Abstract
Our colons harbor trillions of microbes including a prominent archaeon, Methanobrevibacter smithii. To examine the contributions of Archaea to digestive health, we colonized germ-free mice with Bacteroides thetaiotaomicron, an adaptive bacterial forager of the polysaccharides that we consume, with or without M. smithii or the sulfate-reducing bacterium Desulfovibrio piger. Whole-genome transcriptional profiling of B. thetaiotaomicron, combined with mass spectrometry, revealed that, unlike D. piger, M. smithii directs B. thetaiotaomicron to focus on fermentation of dietary fructans to acetate, whereas B. thetaiotaomicron-derived formate is used by M. smithii for methanogenesis. B. thetaiotaomicron–M. smithii cocolonization produces a significant increase in host adiposity compared with monoassociated, or B. thetaiotaomicron–D. piger biassociated, animals. These findings demonstrate a link between this archaeon, prioritized bacterial utilization of polysaccharides commonly encountered in our modern diets, and host energy balance.
Keywords: adiposity, energy homeostasis, gut microbial ecology, polysaccharide metabolism, Methanobrevibacter smithii
The role of Archaea in human health remains unclear (1). One site where their influence may be profound is the gut. Hydrogen-consuming methanogenic archaeons are present in the intestinal tracts of many invertebrate and vertebrate species (2–4). Our adult intestine contains 10 trillion to 100 trillion microbial cells and is dominated by members of just two divisions of Bacteria, the Bacteroidetes and the Firmicutes (5). Archaea, the only known producers of methane, are present at high levels in 50–85% of humans (6–8). Concordance rates for methane production are equivalent for both monozygotic and dizygotic twins, underscoring the importance of family environment (mothers) in acquisition of methanogens (9). The most comprehensive enumeration of the adult human colonic microbiota reported to date (5) found a single predominant archaeal species, Methanobrevibacter smithii. This Euryarchaeote can comprise up to 10% of all anaerobes in the colons of healthy adults, while Methanosphaera stadtmanae and Crenarchaeotes can be minor members (10, 11).
One manifestation of the mutualism between humans and their distal gut microbiota is that the latter extracts energy that would be lost from otherwise indigestible dietary polysaccharides (fiber). Fermentation of dietary fiber is accomplished by syntrophic interactions between microbes linked in a metabolic food web and is a major energy-producing pathway for members of the Bacteroidetes and the Firmicutes. These primary bacterial fermentors generate short-chain fatty acids (SCFAs), principally acetate, propionate, and butyrate, as well as other organic acids (e.g., formate) and gases [e.g., hydrogen (H2) and carbon dioxide (CO2)]. Accumulation of H2 inhibits bacterial NADH dehydrogenases, thereby reducing the yield of ATP. Studies in man-made bioreactors have shown that removal of H2 by means of archaeal methanogenesis improves fermentation efficiency (12).
Little is known about how our distal gut microbial community prioritizes processing of undigested dietary polysaccharides delivered from the small intestine. We do know that microbial fermentation of these polysaccharides to SCFAs accounts for up to 10% of our daily caloric intake (13), although the amount will vary depending on the amount of polysaccharides consumed (14), and that, relative to our own genome, our colonic microbial community genome (microbiome) is significantly enriched in components of metabolic pathways involved in glycan degradation (15). We also know that colonization of adult germ-free (GF) mice with an unfractionated gut microbiota harvested from conventionally raised animals produces a prompt and marked increase in adiposity without a corresponding increase in food consumption (16). Together, these observations raise the question of how archaeons, such as M. smithii, influence the dining habitats of bacteria living in our guts and how differences in their representation between humans could affect host-energy balance.
To our knowledge, there have been no reported in vivo analyses of the impact of a methanogen and a saccharolytic bacterium on one another’s transcriptomes or metabolomes. In the present study, we perform such an analysis by examining the effects of M. smithii in gnotobiotic mice harboring a simplified two-component model of our colonic microbiota. These experiments indicate that M. smithii acts as a “power broker” in the distal gut community, regulating the specificity of polysaccharide fermentation and influencing the amount of calories deposited in fat stores.
Results and Discussion
Cocolonization of Gnotobiotic Mice with Bacteroides thetaiotaomicron and M. smithii Increases Their Population Density in the Distal Gut.
Groups of age-matched adult GF mice belonging to the NMRI inbred strain (17) were colonized with one or more of the following human fecal-derived microbial strains: B. thetaiotaomicron, a saccharolytic bacterium whose genome has been sequenced (5, 17, 18) (alone for 14 or 28 d); M. smithii (alone for 14 d); or B. thetaiotaomicron alone for 14 d followed by M. smithii for 14 d.
