Abstract
The Rac-specific GEF (guanine-nucleotide exchange factor) Tiam1 (T-lymphoma invasion and metastasis 1) regulates migration, cell–matrix and cell–cell adhesion by modulating the actin cytoskeleton through the GTPase, Rac1. Using yeast two-hybrid screening and biochemical assays, we found that Tiam1 interacts with the p21-Arc [Arp (actin-related protein) complex] subunit of the Arp2/3 complex. Association occurred through the N-terminal pleckstrin homology domain and the adjacent coiled-coil region of Tiam1. As a result, Tiam1 co-localizes with the Arp2/3 complex at sites of actin polymerization, such as epithelial cell–cell contacts and membrane ruffles. Deletion of the p21-Arc-binding domain in Tiam1 impairs its subcellular localization and capacity to activate Rac1, suggesting that binding to the Arp2/3 complex is important for the function of Tiam1. Indeed, blocking Arp2/3 activation with a WASP (Wiskott–Aldrich syndrome protein) inhibitor leads to subcellular relocalization of Tiam1 and decreased Rac activation. Conversely, functionally active Tiam1, but not a GEF-deficient mutant, promotes activation of the Arp2/3 complex and its association with cytoskeletal components, indicating that Tiam1 and Arp2/3 are mutually dependent for their correct localization and signalling. Our data suggests a model in which the Arp2/3 complex acts as a scaffold to localize Tiam1, and thereby Rac activity, which are both required for activation of the Arp2/3 complex and further Arp2/3 recruitment. This ‘self-amplifying’ signalling module involving Tiam1, Rac and the Arp2/3 complex could thus drive actin polymerization at specific sites in cells that are required for dynamic morphological changes.
Keywords: actin, Arp2/3 complex, Wiskott–Aldrich syndrome protein (WASP), p21-Arc, Rac1, T-lymphoma invasion and metastasis 1 (Tiam1)
Abbreviations: 3AT, 3-aminotriazole; Arc, Arp complex; Arp, actin-related protein; CC, coiled-coil; Cdc42, cell division cycle 42; CRIB, Cdc42/Rac interacting binding; DH, Dbl homology; Ex, extended; FL, full-length; GEF, guanine-nucleotide exchange factor; GST, glutathione S-transferase; HA, haemagglutinin; IRSp53, insulin receptor substrate p53; JIP, c-Jun N-terminal kinase-interacting protein; MDCK, Madin–Darby canine kidney cells; PAK, p21-activated kinase; PH, pleckstrin homology; PHn, N-terminal PH domain; PIR121, p53-inducible mRNA with an Mr of 140000; Tiam1, T-lymphoma invasion and metastasis; WASP, Wiskott–Aldrich syndrome protein; WAVE, WASP-family verprolin homologous protein
INTRODUCTION
The Rho-like GTPases, Rac1 and Cdc42 (cell division cycle 42), are involved in migration and adhesion by modulating the actin cytoskeleton of cells [1]. In order to accomplish this, these GTPases must be activated by GEFs (guanine-nucleotide exchange factors) such as Dbl, Tiam1 (T-lymphoma invasion and metastasis 1) or β-Pix [2–4]. Consequently, Cdc42 and Rac1 can stimulate actin polymerization mediated by the Arp2/3 complex, resulting in the formation of filopodia or lamellipodia respectively [5–8].
The Arp2/3 complex consists of seven proteins, Arp2 (actinrelated protein 2) and Arp3, and the Arc (Arp complex) proteins, p41-Arc, p34-Arc, p21-Arc, p20-Arc and p16-Arc [9]. The Arp2/3 complex forms an actin nucleation site enabling the de novo polymerization of actin filaments. In order to induce actin polymerization, the Arp2/3 complex has to be activated by WASP (Wiskott–Aldrich syndrome protein) family members such as WASP, neural-WASP and WAVE (WASP-family verprolin homologous protein)1–3 or by PAKs (p21-activated kinases) [7,10–14]. Activated Cdc42 can bind and activate WASP via its effector loop [14], whereas Rac can activate WAVE by binding to PIR121 (p53-inducible mRNA with an Mr of 140000), resulting in the dissociation of WAVE from its trans-inhibitory binding partners NAP125 (Nck-associated protein with an Mr of 125000) and PIR121 [15]. In addition, WAVE has been shown to bind to Rac1 via IRSp53 (insulin receptor substrate p53), forming a trimeric protein complex [16].
The Rac-specific GEF, Tiam1 [4,17], controls E-cadherin-mediated cell–cell adhesion and cell migration by regulating the actin cytoskeleton [18–20]. Like all GEFs for Rho-like GTPases, Tiam1 contains a DH (Dbl homology) domain flanked at its C-terminus by a PH (pleckstrin homology) domain. In contrast with most GEFs, Tiam1 harbours an additional PH domain in the N-terminal part of the protein, PHn (N-terminal PH domain) with an adjacent CC (coiled-coil) and an Ex (extended) region. Both the PHn and CC domains are involved in the membrane/cortical actin targeting of Tiam1 [21,22]. In order to identify proteins that associate with Tiam1 and regulate its activity or intracellular localization, we performed a yeast two-hybrid screen using the PHn-CC-Ex domain of Tiam1 as bait. In the present study, we show that Tiam1 binds to the p21-Arc subunit of the Arp2/3 complex via its PHn-CC domain and co-localizes with the Arp2/3 complex at sites enriched in cortical actin. Our data suggest that the association of Tiam1 with the Arp2/3 complex promotes the local activation of Rac, which is required for the subsequent activation of Arp2/3 complex proteins leading to actin filament assembly.
