. In the present study, we characterize the different properties and mechanism of production of these two forms of PHD1. An alternative initiation AUG, encoding amino acid 34 of the predicted full-length protein, generates a shorter form that has equivalent HIF prolyl hydroxylase activity, but differs in its protein stability.
MATERIALS AND METHODS
Cell culture
The human cell lines MCF7, U2OS, HEK-293T (human embryonic kidney 293T), MEFs (mouse embryonic fibroblasts) and HUVEC (human umbilical-vein endothelial cells) were maintained in Dulbecco's modified Eagle's medium. BT474, ZR751, Susa, OVCAR3 and ND21 were cultured in RPMI 1640 and A549 in Ham's F-12 (Sigma). MEFs were derived from wild-type and Siah1a−/−/Siah2−/− mice [17] and were a gift from David Bowtell (Peter MacCallum Cancer Centre, Melbourne, Australia). Culture medium was supplemented with 10% (v/v) foetal calf serum, 2 mM L-glutamine, 50 i.u. (international units)/ml penicillin and 50 μg/ml streptomycin. HUVECs were additionally supplemented with 50 units/ml heparin and 30 μg/ml endothelial-cell growth supplement and MEFs were additionally supplemented with 0.1 mM non-essential amino acids, 0.15 mM 2-mercaptoethanol and 1 mM sodium pyruvate. Cells were treated with hypoxia in an Invivo2 400 Hypoxic workstation. For exposure to oestrogen, cells were incubated with oestradiol-17β (Sigma) at 10 ng/ml for 48 h with or without 25 μM of the proteasomal inhibitor MG132 (Biomol) for the last 8 h. Cells were examined at 80% confluence except in studies of the effects of confluence when they were examined at confluence (100%), 50% confluence and ‘over confluence’ (100%+) when they were growing beyond a single monolayer of cells.
Plasmids
The plasmid encoding wild-type PHD1 (pcDNA3 PHD1) has been described previously [6]. Mutations of this plasmid were generated using the QuikChange® site-directed mutagenesis kit (Stratagene) to encode alanine instead of methionine at amino acid 1 (pcDNA3 PHD1M1A) and amino acid 34 (pcDNA3 PHD1M34A), and confirmed by DNA sequencing. Plasmids encoding murine Siah2 (seven in absentia homologue 2) with a FLAG epitope tag (pcDNA3-FLAG-Siah2) and a RING (really interesting new gene) finger domain mutant version (pcDNA3-FLAG-Siah2Rm) were gifts from Dr Ze'ev Ronai (The Burnham Institute, La Jolla, CA, U.S.A.) [16]. A plasmid encoding FLAG-tagged human Siah1 (pcDNA3-FLAG-Siah1) was created by PCR using an IMAGE clone (ID: 5224755; MRC Geneservice, Geneservice Ltd, Cambridge, U.K.) as template. The forward and reverse primers used were: 5′-GGAATTCCACCATGGACTACAAGGACGATGACGACAAGAGCCGTCAGACTGCTC-3′ and 5′-GCTCTAGAGCTCAACACATGGAAATAGTTACATTGATGCCTAAATTGCC-3′. IVTT (in vitro transcription/translation) of plasmids was performed using TNT T7 Quick Coupled Reticulocyte or Wheatgerm lysate (Promega).
Immunoblotting
Cells were lysed in urea/SDS buffer [6.7 mM urea, 10 mM Tris/HCl, pH 6.8, 1 mM DTT (dithiothreitol), 10% (v/v) glycerol and 1% (w/v) SDS] supplemented with complete protease inhibitor cocktail (Roche). Protein content was determined using a Detergent Compatible Protein Assay kit (Bio-Rad). Whole cell lysates were resolved by SDS/PAGE, electroblotted on to PVDF membrane (Millipore) and probed by indicated antibodies. Horseradish peroxidase-conjugated anti-rabbit, goat or mouse secondary antibodies (Dako) were used with ECL® Plus system (Amersham Biosciences) to visualize immunoreactive bands. Primary antibodies used were PHD3; mAb (monoclonal antibody) 188e, PHD1; mAb 112 and polyclonal antibody P1.1af [10], Siah1 (sc-5505), Siah2 (sc-5507) (Santa Cruz Biotechnology), β-tubulin (T5293; Sigma), FLAG (M2, F3165; Sigma) and HIF-1α mAb (to detect human HIF-1α, H72320, clone 54; BD Transduction Laboratories; to detect murine HIF-1α, NB100-105; Novus).
Pulse–chase radiolabelling
ZR751 cells were seeded into 75 cm2 plates at 12×106 cells/plate and incubated with oestrogen for PHD1 induction for 2 days prior to radiolabelling. The cells were labelled in fresh culture medium with 200 μCi/ml [35S]methionine (Redivue Pro-mix; Amersham) for 30 min and then washed twice, followed by incubation for 0, 15, 30, 60 or 120 min. Cells were washed once with PBS and lysed in 400 μl of wash buffer (50 mM Tris/HCl, pH 7.5, 120 mM NaCl and 0.5% Nonidet P40) containing 1 mM DTT and complete protease inhibitor cocktails (Roche). Lysates were centrifuged at 14830 g for 10 min at 4 °C and the supernatants were used for immunoprecipitation. PHD1 mAb 112 and PK (an epitope widely used as a sequence tag) mAb (Serotec; as negative control) were prebound to Protein G–Sepharose (Sigma) by incubation at 4 °C rotating for 2 h and then washed. ZR751 lysates were incubated for 2 h on ice (with gentle vortex-mixing at 20 min intervals) with aliquots of the mAb/Protein G beads. The beads were washed with 0.8 ml of wash buffer six times and the immunoprecipitates were eluted by heating at 95 °C for 5 min in Laemmli sample buffer (20% glycerol, 2% SDS, 250 mM Tris, pH 6.8, 10% 2-mercaptoethanol and 0.001% Bromophenol Blue) before analysis by SDS/PAGE. The gel was exposed to a phosphoimager screen and 35S signals were quantified.
Cycloheximide chase
U2OS cells were seeded in 19.6 cm2 dishes at 8×105 cells/dish and incubated overnight, followed by transfection using FuGENE™ reagent (Roche). Plasmids used were pcDNA3 PHD1 (0.1 μg), pcDNA3 PHD1M1A (4 μg) and pcDNA3 PHD1M34A (0.2 μg), the amount of plasmid chosen to obtain equivalent levels of expressed protein, and cells were treated with FuGENE™/DNA mixtures for 10 h. Then cells from each dish were replated and subjected to hypoxic incubation at 0.1% oxygen for 16 h. Cells were treated with 0.1 mM cycloheximide for 0, 30, 60 or 90 min and then lysed in urea/SDS buffer. Protein levels of the transfected PHD1 species were determined by immunoblotting.