Sulfate-reducing bacteria (SRB), and to some extent homoacetogens, serve as alternative consumers of H2 in the human gut (19, 20). These SRBs are almost exclusively Desulfovibrio spp., with Desulfovibrio piger being the most abundant species in healthy adults (5, 21). D. piger, like M. smithii, is nonsaccharolytic; unlike M. smithii, it cannot use formate (22). Therefore, control experiments were performed where GF mice were colonized with D. piger alone or in place of M. smithii in biassociation (cocolonization) experiments.
All mice were fed an autoclaved standard rodent chow diet rich in plant polysaccharides (17), including polyfructose-containing glycans (fructans). Biochemical studies of cecal contents recovered from GF mice fed this diet revealed that fructans were 3.8-fold higher than polyglucose-containing glycans (glucans) (85 ± 6 vs. 25 ± 2 μmol/g dry weight of contents; P < 0.005).
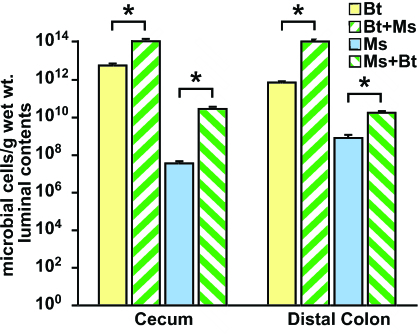
GF mice were reliably and efficiently colonized after a single gavage of 108 M. smithii or B. thetaiotaomicron (mean values: 1012 organisms per g of cecal contents for B. thetaiotaomicron; 107 for M. smithii; Fig. 1). There were no significant differences in cecal B. thetaiotaomicron levels after 14- versus 28-d monoassociations (data not shown). Cocolonization with M. smithii and B. thetaiotaomicron resulted in statistically significant (P < 0.03), 100- to 1,000-fold enhancement in the density of cecal colonization by both organisms (Fig. 1). The levels of colonization achieved by M. smithii in the ceca and colons (Fig. 1) of these biassociated mice were equivalent to those reported in the feces of healthy adult humans (11). In contrast, biassociation of mice with B. thetaiotaomicron and D. piger did not significantly alter cecal or colonic levels of either organism (data not shown). These results suggest that a mutually beneficial relationship is forged between M. smithii and B. thetaiotaomicron in the distal mouse gut that allows them to markedly increase their population size.
Fig. 1.
Cocolonization with M. smithii (Ms) and B. thetaiotaomicron (Bt) enhances the representation of both species in the distal intestines of gnotobiotic mice. The density of colonization was defined by using quantitative PCR of DNA isolated from the cecal contents of monoassociated and biassociated mice (n = 5 per group per experiment; three independent experiments; each sample assayed in triplicate; mean values ± SEM plotted); ∗, P < 0.05 vs. monoassociated controls. Bt and Bt + Ms represent the number of B. thetaiotaomicron cells present in mice colonized with Bt alone or with Bt + Ms, respectively; Ms and Ms + Bt represent the number of M. smithii cells present in mice colonized with Ms alone or with Ms + Bt, respectively.
M. smithii Enhances the Ability of B. thetaiotaomicron to Degrade Polyfructose-Containing Glycans.
We used a combination of whole-genome transcriptional profiling, mass spectrometry (MS), and microanalytic biochemical assays to determine the impact of M. smithii on B. thetaiotaomicron glycan utilization in vivo. Total microbial RNA was isolated from cecal contents of monoassociated and biassociated mice (n = 4–5 per group). cDNA targets, generated from each RNA preparation, were hybridized to individual B. thetaiotaomicron GeneChips containing probe sets representing 4,719 of B. thetaiotaomicron’s 4,779 predicted protein-coding ORFs (17). These probe sets encompass all components of B. thetaiotaomicron’s very prominent “glycobiome” (genes involved in carbohydrate acquisition/metabolism/biosynthesis), including 226 predicted glycoside hydrolases, 15 polysaccharide lyases, and 163 paralogs of two outer membrane proteins (SusC, a malto-oligosaccharide porin, and SusD, which binds starch) (18).