EXPERIMENTAL PROCEDURES
Eukaryotic expression constructs and cell lines
HA (haemagglutinin)-tagged C1199-Tiam1, ΔPHn3-Tiam1, ΔDH-Tiam1 and C580-Tiam1 have been described previously [22]. For generation of Myc-epitope-tagged mouse p21-Arc, a PstI(blunt)/KpnI fragment containing the Myc epitope from pMT2SM-myc was cloned into the HindIII(blunt)/KpnI sites of pCDNA3 (Invitrogen Corp., Carlsbad, CA, U.S.A.) to yield pCDNA3-myc. An EcoRI fragment containing mouse p21-Arc from the yeast expression vector pGAD10 (Clontech Laboratories, Palo Alto, CA, U.S.A.) was cloned into the EcoRI site of pCDNA3-myc to yield pCDNA3-myc-p21-Arc. NIH3T3, MDCK (Madin–Darby canine kidney cells)-f3 and MDCKII cells stably expressing C1199-Tiam1 using a retroviral transduction protocol have been described in [18,23]. All cell lines were cultured in Iscove's modified Dulbecco's medium (Gibco) containing 10% foetal calf serum (Bodinco, Alkmaar, The Netherlands).
Yeast plasmids
PHn-CC-Ex and the deletion mutants PHn-CC-ExΔPHn3 (deletion of amino acids 420–552), PHn-CC-ExΔPHn1 (deletion of amino acids 513–552), PHn-CC-ExΔCC (deletion of amino acids 554–587), PHn-CC-ExΔEx (deletion of amino acids 599–691) were cloned as SalI/NotI fragments originating from the corresponding deletion mutants made in C1199-Tiam1 [21] into the SalI/NotI sites of pMD4, containing the GAL4-DNA binding domain and a tyrosinase related protein-1 marker (a gift from M. van Dijk and L. van‘t Veer, Division of Pathology, The Netherlands Cancer Institute, Amsterdam, The Netherlands). FL (full-length)-Tiam1 and C580-Tiam1 were cloned as SalI/SpeI fragments into the SalI/SpeI sites of pMD4. To generate pMD4-Ex, a PvuII/NotI fragment from Tiam1, containing amino acids 588–853, was shuttled into the SmaI site of a modified pMD4 (containing an additional nucleotide in the polylinker) vector.
Yeast two-hybrid screen
Yeast strain, Y190, containing the ‘bait’ plasmid pMD4-PHn-CC-Ex, encoding the GAL4-DNA binding domain fused with the PHn-CC-Ex region of Tiam1, was transformed with 17-day-old mouse embryo library cDNA, and cloned into pGAD10 (containing the GAL4-transactivation domain; Clontech Laboratories), by the lithium acetate method. Transformants (4×106) were selected for growth on plates lacking histidine, tryptophan and leucine, supplemented with 30 mM 3AT (3-aminotriazole). Colonies obtained were subsequently analysed for β-galactosidase activity. cDNA library plasmids derived from positive yeast colonies were tested for bait specificity by retransformation with pMD4-PHn-CC-Ex, pMD4 containing deletion mutants of PHn-CC-Ex, and pMD4 without an insert. Similarly, 4×106 transformants that were obtained using a human Jurkat cDNA library (a gift from L. van Aelst, Department of Human Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, U.S.A.) were screened for interaction with pMD4-PHn-CC-Ex.
Biochemical fractionation
Fractionation of cellular components was performed as described previously [24]. Briefly, 5×106 cells were cultured for 24 h and then washed twice with ice-cold PBS containing 0.5 mM MgCl2 and 1 mM CaCl2. Cells were lysed on ice in 500 μl of lysis buffer (PBS/0.1% Triton X-100, 10% glycerol, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 1 μg/ml aprotinin). The detergent-soluble fraction was recovered and the dish was washed twice with 1 ml of lysis buffer. The remaining detergent-insoluble fraction was collected in 500 μl of Laemmli sample buffer.
Affinity chromatography
A GST (glutathione S-transferase)-tagged bacterial expression construct for mouse p21-Arc was generated by cloning an EcoRI fragment from pGAD10-p21-Arc into the EcoRI site of pGEX1 (Pharmacia Biotech, Piscataway, NJ, U.S.A.). Bacterial protein was produced as described [23]. For preparation of affinity columns, glutathione-coupled Sepharose 4B beads (Pharmacia Biotech) were loaded with GST–p21-Arc to yield a column harbouring 2.5–3 mg/ml protein. Typically, columns of 150–200 μl were equilibrated with several volumes of loading buffer [50 mM Tris/HCl (pH 7.5), 100 mM NaCl, 1% Triton X-100, 10 mM NaF, 5 mM EDTA, 0.1 mM NaVO3, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 1 μg/ml aprotinin]. COS-7 cell lysates (prepared in loading buffer) with Tiam1 mutants (Cys1199–HA, C580–HA and C1199–ΔPHn3HA) were each loaded three times on to the column. Loaded columns were washed with at least 3 vol. of loading buffer, and then subsequently eluted with 2 vol. of loading buffer containing 250 mM, 400 mM or 1000 mM NaCl. Aliquots of the load and the eluates were analysed for Tiam1 proteins by Western blotting using an antibody against the HA-epitope tag.
GTPase activity assay
Rac activity was assayed as described previously [25]. Briefly, cells were lysed with a lysis buffer containing 2 μg/ml PAK-derived biotinylated CRIB (Cdc42/Rac interacting binding) peptide. Lysates were cleared by centrifugation, and active Rac–CRIB complexes were precipitated with streptavidin-coated beads (Sigma). After washing, Rac was solubilized in Laemmli sample buffer and detected on Western blots with anti-Rac antibodies (clone 23A8; Upstate Biotechnology, Inc.).