PHD1 activity assays
[35S]Methionine-radiolabelled wild-type and mutant PHD1 enzymes were produced in a rabbit reticulocyte lysate IVTT system as described above. Relative abundance of each translate was quantified by SDS/PAGE and autoradiography using a phosphoimager. Equimolar amounts of the enzymes were used in the following reaction. Reaction mixture contained 1 μM of an N-terminal biotinylated peptide corresponding to human HIF-1α residues 556–574, 50 mM Tris/HCl (pH 7.5), 2 mg/ml BSA, 1 mM DTT, 1 mM 2-oxoglutarate, 1 mM ascorbate, 50 μM iron(II) chloride and 0.3 mg/ml catalase. Reactions were performed at 37 °C and commenced by addition of the enzymes. Aliquots were removed at subsequent time points, the reaction was quenched by addition of 150 μM desferrioxamine and the peptide was captured using streptavidin-conjugated magnetic beads in Capture Buffer (150 mM NaCl, 20 mM Tris/HCl, pH 7.5, 0.5% Nonidet P40 and 100 μM desferrioxamine) followed by washing. The peptide-bound beads were then incubated at 4 °C with excess [35S]pVHL (VHL protein) produced by IVTT, diluted in the Capture Buffer. After two further washes, captured [35S]pVHL was quantified by SDS/PAGE and autoradiography. Reactions were performed in triplicate and results are given as means±S.E.M.
Reporter gene assays
MCF7 cells were seeded in 12-well plates (3.8 cm2/well) at 1.5× 105 cells/well and incubated for 16 h. The cells were then co-transfected using FuGENE™ 6 transfection reagent (Roche Applied Science) with plasmids expressing a Gal4-responsive luciferase reporter (pUAS.tk.Luc; 0.4 μg), β-galactosidase (pCMV-βGal; 0.12 μg), PHD1 enzymes (wild-type, 5 ng; M1A, 80 ng) and Gal4–HIF–VP16 fusions (where VP16 is a viral protein derived from herpes simplex virus). HIF domains encoded by the fusion plasmids were: HIF-1α/NODD (amino acids 344–553), HIF-1α/CODD-(554–698), HIF-2α/NODD-(345–517) and HIF-2α/CODD-(517–682) (where NODD is N-terminal oxygen-dependent degradation domain and CODD is C-terminal oxygen-dependent degradation domain). Gal4-HIF-VP16 plasmid doses were in the range 1–4 ng, based on preliminary titration experiments that aimed to generate approximately similar luciferase activities in cells not co-transfected with PHD1 plasmids. The doses of PHD1 plasmids were adjusted to achieve an approximately equimolar concentration of the enzymes in the cell lysates. Cell extracts were prepared 24 h after transfection in Passive Lysis Buffer (Promega) and analysed for β-galactosidase and luciferase activities as previously described [10]. The same cell extracts were used to examine expression levels of the PHD1 proteins by immunoblotting.
Suppression of PHD1 and Siah expression by siRNA
Cells were seeded in antibiotic-free medium into 6-well dishes (9.6 cm2/well) at 0.5×106 cells/dish and incubated overnight. siRNAs were transfected using Oligofectamine (Invitrogen) twice at 24 h intervals at a dose of 100 nM for single siRNA transfection and 50 nM of each oligonucleotide for combined Siah1 and Siah2 suppression. When indicated, hypoxic incubation (1% O2 for 16 h) or oestrogen induction (10 ng/ml oestradiol-17β for 2 days) was performed subsequently. siRNA sequences targeting Drosophila melanogaster HIF (negative control) and PHD1 have been described previously [10]. Sequences designed to suppress expression of Siah1 (M-012598-01) and Siah2 (M-006561-01) were purchased from Dharmacon.
RESULTS
Two species of PHD1
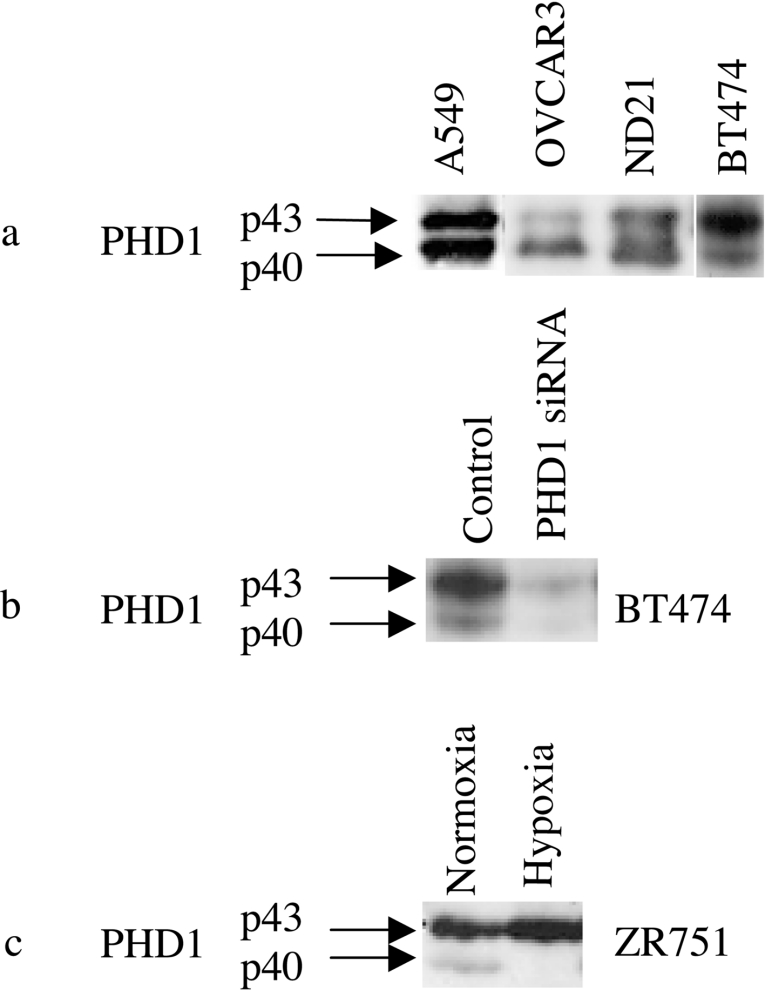
In a previous study, immunoblotting to detect PHD1 in a wide variety of cell lines revealed a predominant species at the predicted molecular mass (PHD1p43) and an additional species running at a slightly increased mobility (PHD1p40) [10]. We surveyed further cell lines to examine for the occurrence of these two forms and representative immunoblots from four different cell lines are shown in Figure 1(a). Both species were seen in cancer cells derived from a variety of different tissues and from HUVEC cells. PHD1p40 was more prominent in certain cell lines (e.g. OVCAR3) but was less abundant than the slower mobility species in others (e.g. BT474). Two different, specific siRNA oligonucleotides suppressed the expression of both forms, indicating that they were products derived from PHD1 mRNA (Figure 1b and results not shown). Under hypoxic conditions in many cells there was reduction in the level of PHD1 protein, which was usually more marked for the faster mobility species (PHD1p40) (see for example Figure 1c).