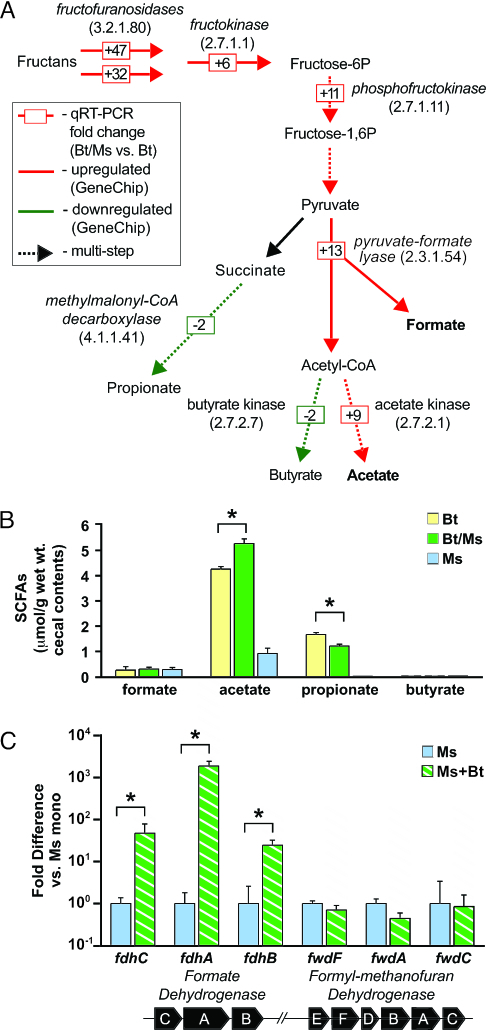
Unsupervised hierarchical clustering of the resulting GeneChip data sets revealed that colonization of the cecal habitat with M. smithii dramatically alters B. thetaiotaomicron’s transcriptome: 638 genes were defined (see Materials and Methods) as significantly up-regulated, and 462 genes were defined as significantly down-regulated compared with their levels of expression during a 14-d B. thetaiotaomicron monoassociation [P < 0.05; see Table 1, which is published as supporting information on the PNAS web site, for a gene list sorted by clusters of orthologous groups (COGs)]. Consistent with the enhanced representation of B. thetaiotaomicron in the distal gut, our COG-based functional classification indicated that cocolonization with M. smithii up-regulates B. thetaiotaomicron genes involved in DNA replication and protein production (see Fig. 5, which is published as supporting information on the PNAS web site). The presence of M. smithii also causes B. thetaiotaomicron to down-regulate expression of many genes involved in carbohydrate metabolism (Fig. 5), including 57 glycoside hydrolases [e.g., arabinosidases, xylosidases, glucosidases, galactosidases, mannosidases, rhamnosidases, and pectate lyases; grouped by carbohydrate-active enzyme families (CAZy; http://afmb.cnrs-mrs.fr/CAZY/acc.html) in Table 2, which is published as supporting information on the PNAS web site]. There is an accompanying induction of three fructofuranosidases (Fig. 2A). Two of these polyfructose-degrading glycoside hydrolases are encoded by ORFs situated in a gene cluster (BT1757–BT1765) that includes a putative sugar transporter, SusC/SusD paralogs, and the organism’s only fructokinase (Fig. 2B). Augmented expression of this cluster was validated by quantitative RT-PCR (qRT-PCR): the results revealed 32 ± 5.8- and 47 ± 5.9-fold increases for the fructofuranosidases (BT1759 and BT1765, respectively) and a 6.4 ± 2.8-fold increase for the fructokinase (BT1757) (see Fig. 6A and Tables 3 and 4, which are published as supporting information on the PNAS web site). Changes in the fructofuranosidases represent the most robust B. thetaiotaomicron transcriptional responses to M. smithii.
Fig. 2.
M. smithii enhances B. thetaiotaomicron polyfructose-containing glycan degradation in the distal gut. Results of GeneChip analysis of RNA isolated from cecal contents of individual mice colonized with B. thetaiotaomicron ± M. smithii (n = 4–5 per group) are shown. (A) Unsupervised hierarchical clustering (dchip) of B. thetaiotaomicron (Bt) glycoside hydrolases (GH) and polysaccharide lysases (PL) that are up-regulated (13 genes) or down-regulated (57 genes) in the presence of M. smithii (Ms). Some GH/PL families represented in this data set are highlighted by using the classification scheme in CAZy (http://afmb.cnrs-mrs.fr/CAZY/acc.html; see Table 2 for a complete list). Each column represents data obtained from a cecal sample harvested from an individual mouse, whereas each row represents a B. thetaiotaomicron GH or PL gene. (B) B. thetaiotaomicron polyfructose degradation cluster induced in the presence of M. smithii (see Table 1 for fold changes defined by GeneChip). (C) Biochemical analysis of fructan and glucan levels in cecal contents (n = 5 mice per group; each sample assayed in duplicate; mean values ± SEM plotted). ∗, P < 0.05.
In contrast, cocolonization with D. piger did not produce a significant change in expression of these fructofuranosidases, or the fructokinase, as judged by GeneChip and qRT-PCR assays (Fig. 6 and Table 3). Overall, D. piger had very modest effects on the B. thetaiotaomicron transcriptome: of the 41 differentially expressed genes, only four were glycoside hydrolases (two α-mannosidases, a β-hexosaminidase, and a glucuronyl hydrolase; all down-regulated; see Fig. 6B).
Fructose is easily shunted into the glycolytic pathway by means of fructokinase, making fructans desirable energy sources. This notion is supported by GeneChip analyses of B. thetaiotaomicron grown in a batch culture fermentor containing glucose and a complex mixture of polysaccharides (TYG medium). Expression of the polyfructose degradation cluster peaks in early log phase with 7.5- to 53.2-fold higher levels for BT1757–BT1765 transcripts compared with late log/stationary phase where B. thetaiotaomicron utilizes less coveted glycans such as mannans (ref. 17; data sets are available from the authors upon request).