Immunofluorescence
Cells grown on glass coverslips were processed as described previously [4]. Briefly, cells were permeabilized with 0.5% (w/v) Triton X-100. C1199-Tiam1 was stained with an anti-DH antibody [4] or a monoclonal antibody against the HA-tag (12CA5; Boehringer Mannheim Corp., Indianapolis, IN, U.S.A.). Antibodies against Arp3 and p34-Arc have been described [26]. F-actin was stained with rhodamine-labelled phalloidin (Molecular Probes).
RESULTS
Tiam1 interacts with p21-Arc of the Arp2/3 complex
In order to identify proteins that interact with Tiam1 we performed a yeast two-hybrid screen. As bait we used a region of Tiam-1 that included the PHn domain, the adjacent CC domain and the EX region (PHn-CC-Ex; see Figures 1A and 1B) and screened a 17-day-old mouse embryo cDNA library. Three independently isolated clones encoding overlapping sequences were obtained. Sequence analysis identified these clones as murine p21-Arc, which showed 98% amino acid sequence identity with a clone found in a similar screen using a human Jurkat T-lymphoma library. Similar to the PHn-CC-Ex region, FL-Tiam1 and the C1199 mutant of Tiam1 (Figure 1B), a truncated Tiam1 version that harbours the PHn-CC-Ex region also binds to p21-Arc in the yeast two-hybrid assay (Figures 1A and 1B). By contrast, C580-Tiam1, a mutant that consists of the DH domain and the adjacent C-terminal PH domain but lacks the PHn-CC-Ex region, did not associate with p21-Arc. From these data we conclude that Tiam1 binds to the p21-Arc subunit of the Arp2/3 complex via its PHn-CC-Ex domain.
Figure 1. Tiam1 interacts with p21-Arc through the PHn domain and the adjacent CC region of Tiam1.
(A) Growth and β-galactosidase expression on 3AT/Trp−/Leu−/His− plates, in yeast transformed with the plasmid encoding p21-Arc, together with plasmids encoding the PHn-CC-Ex region of Tiam1, FLTiam1 or C580-Tiam1. (B) Comparison of Tiam1 domain organization and yeast two-hybrid results. +, An interaction with p21-Arc in the yeast two-hybrid assay, as measured by growth on 3AT/Trp−/Leu−/His− plates and β-galactosidase expression. −, No interaction in the yeast two-hybrid assay. M, myristoylation site; P, PEST (amino acid sequence enriched in proline, glutamic acid, serine and threonine) domain; DHR, discs large homology region; PHc, C-terminal PH domain [22]. PH domains from human Dbl and human β-Ark were used. (C) Crude cell lysates prepared from COS-7 cells (see input) expressing HA-tagged C1199- or C580-Tiam1 were passed over affinity chromatography columns containing bacterially expressed GST–p21-Arc. C1199-Tiam1, but not C580-Tiam1 is specifically retained on the column and is eluted at between 250 and 400 mM NaCl. Tiam1 proteins were detected using the anti-12CA5 antibody against the HA-tag. (D) The PHn-CC-Ex domain of Tiam1 is sufficient for Arp 3 binding. Purified GST and GST–PHn-CC-Ex were used for pull-down assays in COS-7 cell lysates, and Arp3-binding was determined by Western blot analysis.
Further analysis of various Tiam1 mutants showed that complete or partial deletion of either the PHn domain (PHn3-CC-Ex and PHn1-CC-Ex respectively) or the adjacent CC domain (PHn-ΔCC-Ex) abolished the interaction with p21-Arc, whereas a deletion in the Ex region (PHn-CC-ΔEx) did not (Figure 1B). These data indicate that the PHn domain and the adjacent CC region of Tiam1 are required for the interaction with p21-Arc. In addition, mutants in which the PHn domain of Tiam1 was replaced with the PH domain of either β-Ark, Dbl or by the C-terminal PH domain of Tiam1, were no longer able to bind to p21-Arc (Figure 1B). This indicates that the binding of p21-Arc is restricted to the unique N-terminal PH domain of Tiam1.
We subsequently attempted to co-immunoprecipitate Tiam1 in association with the Arp2/3 complex using antibodies specific for Tiam1, Arp2, Arp3 and p34-Arc. However, these studies were unsuccessful because the complex between Tiam1 and Arp2/3 was found to be present in the actin cytoskeleton fraction (see below), which does not allow demonstration of protein association by immunoprecipitation. Therefore we investigated the interaction between p21-Arc and Tiam1 by an alternative biochemical assay using affinity chromatography. For this, various Tiam1 mutant proteins (C1199-Tiam1 and C580-Tiam1) were expressed in COS-7 cells and the respective lysates were passed over an affinity chromatography column containing GST–p21-Arc. Consistent with the yeast two-hybrid data, only the C1199-Tiam1 protein that contains an intact PHn-CC-Ex domain was specifically retained on the p21-Arc affinity column, whereas the C580-Tiam1 mutant did not (Figure 1C). C1199-Tiam1 was eventually eluted at between 250 and 400 mM NaCl. None of the Tiam1 proteins showed interaction with a control column of immobilized GST protein alone (results not shown). These data indicate that Tiam1 can interact with p21-Arc. However, p21-Arc is normally part of the Arp2/3 complex, suggesting that Tiam1 interacts with the entire complex. Therefore we performed pull-down assays using purified GST–PHn-CC-Ex from bacteria in COS-7-derived cell lysates. Indeed, the PHn-CC-Ex domain of Tiam1 interacts with Arp3 (Figure 1D), indicating that Tiam1 interacts not only with p21-Arc but with the entire Arp2/3 complex. These results are consistent with the binding data obtained from yeast and demonstrate that Tiam1 interacts with the p21-Arc subunit of the Arp2/3 complex through its PHn domain and the adjacent CC region.