Figure 1. Expression of two species of PHD1 protein in different cell lines.
(a) Immunoblot of cell extracts from A549, OVCAR3, ND21 and BT474 cells detecting PHD1. A doublet of two species labelled p43 and p40 is seen. (b) Immunoblot detecting PHD1 in cell extracts from BT474 cells treated with control and PHD1 siRNA oligonucleotides, showing suppression of both PHD1 species by PHD1 siRNA. (c) Immunoblot detecting PHD1 in cell extracts from ZR751 cells under normoxic conditions and after 16 h of hypoxia (1% O2), showing a reduction in expression of the faster mobility species (p40) under hypoxia.
Mechanism for generation of two PHD1 species
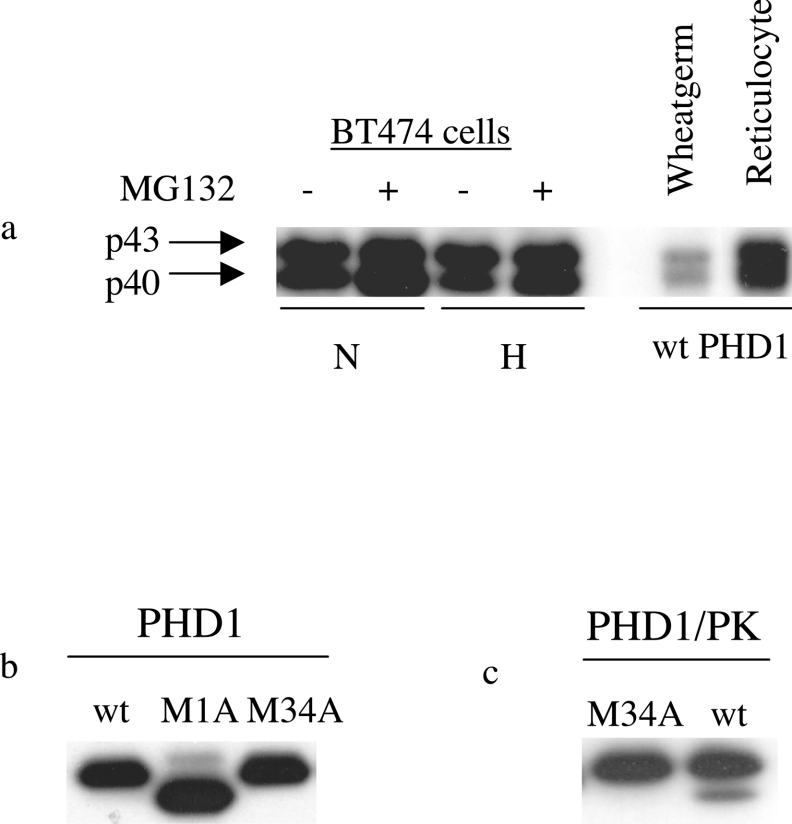
We first considered whether the two protein species might be generated by alternative splicing. However, a bioinformatic analysis of mRNA expression databases did not reveal any commonly expressed mRNA transcripts generated by alternative splicing that would produce protein products of this or other sizes, in keeping with previous analyses [8,9]. Furthermore, expression of the PHD1 cDNA in reticulocyte lysate IVTT systems was capable of generating two forms (see Figure 2a), arguing against a role for splicing in their generation. We also expressed the PHD1 cDNA in wheatgerm lysate and again generated two similarly sized forms, arguing against a role for mammalian post-translational modification in their production. When generated by reticulocyte or wheatgerm IVTT, the PHD1 species were of identical mobility as those detected in BT474 cells by immunoblotting of a single gel (Figure 2a).
Figure 2. Endogenous expression of the two forms of PHD1 and comparison with IVTT syntheses of wild-type, PHD1M1A and PHD1M34A proteins.
(a) Immunoblot detecting PHD1 in cell extracts from BT474 cells treated with normoxia (21% O2) or hypoxia (1% O2) for 16 h with or without the proteasomal inhibitor MG132 (25 μM) and of IVTT syntheses by rabbit reticulocyte lysate or wheatgerm extract programmed with plasmid encoding wild-type PHD1 (pcDNA3 PHD1). Two protein species are detected with similar mobility in cell extract and IVTT syntheses. The faster mobility species (p40) shows increased levels with proteasomal inhibition but reduced levels under hypoxic conditions. (b) Immunoblot detecting PHD1 in rabbit reticulocyte lysate programmed with plasmids encoding wild-type PHD1 (wt, pcDNA3 PHD1) or mutated PHD1 (M1A, pcDNA3 PHD1M1A or M34A, pcDNA3 PHD1M34A). (c) Immunoblot detecting PK in rabbit reticulocyte lysate programmed with plasmids encoding wild-type PHD1 with a C-terminal PK epitope tag (pcDNA3 PHD1PK) and a mutated version (pcDNA3 PHD1M34APK).
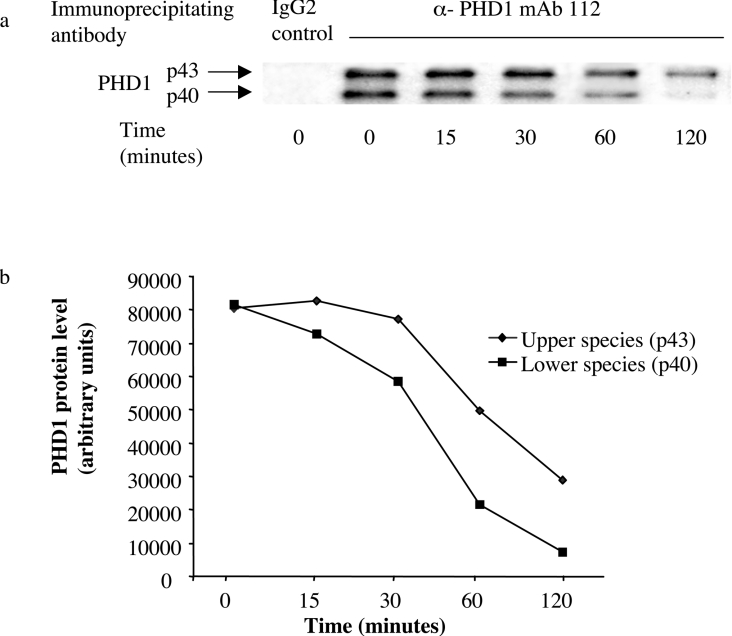
We next considered whether the two species might arise by post-translational processing. In order to examine for a precursor–product relationship between the two species, we undertook pulse–chase labelling studies using [35S]methionine. Both PHD1 species were detected at the earliest time point of the experiment and both showed a monotonic decay of protein level with PHD1p40 showing a more rapid rate of decay (Figure 3), suggesting that there is no precursor–product relationship between the two isoforms.