Consistent with the in vitro and in vivo transcriptional profiling results, biochemical assays demonstrated a statistically significant 52 ± 4% decrease in cecal fructan levels after B. thetaiotaomicron/M. smithii cocolonization, compared with B. thetaiotaomicron monoassociated mice (P < 0.05; Fig. 2C). Glucans increased modestly (15 ± 3%; P < 0.05; Fig. 2C), indicating continued albeit slightly reduced digestion of glucose-containing polysaccharides. In contrast, cocolonization with D. piger did not produce any significant changes in fructan or glucan levels compared with the B. thetaiotaomicron monoassociated state (data not shown).
GC-MS analysis of neutral and amino sugars released by acid hydrolysis of cecal contents, revealed that colonization with B. thetaiotaomicron plus M. smithii produced modest, but not statistically significant, decreases in the consumption of these carbohydrates compared with colonization with B. thetaiotaomicron alone (see Fig. 7, which is published as supporting information on the PNAS web site). These latter results suggest that increased consumption of otherwise inaccessible fructans does not demand forfeiture of the use of other available polysaccharides.
Metabolic Underpinnings of M. smithii–B. thetaiotaomicron Mutualism.
In silico reconstructions of the B. thetaiotaomicron metabolome, obtained by placing the enzymes encoded by bacterial genes responsive to the presence of M. smithii onto Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg) metabolic maps, indicated that cocolonization produces a shift toward increased production of acetate and formate, and reduced production of butyrate and propionate (Fig. 3A and Table 3). These changes in gene expression did not occur with the addition of D. piger (Table 3). Follow-up GC-MS analysis of cecal SCFA levels confirmed a significant increase in acetate, and a significant decrease in propionate in biassociated compared with B. thetaiotaomicron monoassociated mice (P < 0.02; Fig. 3B). Butyrate levels in cecal contents were at the lower limits of detection in all groups (Fig. 3B); thus, we could not measure the extent of its production by B. thetaiotaomicron in vivo or the effects of cocolonization on its generation.
Fig. 3.
Biassociation with B. thetaiotaomicron and M. smithii increases B. thetaiotaomicron production of acetate and formate. (A) qRT-PCR analysis (boxed numbers) of the effects of M. smithii on expression of selected B. thetaiotaomicron genes encoding enzymes involved in fermentation of polyfructose-containing glycans: fructofuranosidases, BT1765/BT1759; fructokinase, BT1757; phosphofructokinase, BT0307; pyruvate:formate lyase, BT4738; acetate kinase, BT3963, methylmalonyl-CoA decarboxylase, BT1688; butyrate kinase, BT2552. Enzyme classification (EC) numbers are provided in parentheses. Dotted lines indicate multistep pathways. (Expression of fructofuranosidases, acetate kinase, puruvate:formate lyase, and butyrate kinase is constant if the colonization period is extended from 14 to 28 d; see Table 3.) (B) GC-MS analyses of cecal SCFAs (n = 5 per group; each sample assayed in duplicate; mean values ± SEM plotted; ∗, P < 0.05). (C) qRT-PCR study of the in vivo expression of M. smithii genes in a cluster (Lower) containing a formate transporter and dehydrogenase (fdhCAB) plus tungsten-containing formylmethanofuran dehydrogenase subunits (fwdEFDBAC) (n = 5 per group; each sample assayed in triplicate; mean values ± SEM plotted; ∗, P < 0.05).
Degradation of glycans by means of glycolysis leads to a buildup of reducing equivalents (e.g., NADH): primary fermentors in monoculture can respond by reducing pyruvate to succinate and lactate, at the expense of acetate (12). We observed enhanced expression of B. thetaiotaomicron glycolytic genes in biassociated compared with monoassociated animals (Fig. 3A and Table 1). However, biassociated mice had significantly lower cecal NADH/NAD+ ratios [0.17 ± 0.01 (B. thetaiotaomicron/M. smithii) vs. 0.28 ± 0.04 (B. thetaioatomicron alone); P < 0.05] and significantly lower levels of succinate [318 ± 48 vs. 517 ± 42 μmol/g dry weight cecal contents; P < 0.05] and lactate [152 ± 3 vs. 206 ± 11 μmol/g; P < 0.005] (n = 5 mice per group). In contrast, biassociation with D. piger had no significant impact on B. thetaiotaomicron fermentation as judged by these parameters (data not shown). Together, our results suggest that in vivo accumulation of reducing equivalents by B. thetaiotaomicron fermentation is relieved by interspecies transfer of electrons to M. smithii rather than by reduction of pyruvate to lactate or succinate, thereby allowing increased acetate production.