Tiam1 co-localizes with Arp2/3 at sites of actin polymerization
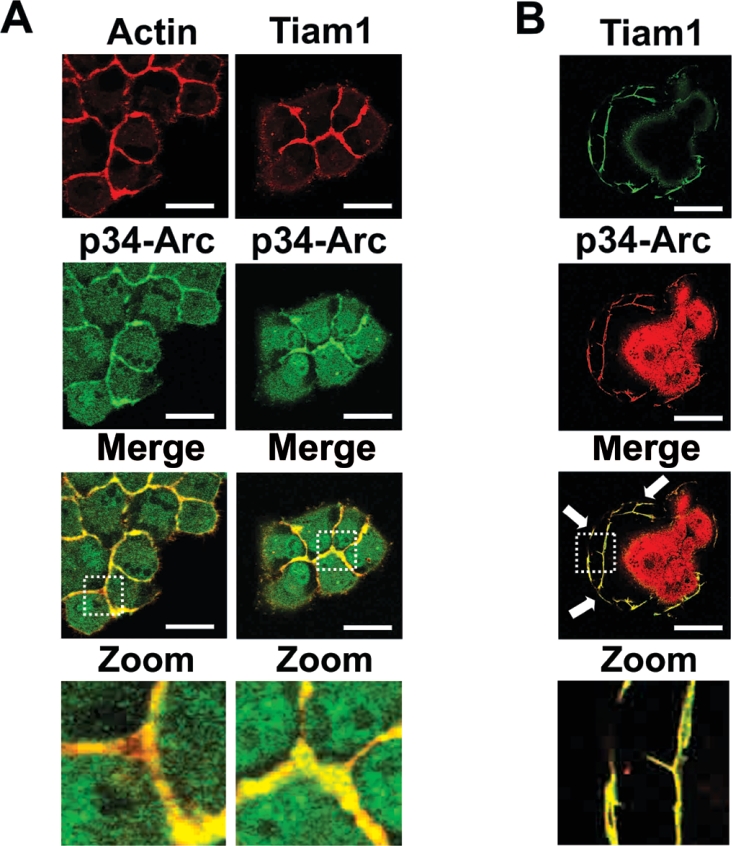
To further substantiate the possibility of an interaction between Tiam1 and p21-Arc, we analysed the subcellular localization of both proteins immunocytochemically. Since endogenous Tiam1 is very difficult to visualize, MDCK-f3 cells were transfected with Tiam1 [18] and subsequently immunostained for exogenous Tiam1 and for endogenous p34-Arc and actin. As shown in Figure 2(A), Tiam1 co-localizes with the Arp2/3 complex in F-actinrich structures at cell–cell contacts. Similar experiments performed using NIH3T3 fibroblasts showed that exogenous Tiam1 co-localized with endogenous p34-Arc in actin-rich membrane ruffles (Figure 2B). These localization data are consistent with the yeast-two hybrid and biochemical data, and strongly suggest that Tiam1 also associates with the Arp2/3 complex in cells in vivo.
Figure 2. Tiam1 co-localizes with Arp2/3 in actin-rich regions in cells.
(A) Tiam1 co-localizes with the Arp2/3 complex in adherens junctions of MDCK-f3 cells. Tiam1-expressing MDCK-f3 cells were immunostained for both Tiam1 and p34-Arc. Tiam1 and actin are shown in red, endogenous p34-Arc in green and co-localization in yellow. Scale bar, 10 μm. (B) Co-localization of Tiam1 with the endogenous Arp2/3 complex in membrane ruffles. NIH3T3 cells expressing HA-tagged Tiam1. Tiam1 is shown in green and endogenous p34-Arc in red and co-localization in yellow. Scale bar, 10 μm. Arrows indicate co-localization of Tiam1 with p34-Arc in membrane ruffles.
Specific intracellular localization of Tiam1 is promoted by the Arp2/3 complex
In order to study the functional consequences of the interaction between Tiam1 and the Arp2/3 complex we generated a mutant of Tiam1 with a deletion in the PHn domain (ΔPHn3-Tiam1) or with a mutation in the CC region adjacent to the PHn domain (ΔCC-Tiam1). Of note, these mutants can no longer interact with p21-Arc in the yeast-two hybrid screen (Figure 1B) or in biochemical assays (results not shown). Ectopic expression of Tiam1, ΔPHn3-Tiam1 or ΔCC-Tiam1 in MDCKII cells revealed that both the PHn and the CC domain are necessary for the localization of Tiam1 to actin-rich regions at the plasma membrane, in particular at cell–cell contacts (Figure 3A). Interestingly, the PHn domain has been shown to bind polyphosphorylated inositol lipids [27], suggesting that the observed plasma-membrane localization might also be due to lipid binding. However, the ΔCC-Tiam1 mutant has an intact PHn domain but can no longer be found at the cell–cell junctions, suggesting that the localization of Tiam1 to these actin-rich structures is mainly due to the PHn–p21Arc interaction and not a PHn–lipid interaction.
Figure 3. Localization of Tiam1 in cell–cell junctions is dependent on the Arp2/3 complex.