Figure 3. Pulse–chase study examining for a precursor–product relationship between the two species of PHD1.
(a) Pulse–chase radiolabelling studies of PHD1. ZR751 cells were treated with [35S]methionine/cysteine for 30 min followed by incubation for 0, 15, 30, 60 or 120 min and immunoprecipitation of cell extract was then performed with the anti-PHD1 antibody mAb 112 or a control antibody. The immunoprecipitates were separated by SDS/PAGE, and 35S signals of the upper (p43) and lower (p40) species were quantified by a phosphoimager screen and are displayed graphically in (b).
An inspection of the PHD1 mRNA sequence revealed a plausible alternative initiation codon corresponding to methionine amino acid 34 and which is conserved in rat and mouse PHD1 cDNA sequences. This mRNA sequence GCUGCCAUGGA corresponds precisely to an optimum Kozak initiation sequence, with preservation of a guanine at position +4 and a purine at −3 [18]. Initiation at this AUG would produce a protein of predicted molecular mass 40.2 kDa, in comparison with 43.7 kDa for a protein generated by initiation at the upstream ATG codon, consistent with the sizes of the two species we had observed.
In order to test the hypothesis that this sequence can act as an alternative initiating ATG codon, we introduced mutations of the PHD1 cDNA by site-directed mutagenesis at the first initiating ATG (M1A) and at the potential alternative initiation site (M34A). We examined the production of proteins by these DNA sequences in rabbit reticulocyte lysate IVTT syntheses. As seen in Figure 2(b), mutation of the first putative initiating ATG codon led to the production of a shorter form of PHD1 protein of a mobility consistent with that of PHD1p40 seen in cell extracts, while abolishing the generation of the slower mobility species. This was seen when protein was detected both by immunoblotting (Figure 2b) and by incorporation of [35S]methionine (results not shown). Consistent with this result, mutation of the putative internal initiating codon (M34A) generated the slower mobility form only, with no faster mobility form being generated (Figure 2b). This was also true for proteins similarly generated to contain a C-terminal PK tag, with both species retaining PK immunogenicity (Figure 2c). These results support the hypothesis that the faster mobility species of PHD1 is generated by alternative internal translational initiation. The production of the faster mobility form of PHD1 from a plasmid encoding wild-type PHD1 by IVTT was variable, probably reflecting strong initiation at the initial ATG codon under these conditions. Furthermore, transient transfection of plasmids encoding these mutated sequences into MCF7 cells produced the same pattern of proteins with similar mobilities to the endogenous species (see for example Figures 2d and 6). These results further reinforce the hypothesis that the two differing mobility species of PHD1 arise from alternative translational initiation.
Figure 6. Prolyl hydroxylase activity of wild-type, PHD1M1A and PHD1M34A enzymes.
Proteins synthesized in rabbit reticulocyte lysate were assayed for their ability to convert a HIF-1α peptide (HIF-1α residues 556–574) into a VHL-binding form. PHD1 enzymes were produced in a rabbit reticulocyte lysate IVTT and relative abundance of each protein was quantified by SDS/PAGE and autoradiography using a phosphoimager and equimolar amounts of the enzymes were then used in the assay. The N-terminal biotinylated peptide corresponding to human HIF-1α residues 556–574 was treated with the enzymes and subsequent binding to 35S-radiolabelled VHL protein was quantified by SDS/PAGE and autoradiography. Reactions were performed in triplicate and results are given as means±S.D. No statistically significant differences were seen between the enzymatic activities of the different forms of PHD1.
Regulation of the different PHD1 species
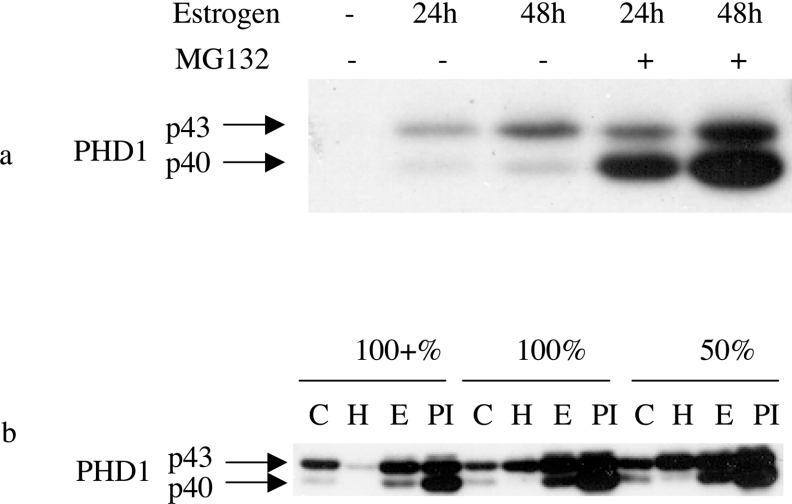
We next wished to examine the influence of different conditions on the relative abundance of the PHD1 isoforms. We and others have previously described induction of PHD1 levels by oestrogen [10,13]. Oestrogen exposure caused a substantial induction of both isoforms of PHD1 in both ZR751 (Figure 4a) and BT474 cells (results not shown). During the course of these experiments, we noticed that the shorter isoform PHD1p40 was more readily detected under conditions of decreased cell confluence. We therefore immunoblotted equivalent amounts of protein extracts from cells that were at varying confluence (Figure 4b). Both PHD1 forms were increased at lower confluence but the proportion of the PHD1p40 was increased. As had been seen in previous experiments (see for example Figure 1c), there was repression of expression of PHD1p40 by hypoxia and enhancement of the level of PHD1p43. Proteasomal inhibition with MG132 also enhanced the levels of both forms of PHD1 but was more marked for the faster mobility form, p40, and had an additive effect when combined with oestrogen exposure (see Figure 4a).
Figure 4. Expression of the two species of PHD1 in response to oestrogen, proteasomal inhibition, hypoxia and cellular confluence.