Although H2 is generally viewed as the principal currency for bacterial–archaeal electron transfer (12), formate can serve an analogous role: it has greater solubility than H2 in aqueous environments (23) and has energetically equivalent couples [−420 mV for CO2/formate and −414 mV for H+/H2 (24, 25)]. Cecal formate levels were not significantly different between biassociated and monoassociated animals despite significant up-regulation of bacterial pyruvate:formate lyase expression in mice harboring the archaeon and B. thetaiotaomicron (Fig. 3 A and B). However, we found that during in vitro growth in acetate- and formate-supplemented rich medium, M. smithii preferentially consumes formate (see Fig. 8, which is published as supporting information on the PNAS web site), raising the possibility that augmented formate production by B. thetaiotaomicron in vivo is masked by its utilization by M. smithii. Evidence for in vivo formate consumption by M. smithii came from additional experiments based on our current draft sequence of its genome, which revealed a gene cluster consisting of a formate transporter (fdhC), formate dehydrogenase subunits (fdhAB), and the subunits of tungsten-containing formylmethanofuran dehydrogenase (fwdEFDBAC; the first enzyme in the methanogenesis pathway) (Fig. 3C). qRT-PCR established that M. smithii transcripts encoding FdhC, FdhA, and FdhB were expressed at 48 ± 3-fold, 1,882 ± 559-fold, and 25 ± 8-fold higher levels, respectively, when B. thetaiotaomicron was present. Formyl-methanofuran dehydrogenase was constitutively expressed and not affected by biassociation (Fig. 3C).
These findings reveal some of the underpinnings of M. smithii–B. thetaiotaomicron mutualism. B. thetaiotaomicron obtains energy from facilitated fermentation of coveted glycans (fructans) and increased production of acetate (yields more ATP than other end products of fermentation; ref. 12). This process allows a larger population of B. thetaiotaomicron to be supported (Fig. 1). M. smithii, in turn, benefits by obtaining formate from B. thetaiotaomicron for methanogenesis, and its population expands accordingly (Fig. 1).
Ability of the Host to Harvest and Store Calories from the Diet Is Enhanced by M. smithii.
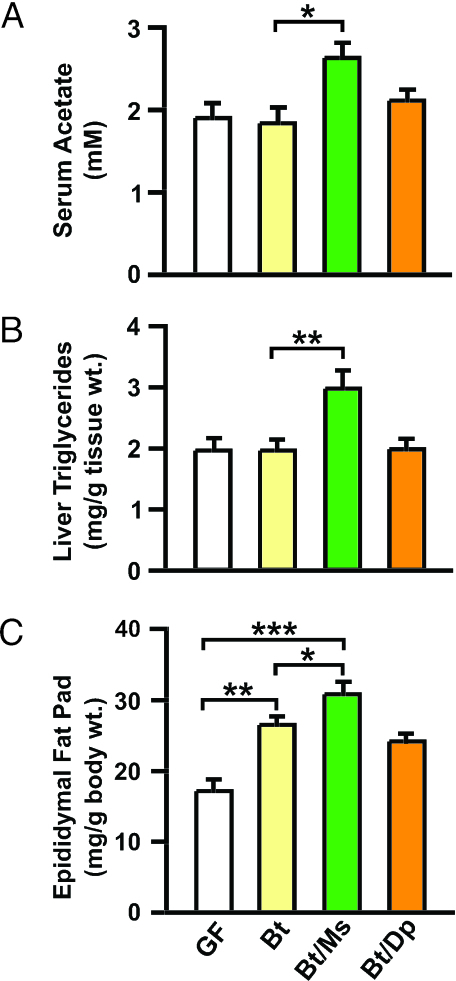
The host also benefits. As noted above, colonic absorption of SCFAs generated during fermentation represents ≈10% of the daily caloric intake from a Western diet (13). B. thetaiotaomicron/M. smithii biassociated mice exhibit increased production and absorption of SCFAs. As in the cecum, addition of M. smithii produced significantly greater serum acetate levels compared with B. thetaiotaomicron monoassociated controls; in contrast, no significant increases occurred with addition of D. piger (Fig. 4A). Distal gut-derived SCFAs are transported, via the portal vein, to the liver where they stimulate de novo lipogenesis. Fatty acid synthase (Fas) is a key enzyme in this pathway; qRT-PCR studies showed that hepatic Fas gene expression was increased 142 ± 13% in B. thetaiotaomicron/M. smithii vs. 61 ± 9% in B. thetaiotaomicron monoassociated mice (P < 0.03). Biochemical assays confirmed that addition of M. smithii, but not D. piger, to B. thetaiotaomicron-colonized animals produced significant increases in total liver triglyceride levels (Fig. 4B).
Fig. 4.
Cocolonization of mice with M. smithii and B. thetaiotaomicron enhances host energy storage. (A) GC-MS analyses of acetate in sera obtained by retro-orbital phlebotomy from fasted (4 h) 12-week-old male GF, B. thetaiotaomicron monoassociated, and biassociated [B. thetaiotaomicron/M. smithii or B. thetaiotaomicron/D. piger (Dp)] gnotobiotic mice (n = 5 per group per experiment; two independent experiments; mean ± SEM are plotted). (B) Liver triglyceride levels (n = 5 per group; each assayed in duplicate; mean ± SEM plotted). (C) Epididymal fat pad weights (n = 5 per group per experiment; two independent experiments; mean ± SEM plotted). ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.005.