(A) The N-terminal PH domain and the CC region of Tiam1 are essential for targeting Tiam1 to actin-rich structures at cell–cell contacts. MDCKII cells were transfected with either Tiam1, ΔPHn3-Tiam1 or ΔCC-Tiam1. Cells were immunostained for Tiam1 (green) and F-actin (red). Co-localization is shown in yellow. Scale bar, 5 μm. Note that ΔPHn3-Tiam1 does not localize at adherens junctions. WT, wild-type. (B) Tiam1-induced Rac1 activation is dependent on its p21-Arc binding domain. Rac1-activity was determined in MDCKII cells expressing HA-tagged Tiam1 or ΔPHn3-Tiam1 by precipitating active Rac1 with a biotinylated-PAK-CRIB peptide. The PAK-CRIB bound Rac1 was detected by Western blotting using an anti-Rac1 antibody (clone 23A8; Upstate Biotechnology, Inc.). Total cell lysates were stained with an anti-Rac antibody to determine the level of Rac1 in cells. The expression of Tiam1 mutants was determined by Western blot analysis using an anti-HA antibody. Rac1-activity was quantified relative to empty vector (E.V.) transfected cells (bar diagram). The experiment shown here is a representative example from three independent experiments. (C) The WASP inhibitor, wiskostatin, inhibits both Arp2/3 and Tiam1 localization in cell–cell junctions. MDCK-f3 cells expressing FL-Tiam1 were treated with wiskostatin for 1 h (15 μm). Control and treated cells were subsequently immunostained with anti-HA and anti-Arp3 antibodies. Arp3 is shown in green and Tiam1 in red. Scale bar, 10 μm. (D) Tiam1-induced Rac1 activation is dependent on the Arp2/3 complex. MDCK-f3 cells expressing HA-tagged FL-Tiam1 were treated either with DMSO or wiskostatin (WIS; 15 μm) for 1 h. Subsequently, Rac1 activity was determined by precipitating active Rac with the biotinylated-PAK-CRIB peptide. The PAK-CRIB bound Rac1 was detected by Western blotting using an anti-Rac1 antibody. Total cell lysates were stained with an anti-Rac1 antibody to determine the level of Rac1 in cells. The expression of Tiam1 was determined by Western blotting using an anti-HA antibody. Rac1 level was quantified relative to E.V. transfected cells (bar diagram). Values (arbitrary units) are the means±S.E.M for three different experiments.
When analysing Rac activity in these cells by performing pull-down assays with the CRIB domain of PAK1 [25], we found that the highly expressed ΔPHn3-Tiam1 mutant is unable to activate Rac1 in cells, whereas Tiam1 is able to do so (Figure 3B). These data suggest that the specific localization of Tiam1 to actin-rich structures at the plasma membrane through association with the Arp2/3 complex promotes the activation of Rac. To further support this notion, MDCK-f3 cells that expressed FL-Tiam1 were treated with the cell-permeable WASP inhibitor, wiskostatin [28]. This inhibitor blocks the interaction between WASP and Cdc42, thereby preventing the activation of WASP and, as a consequence, the Arp2/3 complex. Inactivation of Arp2/3 by the WASP inhibitor resulted in the release of the Arp2/3 complex from adherens junctions (Figure 3C, left panels). Moreover, Tiam1 also dissociated from the adherens junctions and accumulated in the cytoplasma after the inhibition of WASP (Figure 3C, right panels). These data suggest that Tiam1 is dependent on the activated Arp2/3 complex for localization to regions where actin polymerization occurs. Consistent with these conclusions are the findings that Rac activation is inhibited in MDCK-f3 cells, and MDCK-f3 cells that express FL-Tiam1, by treatment of these cells with wiskostatin (Figure 3D). Together, these data suggest that the ability of Tiam1 to activate Rac is stimulated by the localization and activity of the Arp2/3 complex.
Activated Arp2/3 complex translocates to the cytoskeleton fraction
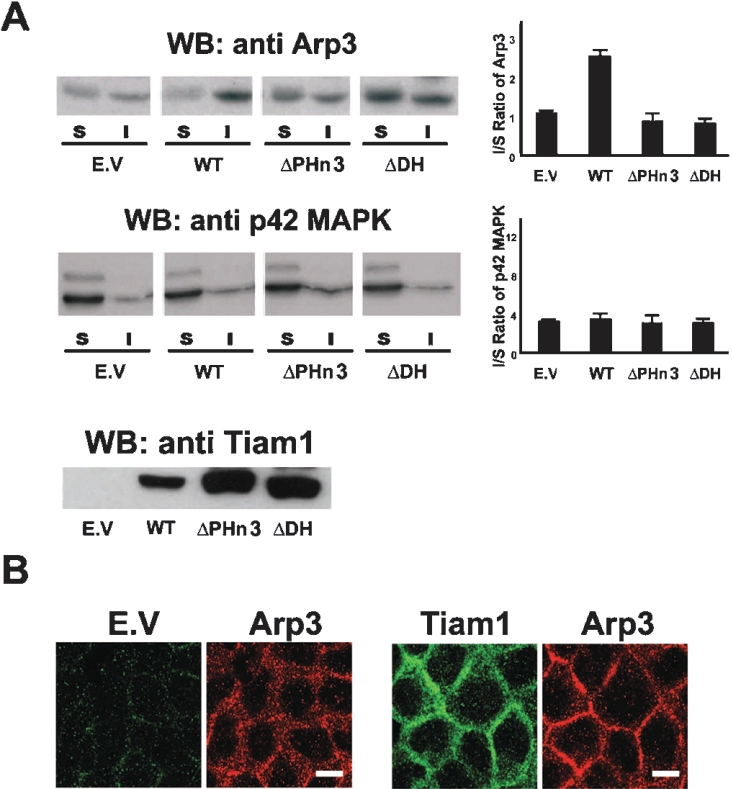
The Rac effectors, WAVE and PAK, can induce actin polymerization by activating the Arp2/3 complex [11,13]. The targeting of Tiam1 to the Arp2/3 complex might thus provide a mechanism by which to activate the complex via the Rac1–WAVE or Rac1–PAK pathway. To support this hypothesis we determined the distribution of the Arp2/3 complex in a membrane/cytosol (S) and an actin cytoskeleton (I) fraction in MDCKII cells that express the various Tiam1 mutants, ΔPHn3 or ΔDH. The latter mutant contains a deletion in the DH domain, thereby blocking the ability of Tiam1 to activate Rac. In control MDCKII cells, most of the Arp2/3 complex was equally distributed between the membrane/cytosol fraction and the cytoskeleton fraction, as visualized by the detection of Arp3 (Figure 4A) and p34-Arc (results not shown). However, upon ectopic expression of Tiam1, most of the Arp2/3 complex re-localized to the cytoskeleton fraction (Figure 4A), where it is thought to play a role in actin filament assembly. The re-localization of Arp2/3 was not induced by the expression of ΔPHn3-Tiam1 or ΔDH-Tiam1 (Figure 4A). Both mutants are unable to activate Rac1 either by dislocation or mutation of the catalytic domain respectively, suggesting that the ability of Tiam1 to localize the Arp2/3 complex to the actin cytoskeleton fraction is dependent on its capacity to activate Rac. As a control we determined the distribution of the p42 MAPK (mitogen-activated protein kinase), which was not affected by the expression of the various Tiam1 mutants (Figure 4A). Similar results for the redistribution of Arp2/3 to the cytoskeleton fraction under the control of Tiam1 were obtained in NIH3T3 cells (results not shown). Confocal imaging of MDCKII cells expressing either empty vector or Tiam1 further indicated that the localization of Arp2/3 to adherens junctions was promoted by the expression of Tiam1 (Figure 4B).