(a) Immunoblot detecting the two species of PHD1 (p43 and p40) in cell extracts from ZR751 cells treated with oestrogen (10 ng/ml oestradiol-17β) for 24 or 48 h as indicated and/or the proteasomal inhibitor MG132 (25 μM) for the last 4 h. (b) Immunoblot detecting the two species of PHD1 (p43 and p40) in cell extracts from ZR751 cells cultured at varying levels of confluence (50%, 100% and 100%+) and treated with hypoxia (‘H’; 1% O2) for 16 h, oestrogen (‘E’; 10 ng/ml oestradiol-17β) for 48 h, or oestrogen for 48 h together with proteasomal inhibition for the last 4 h of incubation (‘PI’; 25 μM MG132). ‘C’, control.
There are several examples of alternative initiation of translation, generating protein products with differing cellular localization. One such example occurs in the HIF system itself where the alternate internal initiation of VHL produces a protein product with enhanced nuclear localization [19]. However, we were unable to see any significant differences in localization between the two forms of PHD1 (results not shown).
Differing protein stability of the two PHD1 species
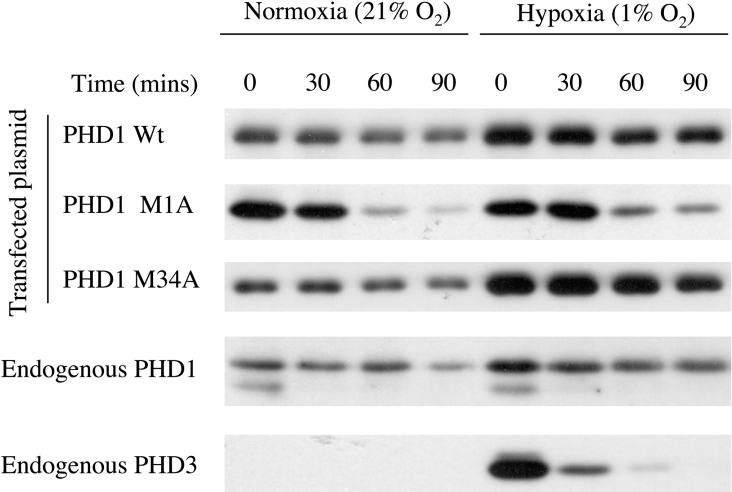
Since pulse–chase experiments had suggested differential half-life, we examined the stability of the two forms of PHD1 under normoxia and hypoxia by transfecting U2OS cells with plasmids encoding M1A and M34A and examining decay of product level in cycloheximide chase. In keeping with the observations on endogenous PHD1, the slower mobility PHD1 form p43 generated by transfection of M34A showed a significantly enhanced stability when compared with the faster mobility form generated by M1A and mirrored the degradation of the endogenous forms of PHD1 (see Figure 5). The difference between the stability of the two isoforms of PHD1 was not significantly affected by hypoxia. However, the greater stability of the slower mobility form of PHD1 when combined with reduction in mRNA expression in hypoxia can provide an explanation for the differential regulation of the two forms under hypoxic conditions.
Figure 5. Cycloheximide chase examining the protein stability of the two species of PHD1 (p43 and p40) under hypoxic and normoxic conditions.
U2OS cells were transfected with plasmids encoding wild-type or mutant versions of PHD1 (Wt, pcDNA3 PHD1; M1A, pcDNA3 PHD1M1A; or M34A, pcDNA3 PHD1M34A) or no plasmid and treated with normoxia (21% O2) or hypoxia (1% O2) for 16 h prior to treatment with 0.1 mM cycloheximide for 0, 30, 60 or 90 min. Endogenous and transfected protein levels of PHD1 and PHD3 were detected by immunoblotting.
Biological activity of the two species of PHD1
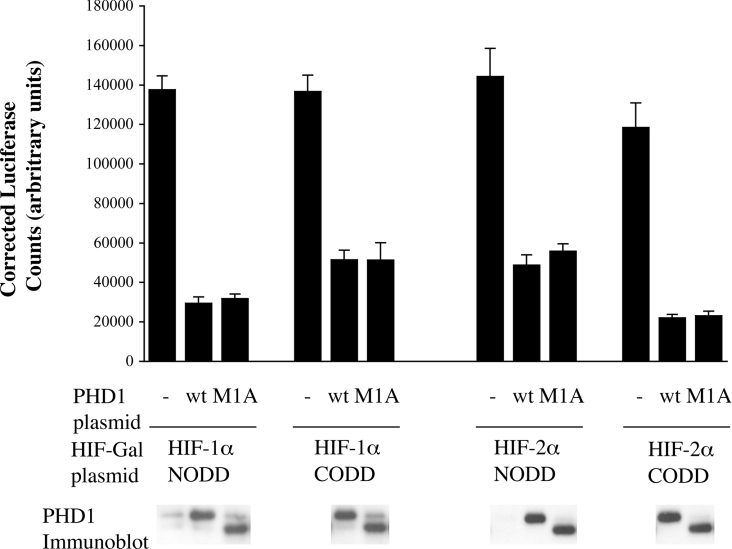
Given the different protein sequences of the two PHD1 isoforms, we wished to establish whether they possessed differing HIF hydroxylase activity. To analyse the HIF prolyl hydroxylase activity of the different PHD forms, we undertook in vitro assays of enzyme activity utilizing a VHL capture assay [5,20]. The different forms of PHD1 were generated in rabbit reticulocyte lysate IVTT, programmed with the PHD1M1A- and PHD1M34A-encoding plasmids. Both forms of the enzyme were able to convert the 19-residue HIF-1α CODD peptide into a hydroxylated VHL binding form with similar efficacy (see Figure 6). Enzyme generated by PHD1M1A had a slightly greater specific activity but this did not reach statistical significance. However, measurements of in vitro activity might not capture all influences on enzyme activity that could occur in vivo between different isoforms due to differing intracellular localization or association with other proteins. Furthermore, different activities might be seen with different HIF isoform substrates. To examine whether the different forms of PHD1 had differing effects on HIF-1α and HIF-2α in cells, we undertook transient transfection assays to examine the ability of the enzymes to suppress the activity of a GAL–HIF-α fusion protein on the expression of a UAS-TK luciferase reporter (Gal4-responsive luciferase reporter as described in the Materials and methods section) gene. Again, both isoforms proved equally active in regulating the different HIF isoforms and their different domains (see Figure 7). They had equivalent levels of activity on peptides derived from HIF-1α and HIF-2α which contained either of the two regulated proline residues.
Figure 7. Effect of PHD1 expression on the activity of Gal4 fusion proteins bearing isolated HIF-α degradation domains.