The increase in hepatic de novo lipogenesis was accompanied by increased storage of energy in fat cells. Epididymal fat pad weights were significantly greater in B. thetaiotaomicron/M. smithii biassociated mice compared with B. thetaiotaomicron monoassociated controls [80 ± 6% increase over GF vs. 54 ± 7%; P < 0.05; Fig. 4C]. In contrast, there was no significant difference in fat pad weights between the B. thetaiotaomicron/D. piger and B. thetaiotaomicron groups (Fig. 4C). Dual-energy x-ray absorptiometry independently confirmed these findings: compared with GF mice, total body fat stores were increased by 47 ± 4% in B. thetaiotaomicron/M. smithii biassociated vs. 34 ± 3% in B. thetaiotaomicron monoassociated animals (n = 5 per group; P < 0.05). The increase in adiposity was not accompanied by any statistically significant differences in chow consumption (data not shown). Total body weight did not change significantly (data not shown), a finding explained by the reduction in cecal weight that is well known to occur after colonization of gnotobiotic animals (26).
Prospectus
The present work indicates that the representation of methanogenic archaea in an individual’s gut microbiota affects energy harvest from dietary glycans as well as host energy storage. The caloric content of food items is typically portrayed as a fixed value on package labels irrespective of differences that exist in the microbial ecology of our intestines (5, 27). Moreover, although metabolic studies indicate that high-fat diets are more likely to result in increased adiposity than high-carbohydrate diets, a common view is that different types of carbohydrates do not differ significantly with respect to their impact on energy balance (28). Our findings suggest that these views should be revisited. We are approaching an era of personalized nutrition where the energy content of our diet should and can be matched to the fermentative capacity of our intestinal microbiota. For example, fructans are commonly consumed as wheat products (29) and, increasingly, as sweeteners (30). Our simplified gnotobiotic model of the human gut ecosystem indicates that the presence of M. smithii may enhance the ability to extract calories from this class of polysaccharides.
Further studies are needed to understand how to manipulate the representation of M. smithii and/or other archaeons in our gut microbiota: the results could lead to a novel means for preventing obesity in the overfed or increasing caloric harvest in the underfed. Gnotobiotic mice harboring intentionally created communities of bacteria and archaea may be helpful in identifying interventions that could be used to test this concept in humans.
Materials and Methods
Colonization of GF Mice.
Mice belonging to the NMRI/KI inbred strain (31) were housed in gnotobiotic isolators (32) where they were maintained on a strict 12-h light cycle (lights on at 0600 h) and fed a standard autoclaved polysaccharide-rich chow diet (B & K Universal, East Yorkshire, U.K.) ad libitum. Each mouse was inoculated with a single gavage with 108 microbes per strain (harvested from overnight stationary phase cultures in the case of B. thetaiotaomicron, 4-d cultures for D. piger, and from serum bottles after a 5-d incubation for M. smithii). Within a given experiment, the same preparation of cultured microbes was used for biassociation and monoassociation. Details of how colonization levels were determined are described in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
RNA Isolation and GeneChip Analysis.
A total of 100–300 mg of frozen cecal contents from each gnotobiotic mouse was added to 2-ml tubes containing 250 μl of 212- to 300-μm-diameter acid-washed glass beads (Sigma), 500 μl of Buffer A (200 mM NaCl/20 mM EDTA), 210 μl of 20% SDS, and 500 μl of a mixture of phenol:chloroform:isoamyl alcohol (25:24:1; pH 4.5; Ambion, Austin, TX). Samples were lysed by using a bead beater (BioSpec Products, Bartlesville, OK; “high” setting for 5 min at room temperature). Cellular debris was pelleted by centrifugation (10,000 × g at 4°C for 3 min). The extraction was repeated by adding another 500 μl of phenol:chloroform:isoamyl alcohol to the aqueous supernatant. RNA was precipitated and resuspended in 100 μl of nuclease-free water (Ambion); 350 μl of Buffer RLT (Qiagen, Valencia, CA) was added, and RNA was further purified by using the Qiagen RNeasy mini kit. cDNA targets for GeneChip hybridization were prepared, according to the manufacturer’s instructions (Affymetrix, Santa Clara, CA), from cecal microbial RNA samples isolated from each mouse in each treatment group and then hybridized to individual custom Affymetrix B. thetaiotaomicron GeneChips (17). Details of qRT-PCR-based analyses of selected transcripts are described in Supporting Materials and Methods.