Figure 4. Tiam1 signalling recruits the Arp2/3 complex to the cytoskeletal fraction.
(A) Tiam1 promotes Arp2/3 localization to the actin cytoskeleton fraction. MDCKII cells expressing either empty vector (E.V.) or HA-tagged Tiam1, ΔPHn3-Tiam1 or ΔDH-Tiam1 were fractionated in a Triton X-100 soluble (S fraction) and insoluble actin cytoskeleton fraction (I fraction). Fractions were either stained for Arp3 or p42 MAPK. The bar diagrams indicate the ratio of Arp3 or p42 MAPK of the insoluble (I) over the soluble (S) fraction. Total cell lysates were stained with an anti-HA antibody to detect the different Tiam1 mutants. Arp3 and p42 MAPK levels were quantified relative to E.V. transfected cells (bar diagram). Values (arbitrary units) are the means±S.E.M for three different experiments. WB, Western blot; WT, wild-type. (B) Tiam1 promotes Arp2/3 localization to cell–cell junctions. MDCKII cells transfected with E.V. or C1199-Tiam1 were immunostained for exogenous Tiam1 (green) and endogenous Arp3 (red). Scale bar, 10 μm.
Together, these data suggest that the initial association of Tiam1 with the Arp2/3 complex can activate Rac1 and can thereby activate the Arp2/3 complex and/or recruit additional Arp2/3 complexes for increased actin polymerization. Tiam1 and the Arp2/3 complex may thus collaborate in a positive-feedback loop to induce rapid and abundant actin polymerization at specific sites in cells.
DISCUSSION
Rho proteins, which include Cdc42, Rac and RhoA, control a wide range of signalling pathways that regulate various biological processes, including the reorganization of the actin cytoskeleton in response to receptor stimulation. Cell-surface receptors use different GEFs to activate specific Rho GTPases, and more than 60 Rho GEFs have been identified within the mammalian genome [29]. GEFs not only activate Rho GTPases, but also participate in signalling to downstream effectors by either binding to these effectors directly or to scaffold proteins that can form complexes with components of the effector pathways [30–32]. Using yeast two-hybrid screening and biochemical assays, we found that the Rac-specific GEF, Tiam1, associates with p21-Arc, a subunit of the Arp2/3 complex. The Arp2/3 complex forms actin nucleation sites enabling the de novo polymerization of actin filaments in response to extracellular stimuli, suggesting that Tiam1 can target activated Rac to effector pathways involving Arp2/3-mediated actin polymerization.
Both the Arp2/3 complex and Rho proteins regulate the reorganization of the actin cytoskeleton, which is required for morphological changes and migration. In particular, Rac regulates the formation of membrane protrusions at the leading edge of cells, which control forward movement. Both Tiam1 and the Arp2/3 complex have been shown to play a role in the formation of cell–cell adhesions, membrane protrusions and lamellipodia [4,18,33]. The intriguing finding that Tiam1 associates with a component (p21-Arc) of the Arp2/3 complex, suggests that Tiam1 acts in complex with Arp2/3 to stimulate Rac and thereby the Arp2/3 complex leading to actin polymerization. This hypothesis is supported by the findings that Tiam1 co-localizes with the Arp2/3 complex at sites of actin polymerization such as membrane ruffles and cell–cell contacts, as well as that Tiam1 and Arp2/3 are mutually dependent for correct subcellular localization and signalling.
Prior to the formation of Rac-dependent lamellipodia, migrating cells have to form small membrane protrusions known as filopodia, which is regulated by Cdc42 [34]. The Cdc42-mediated formation of filopodia is dependent on WASP-mediated activation of the Arp2/3 complex [6], which may thus form the first pool of Arp2/3 present at cell protrusions. This initial pool of Arp2/3 complex proteins might subsequently act as the initial docking sites for Tiam1 to promote Rac-dependent lamellipodia formation. Once Tiam1 is recruited to the Arp2/3 complex, it can locally activate Rac1 and recruit new Arp2/3 complexes, thereby inducing further actin nucleation and branching. Tiam1 might thus be one of the Rac GEFs that is involved in the evolution of filopodia into lamellipodia and thereby forms a link between Cdc42- and Rac-mediated actin filament assembly.