MCF7 cells were co-transfected with plasmids expressing (i) a Gal4 reporter gene, (ii) β-galactosidase, (iii) the indicated HIF-α ODD (oxygen-dependent degradation domain) sequence as a Gal4–HIF–VP16 fusion protein [HIF-1α/NODD (amino acids 344–553), HIF-1α/CODD-(554–698), HIF-2α/NODD-(345–517) or HIF-2α/CODD-(517–682)] and (iv) wild-type PHD1 (wt, pcDNA3 PHD1) or a mutated version of PHD1 (M1A, pcDNA3 PHD1M1A) as indicated. Luciferase activities were analysed and corrected for transfection efficiency by β-galactosidase activity. Immunoblot analysis of expressed PHD1 protein levels are shown. Experiments were performed in triplicate and results are given as means±S.D. No statistically significant differences were seen between the actions of the different forms of PHD1.
Regulation of PHD1 by Siah E3 ligases
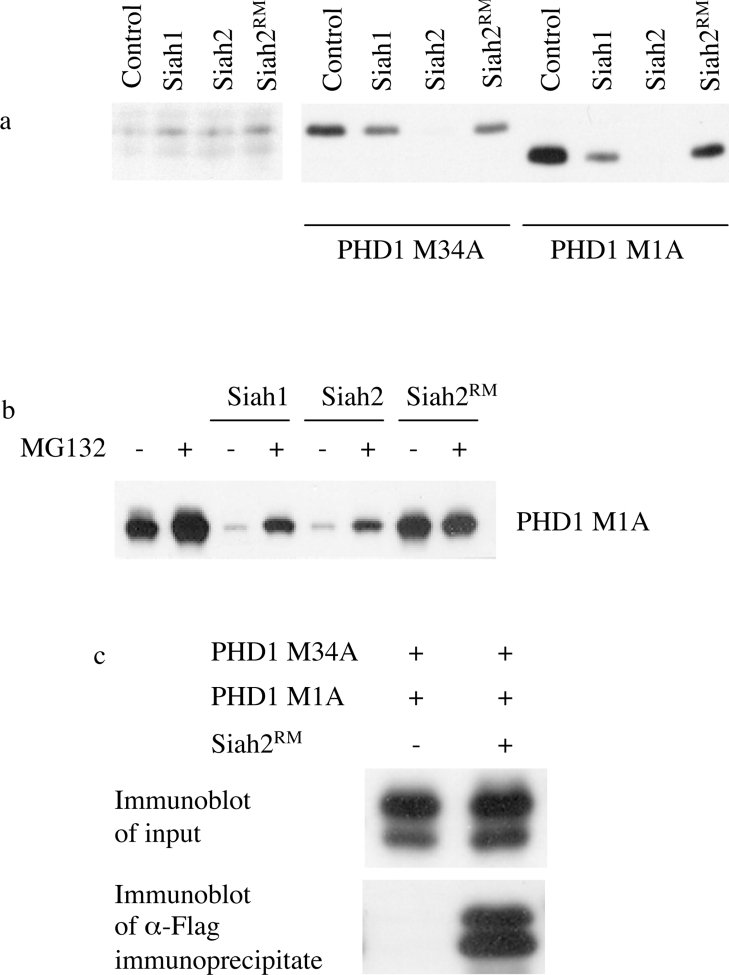
Recently, the Siah ubiquitin ligases have been implicated in the regulation of protein stability of the homologous HIF hydroxylase, PHD3 [16]. In this work, the authors also reported an association between exogenously expressed PHD1 and Siah2 and suggested enhanced degradation of exogenously generated PHD1 by exogenous Siah2 under hypoxic conditions, although effects on endogenous PHD1 were not examined. To examine for the role of the Siah ubiquitin ligases in endogenous PHD stability and to explore the mechanism of greater instability of PHD1p40, we studied the effects of Siah1 and Siah2 on the levels of expression of PHD1. Plasmids encoding PHD1M1A or PHD1M34A were transfected into MCF7 cells with FLAG-tagged, Siah-encoding expression plasmids. We found a marked reduction in the expression of both forms of PHD1 (PHD1M1A and PHD1M34A) when co-transfected with Siah2. Siah1 produced a small reduction in PHD1 expression, whereas the inactive RING finger domain mutant form Siah2Rm had little or no effect (see Figure 8a). This reduction in expression of both species of PHD1 was significantly abrogated in the presence of proteasomal inhibition (see Figure 8b). However, in contrast, the endogenous levels of PHD1p40 and PHD1p43 were unaffected. This result is consistent with a role for Siah1 and Siah2 as E3 ligases capable of contributing to the proteasomal destruction of overexpressed PHD1. As reported by Nakayama et al. [16], we found it difficult to detect the expression of Siah2 following transfection and therefore utilized a plasmid encoding the RING finger domain mutant Siah2Rm that lacks E3 ligase activity and self-ubiquitination in HEK-293T cells for further studies of associations between PHD1 and Siah2. To examine for the association between PHD1 and Siah2, HEK-293T cells were transfected with plasmids encoding PHD1M1A and PHD1M34A and with plasmid encoding FLAG–Siah2Rm. Cell extracts were immunoprecipitated with an anti-FLAG antibody and the immunoprecipitate was immunoblotted with a PHD1 antibody. A specific association was seen between Siah2Rm and PHD1 (see Figure 8b). However, the slower mobility form of PHD1 was predominant in the input, while the faster mobility form dominated in the immunoprecipitate, indicating a preferential association of Siah2 with the faster mobility form of PHD1 under these conditions and a potential mechanism for the greater instability of PHD1p40 when overexpressed.
Figure 8. Effect of the Siah E3 ligase overexpression on PHD1 abundance.
(a) Plasmids encoding PHD1M1A (pcDNA3 PHD1M1A; 1600 ng) or PHD1M34A (pcDNA3 PHD1M34A; 100 ng) were transfected into MCF7 cells with FLAG-tagged, Siah-encoding expression plasmids (pcDNA3-FLAG-Siah1, pcDNA3-FLAG-Siah2 or pcDNA3-FLAG-Siah2Rm; 100 ng). The abundance of the different forms of endogenous and transfected PHD1 was detected by immunoblotting. Siah2 (and to a lesser extent Siah1) decreased the protein levels of both forms of transfected PHD1. (b) Similar experiments were undertaken to examine effects of Siah expression on PHD1M1A in the absence and presence of the proteasomal inhibitor, MG132, in U2OS cells. (c) HEK-293T cells were co-transfected with plasmids encoding PHD1M1A (pcDNA3 PHD1M1A) or PHD1M34A (pcDNA3 PHD1M34A) and with or without plasmid encoding FLAG–Siah2Rm (pcDNA3-FLAG-Siah2Rm). A specific association was seen between Siah2Rm and PHD1. However, the slower mobility species of PHD1 was predominant in the input, while the faster mobility form dominated in the immunoprecipitate.