All GeneChip data sets were analyzed by using dna-chip analyzer v1.3 (dchip; www.biostat.harvard.edu/complab/dchip) and methods described by Sonnenburg et al. (17). Normalized and modeled (PM-MM) data sets were generated and used to identify differentially expressed genes between the experimental (E) and baseline (B) groups based on the following criteria: E − B > 50; E = B; P < 0.05; ≥33% “Present” call in B; ≥66% “Present” call in E; false discovery rate < 3%. In silico metabolic reconstructions were performed by using metaview (33) as described (17). Reconstructions based on the data sets referenced in this study are available from the authors upon request.
Analysis of Cecal Glycans.
GC-MS was used for determining neutral and amino sugars in cecal glycans (16). Fructan levels were assayed by using a different microanalytic approach (34). Cecal samples were collected, freeze dried at −35°C for 4 d, weighed, and stored under vacuum at −80°C until use (stable for at least 1 month). Samples (10–15 mg) then were homogenized at 1°C in 0.25 ml of 1% oxalic acid (prepared in H2O) and divided into two equal-sized aliquots, one of which was heated to 100°C for 30 min (acid hydrolysis sample), whereas the other was maintained at 1°C (control sample). A 10-μl aliquot of each sample was added to a l-ml solution containing 50 mM Tris·HCl (pH 8.1), 1 mM MgCl2, 0.02% BSA, 0.5 mM ATP, 0.1 mM NADP+, 2 μg/ml Leuconostoc mesenteroides glucose-6 phosphate dehydrogenase (253 units/mg protein; Calbiochem), 10 μg/ml yeast hexokinase (50 units/mg protein; Sigma), and 10 μg/ml yeast phosphoglucose isomerase (500 units/mg protein; Sigma). The mixture subsequently was incubated for 30 min at 24°C. The resulting NADPH product was detected by using a fluorimeter. Glucan levels were measured in a similar manner as that for fructans, except that phosphoglucose isomerase was omitted from the reactions. Fructose and glucose standards (5–10 nmol) were carried through all steps.
Assays of Organic Acid Levels.
SCFAs in mouse serum and cecal samples were assayed by using a modification of the method of Moreau et al. (35). For analysis of sera, mice were fasted for 4 h; blood was collected by retro-orbital phlebotomy into serum separation tubes (Becton Dickinson) and spun; and the supernatant (serum) was stored at −80°C before assay. Fifty microliters of serum, or 100–200 mg of frozen cecal contents, was transferred to a 4-ml glass vial fitted with a septum cap polytetrafluoroethylene (PTFE) liner (National Scientific, Rockwood, TN) and containing 10 μl of stock solution of internal standards (Isotec; each of the following components at 20 mM: [2H2]- and [1-13C]acetate, [2H5]propionate, and [13C4]butyrate). After acidification with 10 μl of 37% HCl, SCFAs were extracted (2 ml diethyl ether/extraction; 2 cycles). An aliquot of each sample then was derivatized with N-tert-butyldimethylsilyl-N-methyltrifluoracetamide (MTBSTFA; Sigma), and SCFAs were quantified by using a gas chromatograph (Model 6890; Hewlett–Packard) coupled to a mass spectrometer detector (Model 5973; Agilent Technologies, Palo Alto, CA) as described in Supporting Materials and Methods.
Analysis of Host Energy Storage.
Total body fat content was measured in 12-week-old male NMRI mice by using dual-energy x-ray absorptiometry (Lunar PIXImus Mouse; GE Medical Systems, Waukesha, WI) as described (16, 36). Epididymal fat pads and livers were removed and weighed. A portion of the liver was assayed for triglyceride content according to a standard biochemical method described by Marshall et al. (37).
Supplementary Material
Acknowledgments
We thank W. Barny Whitman (University of Georgia) and our colleagues at Washington University (Jan Amend, Jan Crowley, Fredrik Bäckhed, Jill Manchester, Sabrina Wagoner, Maria Karlsson, David O’Donnell, Justin Sonnenburg, Ruth Ley, Eric Martens, and Laura Kyro) for invaluable assistance at various stages during this project and Clay Semenkovich and Lars Angenent for critical reading of this manuscript. B.S.S. was supported by National Science Foundation Graduate Research Fellowship DGE-0202737. This work was supported by National Institutes of Health Grants DK70977 and DK30292 and the W. M. Keck Foundation.
Abbreviations
- GF
germ-free
- SCFA
short-chain fatty acid
- qRT-PCR
quantitative RT-PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The M. smithii gene sequences reported in this paper have been deposited in the GenBank database (accession no. DQ419923).