Similar to the relationship between filopodia and lamellipodia, initial cell–cell adhesions involve the formation of primordial cell–cell contacts that mature into more rigid adhesion structures that are dependent on the activity of Rac1 [35]. Mature cell–cell adhesions are predominantly formed by the transmembrane protein, E-cadherin, which forms homophilic interactions between neighbouring epithelial cells [36]. In order to form stable contacts, E-cadherins must associate with the actin cytoskeleton via β-catenin and α-catenin [37]. As shown previously, this junctional stability is maintained by Tiam1-mediated Rac activation [18,38] and is also dependent on the Arp2/3 complex [39]. Our finding that targeting of Tiam1 to adherens junctions is dependent on the presence of the Arp2/3 complex, suggests that Tiam1 and the Arp2/3 complex in concert regulate the cortical actin polymerization that is required for the formation of stable cell–cell junctions.
Besides interacting with p21-Arc, Tiam1 has been suggested to interact with a number of other (scaffold) proteins, including spinophillin, JIP (c-Jun N-terminal kinase-interacting protein), PAR-3 (partitioning-defective-3), a PKC (protein kinase C), and recently IRSp53 [30,40–43]. In general, the interaction of Tiam1 with various scaffold proteins directs Rac signalling towards specific downstream signalling. For instance, the Tiam1–JIP complex results in increased p38-mediated gene transcription [30]. Interestingly, the scaffold protein, IRSp53, has been found to form a trimeric complex with WAVE and Rac1 [16]. IRSp53 might thus interact with the Arp2/3 complex through WAVE or through Tiam1 [43] making it probable that Tiam1, IRSp53, WAVE, Rac and the Arp2/3 complex are all part of a larger protein complex (see Figure 5) which is involved in Rac-controlled actin polymerization during processes such as the formation of lamellipodia, membrane ruffles and adherens junctions.
Figure 5. Tiam1 regulates Rac-mediated Arp2/3-controlled actin polymerization.
The localization of Tiam1 to actin-rich structures at the plasma membrane could be accomplished by association of Tiam1 through the PHn-CC domain with the p21-Arc subunit of the Arp2/3 complex (1). Tiam1 activates Rac (2) that influences the actin nucleation activity of Arp2/3 by activating either WAVE or PAK1 [11,15] (3,4). As a result, the activated Arp2/3 complex associates with the actin cytoskeleton and induces actin polymerization (5). In this way Tiam1 and the Arp2/3 complex may collaborate in a positive-feedback loop to induce rapid and abundant actin polymerization at specific sites in cells.
Acknowledgments
We thank Annemiek Beverdam and Rob A. van der Kammen for technical assistance. P.L.H. is a fellow of the Landsteiner Foundation for Blood Transfusion Research. This work was supported by grants from the Dutch Cancer Society to J.G.C.
References
- 1.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 2.Hart M. J., Eva A., Evans T., Aaronson S. A., Cerione R. A. Catalysis of guanine nucleotide exchange on the CDC42Hs protein by the dbl oncogene product. Nature (London) 1991;354:311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 3.Manser E., Loo T. H., Koh C. G., Zhao Z. S., Chen X. Q., Tan L., Tan I., Leung T., Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 4.Michiels F., Habets G. G., Stam J. C., van der Kammen R. A., Collard J. G. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature (London) 1995;375:338–340. doi: 10.1038/375338a0. [DOI] [PubMed] [Google Scholar]
- 5.Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 6.Miki H., Sasaki T., Takai Y., Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature (London) 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 7.Machesky L. M., Insall R. H. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 8.Kunda P., Craig G., Dominguez V., Baum B. Abi, Sra1, and Kette control the stability and localization of SCAR/WAVE to regulate the formation of actin-based protrusions. Curr. Biol. 2003;13:1867–1875. doi: 10.1016/j.cub.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Robinson R. C., Turbedsky K., Kaiser D. A., Marchand J. B., Higgs H. N., Choe S., Pollard T. D. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- 10.Derry J. M., Ochs H. D., Francke U. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell. 1994;79:in the press. [PubMed] [Google Scholar]
- 11.Vadlamudi R. K., Li F., Barnes C. J., Bagheri-Yarmand R., Kumar R. p41-Arc subunit of human Arp2/3 complex is a p21-activated kinase-1-interacting substrate. EMBO J. 2004;5:154–160. doi: 10.1038/sj.embor.7400079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miki H., Miura K., Takenawa T. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 1996;15:5326–5335. [PMC free article] [PubMed] [Google Scholar]
- 13.Miki H., Suetsugu S., Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prehoda K. E., Scott J. A., Mullins R. D., Lim W. A. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- 15.Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature (London) 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 16.Miki H., Yamaguchi H., Suetsugu S., Takenawa T. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature (London) 2000;408:732–735. doi: 10.1038/35047107. [DOI] [PubMed] [Google Scholar]
- 17.Habets G. G., Scholtes E. H., Zuydgeest D., van der Kammen R. A., Stam J. C., Berns A., Collard J. G. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–549. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 18.Hordijk P. L., ten Klooster J. P., van der Kammen R. A., Michiels F., Oomen L. C., Collard J. G. Inhibition of invasion of epithelial cells by Tiam1-Rac signaling. Science. 1997;278:1464–1466. doi: 10.1126/science.278.5342.1464. [DOI] [PubMed] [Google Scholar]