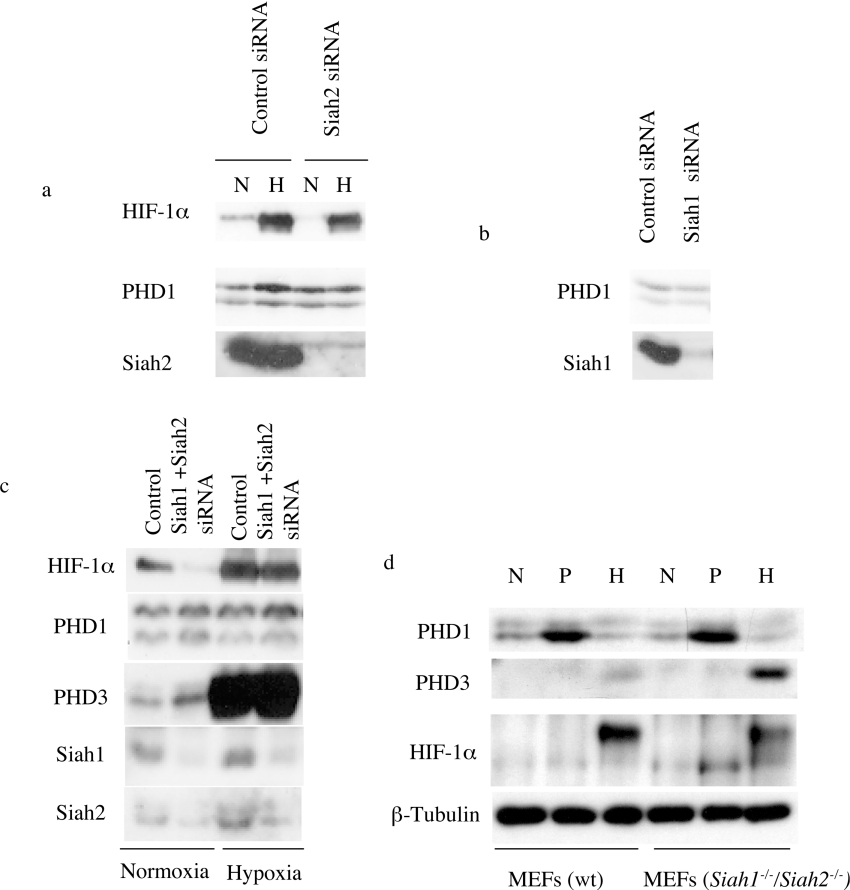
To examine whether this enhanced association could mediate the increased instability and proteasomal degradation of PHD1p40, we undertook suppression of Siah2 with siRNA transfection and examined the endogenous levels of the PHD proteins. Despite achieving substantial reduction in Siah2 protein levels, no effect was seen on the abundance of either PHD1 isoform under hypoxic or normoxic conditions in MCF7 (see Figure 9a), U2OS or ZR731 cells (results not shown). Given the absence of effect of Siah2 on PHD abundance under these conditions, we undertook similar experiments to examine for a role of Siah1. Again, despite significant suppression of Siah1, no effect was seen on the levels of either form of PHD1 (see Figure 9b). In case of functional redundancy in the action of the Siah E3 ligases, we undertook combined siRNA-mediated suppression of Siah1 and Siah2 and observed a moderate increase in endogenous PHD3, a decrease in the levels of HIF-1α but no convincing effect on levels of PHD1p40 (Figure 9c).
Figure 9. Effect of Siah suppression or deficiency on PHD1 expression.
MCF7 cells were treated with Siah-targeted siRNA oligonucleotides to suppress the level of Siah1 (a), Siah2 (b) or both Siah1 and Siah2 (c) and extracts were examined for PHD1, HIF-1α, PHD3 and Siah protein expression as indicated. (d) MEFs derived from Siah1a−/−/Siah2−/− or wild-type (wt) mice were treated with normoxia (‘N’; 21% O2), proteasomal inhibition (‘P’; 25 μM MG132) or hypoxia (‘H’; 5% O2) for 5 h. Cell extracts were examined for PHD1, HIF-1α, PHD3 and β-tubulin expression by immunoblotting.
To explore further the role of Siah E3 ligases in the regulation of endogenous PHD1 and PHD3, we examined the MEFs studied by Nakayama et al. [16] that are genetically deficient in both Siah1a and Siah2. Consistent with their results, we again observed greater levels of PHD3 and reduced hypoxic induction of HIF-1α in the Siah1a−/−/Siah2−/− cells when compared with wild-type MEFs (see Figure 9d) (although in contrast with their report, some hypoxic induction of HIF-1α was preserved in the double-knockout cells). In this murine cell, we could also detect a doublet corresponding to PHD1, consistent with the generation of two forms of PHD1 by alternative initiation from the conserved internal methionine and a marked increase in the levels of the faster mobility species with proteasomal inhibition (Figure 9d). However, there was no substantial difference in the relative or absolute amounts of the two forms between the wild-type and Siah1a−/−/Siah2−/− cells, arguing against a role for Siah1a or Siah2 in the control of levels of the two species of PHD1 in these cells under these conditions.
DISCUSSION
The identification of three enzymes, PHD1, PHD2 and PHD3, as prolyl hydroxylases that promote VHL-mediated proteolytic destruction of HIF-α has provided important insights into the mechanisms by which oxygen regulates gene expression. Of particular interest is developing an understanding of processes that might modulate the rate of HIF hydroxylation so as to enable a flexible response to hypoxia. In this respect, the regulation of the abundance of the PHD enzymes has been a focus of attention, since changes in the amount of available enzyme will alter the rate of hydroxylation at a given oxygen concentration. To date, two processes have been identified that regulate the levels of these HIF prolyl hydroxylases themselves: transcriptional regulation and proteolytic regulation. Thus PHD2 and PHD3 are subject to transcriptional induction by hypoxia that is mediated at least in part by the HIF system [6,14,21–24]. Studies of proteolysis have mainly focused on PHD3, and have demonstrated important regulation by the Siah1/2 ubiquitin ligases [16]. Here we extend these insights by demonstrating that PHD1 is produced as two isoforms by alternative translational initiation and that it is also subject to proteolytic regulation.