- 19.Zondag G. C., Evers E. E., ten Klooster J. P., Janssen L., van der Kammen R. A., Collard J. G. Oncogenic Ras downregulates Rac activity, which leads to increased Rho activity and epithelial-mesenchymal transition. J. Cell Biol. 2000;149:775–782. doi: 10.1083/jcb.149.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sander E. E., van Delft S., ten Klooster J. P., Reid T., van der Kammen R. A., Michiels F., Collard J. G. Matrix-dependent Tiam1/Rac signaling in epithelial cells promotes either cell-cell adhesion or cell migration and is regulated by phosphatidylinositol 3-kinase. J. Cell Biol. 1998;143:1385–1398. doi: 10.1083/jcb.143.5.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michiels F., Stam J. C., Hordijk P. L., van der Kammen R. A., Ruuls-Van Stalle L., Feltkamp C. A., Collard J. G. Regulated membrane localization of Tiam1, mediated by the NH2-terminal pleckstrin homology domain, is required for Rac-dependent membrane ruffling and C-Jun NH2-terminal kinase activation. J. Cell Biol. 1997;137:387–398. doi: 10.1083/jcb.137.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stam J. C., Sander E. E., Michiels F., Van Leeuwen F. N., Kain H. E., van der Kammen R. A., Collard J. G. Targeting of Tiam1 to the plasma membrane requires the cooperative function of the N-terminal pleckstrin homology domain and an adjacent protein interaction domain. J. Biol. Chem. 1997;272:28447–28454. doi: 10.1074/jbc.272.45.28447. [DOI] [PubMed] [Google Scholar]
- 23.Sander E. E., ten Klooster J. P., van Delft S., van der Kammen R. A., Collard J. G. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinck L., Nathke I. S., Papkoff J., Nelson W. J. Dynamics of cadherin/catenin complex formation: novel protein interactions and pathways of complex assembly. J. Cell Biol. 1994;125:1327–1340. doi: 10.1083/jcb.125.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Price L. S., Langeslag M., ten Klooster J. P., Hordijk P. L., Jalink K., Collard J. G. Calcium signaling regulates translocation and activation of Rac. J. Biol. Chem. 2003;278:39413–39421. doi: 10.1074/jbc.M302083200. [DOI] [PubMed] [Google Scholar]
- 26.Machesky L. M., Reeves E., Wientjes F., Mattheyse F. J., Grogan A., Totty N. F., Burlingame A. L., Hsuan J. J., Segal A. W. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem. J. 1997;328:105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fleming I. N., Gray A., Downes C. P. Regulation of the Rac1-specific exchange factor Tiam1 involves both phosphoinositide 3-kinase-dependent and -independent components. Biochem. J. 2000;351:173–182. doi: 10.1042/0264-6021:3510173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson J. R., Bickford L. C., Morgan D., Kim A. S., Ouerfelli O., Kirschner M. W., Rosen M. K. Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation. Nat. Struct. Mol. Biol. 2004;11:747–755. doi: 10.1038/nsmb796. [DOI] [PubMed] [Google Scholar]
- 29.Schmidt A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- 30.Buchsbaum R. J., Connolly B. A., Feig L. A. Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol. Cell Biol. 2002;22:4073–4085. doi: 10.1128/MCB.22.12.4073-4085.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Zhu K., Zheng Y. Oncogenic Dbl, Cdc42, and p21-activated kinase form a ternary signaling intermediate through the minimum interactive domains. Biochemistry. 2004;43:14584–14593. doi: 10.1021/bi048574u. [DOI] [PubMed] [Google Scholar]
- 32.Mertens A. E., Roovers R. C., Collard J. G. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003;546:11–16. doi: 10.1016/s0014-5793(03)00435-6. [DOI] [PubMed] [Google Scholar]
- 33.Svitkina T. M., Borisy G. G. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nobes C. D., Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 35.Vasioukhin V., Bauer C., Yin M., Fuchs E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell. 2000;100:209–219. doi: 10.1016/s0092-8674(00)81559-7. [DOI] [PubMed] [Google Scholar]
- 36.Hyafil F., Babinet C., Jacob F. Cell-cell interactions in early embryogenesis: a molecular approach to the role of calcium. Cell. 1981;26:447–454. doi: 10.1016/0092-8674(81)90214-2. [DOI] [PubMed] [Google Scholar]
- 37.Ozawa M., Baribault H., Kemler R. The cytoplasmic domain of the cell adhesion molecule uvomorulin associates with three independent proteins structurally related in different species. EMBO J. 1989;8:1711–1717. doi: 10.1002/j.1460-2075.1989.tb03563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malliri A., van Es S., Huveneers S., Collard J. G. The Rac exchange factor Tiam1 is required for the establishment and maintenance of cadherin-based adhesions. J. Biol. Chem. 2004;279:30092–30098. doi: 10.1074/jbc.M401192200. [DOI] [PubMed] [Google Scholar]
- 39.Kovacs E. M., Goodwin M., Ali R. G., Paterson A. D., Yap A. S. Cadherin-directed actin assembly: E-cadherin physically associates with the Arp2/3 complex to direct actin assembly in nascent adhesive contacts. Curr. Biol. 2002;12:379–382. doi: 10.1016/s0960-9822(02)00661-9. [DOI] [PubMed] [Google Scholar]
- 40.Buchsbaum R. J., Connolly B. A., Feig L. A. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J. Biol. Chem. 2003;278:18833–18841. doi: 10.1074/jbc.M207876200. [DOI] [PubMed] [Google Scholar]
- 41.Chen X., Macara I. G. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat. Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 42.Nishimura T., Yamaguchi T., Kato K., Yoshizawa M., Nabeshima Y., Ohno S., Hoshino M., Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat. Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- 43.Connolly B. A., Rice J., Feig L. A., Buchsbaum R. J. Tiam1-IRSp53 Complex Formation Directs Specificity of Rac-Mediated Actin Cytoskeleton Regulation. Mol. Cell Biol. 2005;25:4602–4614. doi: 10.1128/MCB.25.11.4602-4614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]