The shorter species of PHD1 (PHD1p40) produced by internal initiation at M34 had a substantially shorter half-life than the longer species (PHD1p43) (∼50 versus 100 min). Although the half-life of PHD1 protein isoforms was not itself regulated by hypoxia, their differential stability, combined with suppression of PHD1 mRNA expression in hypoxia, resulted in enhanced suppression of PHD1p40 following exposure of cells to hypoxia. Interestingly, overexpression of either Siah1 or Siah2 ubiquitin ligases but not a RING finger domain mutant form of Siah2 (Siah2Rm) reduced the levels of both species of transfected PHD1. Furthermore, association studies of transfected proteins demonstrated preferential association of the stabilized RING finger domain mutant Siah2 (Siah2Rm) with the shorter species of PHD1. Nevertheless, steady-state levels of PHD1 isoforms were identical in wild-type and Siah1a−/−/Siah2−/− MEFs, though, as previously reported [16], PHD3 levels were substantially enhanced in Siah1/2 deficient cells. We were also unable to demonstrate alterations in levels of either PHD1 isoform in human cell lines subject to efficient siRNA-mediated suppression of Siah1/2. Taken together, this suggests that although both PHD1 isoforms can potentially be targeted by Siah1/2 ubiquitin ligases, these particular pathways do not contribute to the setting of steady-state levels of PHD1 under the conditions of our experiments, and that other proteolytic pathways are probably responsible for the differential stability of the PHD1 isoforms under these conditions.
Despite different stability and differing regulation by conditions such as changes in cell confluence, the PHD1 isoforms had very similar activity on the HIF system, raising the intriguing possibility that differential isoform regulation might influence activity on another, non-HIF, substrate. The functional relevance of the different PHD1 isoforms is supported by the high degree of mammalian conservation of both the internal initiation site and the N-terminal sequences of PHD1. In this respect, it is also interesting that whereas PHD2 and PHD3 are strongly induced by hypoxia, PHD1 (and PHD1p40 in particular) is suppressed by hypoxia, suggesting a fundamentally different role in the hypoxic response. Whether or not this is the case, the current identification and characterization of two different isoforms of the PHD1 protein indicates the existence of a further level of control acting on the cellular response to hypoxia, and opens new avenues for analysis.
Acknowledgments
We thank Christopher Pugh (University of Oxford, Oxford, U.K.) for helpful discussions, Dr Ze'ev Ronai and David Bowtell for generous provision of reagents and the Wellcome Trust for funding this work.
References
- 1.Kaelin W. G. J. How oxygen makes its presence felt. Genes Dev. 2002;16:1441–1445. doi: 10.1101/gad.1003602. [DOI] [PubMed] [Google Scholar]
- 2.Wang G. L., Semenza G. L. Purification and characterisation of hypoxiainducible factor 1. J. Biol. Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell P. H., Wiesener M. S., Chang G.-W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature (London) 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 4.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G. J. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P., Mole D. R., Tian Y.-M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A. V., Hebestreit H. F., Mukherji M., Schofield C. J., et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 6.Epstein A. C. R., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., et al. C. elegans EGL-9 and mammalian homologues define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 7.Bruick R. K., McKnight S. L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 8.Taylor M. S. Characterization and comparative analysis of the EGLN gene family. Gene. 2001;275:125–132. doi: 10.1016/s0378-1119(01)00633-3. [DOI] [PubMed] [Google Scholar]
- 9.Hirsila M., Koivunen P., Gunzler V., Kivirikko K. I., Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor HIF. J. Biol. Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 10.Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 11.Dupuy D., Aubert I., Duperat V. G., Petit J., Taine L., Stef M., Bloch B., Arveiler B. Mapping, characterization, and expression analysis of the SM-20 human homologue, c1orf12, and identification of a novel related gene, SCAND2. Genomics. 2000;69:348–354. doi: 10.1006/geno.2000.6343. [DOI] [PubMed] [Google Scholar]
- 12.Wax S. D., Tsao L., Lieb M. E., Fallon J. T., Taubman M. B. SM-20 is a novel 40-kd protein whose expression in the arterial wall is restricted to smooth muscle. Lab. Invest. 1996;74:797–808. [PubMed] [Google Scholar]
- 13.Erez N., Milyavsky M., Goldfinger N., Peles E., Gudkov A. V., Rotter V. Falkor, a novel cell growth regulator isolated by a functional genetic screen. Oncogene. 2002;21:6713–6721. doi: 10.1038/sj.onc.1205867. [DOI] [PubMed] [Google Scholar]
- 14.Cioffi C. L., Liu X. Q., Kosinski P. A., Garay M., Bowen B. R. Differential regulation of HIF-1α prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells. Biochem. Biophys. Res. Commun. 2003;303:947–953. doi: 10.1016/s0006-291x(03)00453-4. [DOI] [PubMed] [Google Scholar]
- 15.Oehme F., Ellinghaus P., Kolkhof P., Smith T. J., Ramakrishnan S., Hutter J., Schramm M., Flamme I. Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem. Biophys. Res. Commun. 2002;296:343–349. doi: 10.1016/s0006-291x(02)00862-8. [DOI] [PubMed] [Google Scholar]
- 16.Nakayama K., Frew I. J., Hagensen M., Skals M., Habelhah H., Bhoumik A., Kadoya T., Erdjument-Bromage H., Tempst P., Frappell P. B., et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell (Cambridge, Mass.) 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Frew I. J., Hammond V. E., Dickins R. A., Quinn J. M., Walkley C. R., Sims N. A., Schnall R., Della N. G., Holloway A. J., Digby M. R., et al. Generation and analysis of Siah2 mutant mice. Mol. Cell. Biol. 2003;23:9150–9161. doi: 10.1128/MCB.23.24.9150-9161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 19.Iliopoulos O., Ohh M., Kaelin W. G., Jr pVHL19 is a biologically active product of the von Hippel-Lindau gene arising from internal translation initiation. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11661–11666. doi: 10.1073/pnas.95.20.11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuckerman J. R., Zhao Y., Hewitson K. S., Tian Y. M., Pugh C. W., Ratcliffe P. J., Mole D. R. Determination and comparison of specific activity of the HIF-prolyl hydroxylases. FEBS Lett. 2004;576:145–150. doi: 10.1016/j.febslet.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 21.D'Angelo G., Duplan E., Boyer N., Vigne P., Frelin C. Hypoxia up-regulates prolyl hydroxylase activity. J. Biol. Chem. 2003;278:38183–38187. doi: 10.1074/jbc.M302244200. [DOI] [PubMed] [Google Scholar]
- 22.del Peso L., Castellanos M. C., Temes E., Martin-Puig S., Cuevas Y., Olmos G., Landazuri M. O. The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J. Biol. Chem. 2003;278:48690–48695. doi: 10.1074/jbc.M308862200. [DOI] [PubMed] [Google Scholar]
- 23.Metzen E., Stiehl D. P., Doege K., Marxsen J. H., Hellwig-Burgel T., Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem. J. 2005;387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landazuri M. O., Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]