References
- 1.Eckburg P. B., Lepp P. W., Relman D. A. Infect. Immun. 2003;71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin C., Miller T. L. Arch. Microbiol. 1998;169:397–403. doi: 10.1007/s002030050589. [DOI] [PubMed] [Google Scholar]
- 3.Morvan B., Bonnemoy F., Fonty G., Gouet P. Curr. Microbiol. 1996;32:129–133. doi: 10.1007/s002849900023. [DOI] [PubMed] [Google Scholar]
- 4.Hackstein J. H. P., Van Alen T. A. Evolution (Lawrence, Kans.) 1996;50:559–572. doi: 10.1111/j.1558-5646.1996.tb03868.x. [DOI] [PubMed] [Google Scholar]
- 5.Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., Relman D. A. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segal I., Walker A. R., Lord S., Cummings J. H. Gut. 1988;29:608–613. doi: 10.1136/gut.29.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hudson M. J., Tomkins A. M., Wiggins H. S., Drasar B. S. Scand. J. Gastroenterol. 1993;28:993–998. doi: 10.3109/00365529309098298. [DOI] [PubMed] [Google Scholar]
- 8.Hackstein J. H., Van Alen T. A., Op Den Camp H., Smits A., Mariman E. Dtsch. Tierarztl. Wochenschr. 1995;102:152–154. [PubMed] [Google Scholar]
- 9.Florin T. H., Zhu G., Kirk K. M., Martin N. G. Am. J. Gastroenterol. 2000;95:2872–2879. doi: 10.1111/j.1572-0241.2000.02319.x. [DOI] [PubMed] [Google Scholar]
- 10.Rieu-Lesme F., Delbes C., Sollelis L. Curr. Microbiol. 2005;51:317–321. doi: 10.1007/s00284-005-0036-8. [DOI] [PubMed] [Google Scholar]
- 11.Miller T. L., Wolin M. J. Syst. Appl. Microbiol. 1986;7:223–229. [Google Scholar]
- 12.Stams A. J. Antonie Leeuwenhoek. 1994;66:271–294. doi: 10.1007/BF00871644. [DOI] [PubMed] [Google Scholar]
- 13.McNeil N. I. Am. J. Clin. Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 14.Topping D. L., Clifton P. M. Physiol. Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 15.Gill S. R., Pop M., Deboy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., Gordon J. I., Relman D. A., Fraser-Liggett C. M., Nelson K. E. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Backhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., Semenkovich C. F., Gordon J. I. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnenburg J. L., Xu J., Leip D. D., Chen C. H., Westover B. P., Weatherford J., Buhler J. D., Gordon J. I. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 18.Xu J., Bjursell M. K., Himrod J., Deng S., Carmichael L. K., Chiang H. C., Hooper L. V., Gordon J. I. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 19.Strocchi A., Furne J., Ellis C., Levitt M. D. Gut. 1994;35:1098–1101. doi: 10.1136/gut.35.8.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christl S. U., Murgatroyd P. R., Gibson G. R., Cummings J. H. Gastroenterology. 1992;102:1269–1277. [PubMed] [Google Scholar]
- 21.Loubinoux J., Valente F. M., Pereira I. A., Costa A., Grimont P. A., Le Faou A. E. Int. J. Syst. Evol. Microbiol. 2002;52:1305–1308. doi: 10.1099/00207713-52-4-1305. [DOI] [PubMed] [Google Scholar]
- 22.Moore W. E., Johnson J. L., Holdeman L. V. Int. J. Syst. Bacteriol. 1976;26:238–252. [Google Scholar]
- 23.Boone D. R., Johnson R. L., Liu Y. Appl. Environ. Microbiol. 1989;55:1735–1741. doi: 10.1128/aem.55.7.1735-1741.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bleicher K., Winter J. Eur. J. Biochem. 1991;200:43–51. doi: 10.1111/j.1432-1033.1991.tb21046.x. [DOI] [PubMed] [Google Scholar]
- 25.Wu W. M., Bhatnagar L., Zeikus J. G. Appl. Environ. Microbiol. 1993;59:389–397. doi: 10.1128/aem.59.2.389-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wostmann B., Bruckner-Kardoss E. Am. J. Physiol. 1959;197:1345–1346. doi: 10.1152/ajplegacy.1959.197.6.1345. [DOI] [PubMed] [Google Scholar]
- 27.Ley R. E., Peterson D. A., Gordon J. I. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 28.Saris W. H. Am. J. Clin. Nutr. 2003;78:850S–857S. doi: 10.1093/ajcn/78.4.850S. [DOI] [PubMed] [Google Scholar]
- 29.Moshfegh A. J., Friday J. E., Goldman J. P., Ahuja J. K. J. Nutr. 1999;129:1407S–1411S. doi: 10.1093/jn/129.7.1407S. [DOI] [PubMed] [Google Scholar]
- 30.Gibson G. R. J. Nutr. 1999;129:1438S–1441S. doi: 10.1093/jn/129.7.1438S. [DOI] [PubMed] [Google Scholar]
- 31.Bry L., Falk P. G., Midtvedt T., Gordon J. I. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 32.Hooper L. V., Mills J. C., Roth K. A., Stappenbeck T. S., Wong M. H., Gordon J. I. In: Molecular Cellular Microbiology. Sansonetti P., Zychlinsky A., editors. Vol. 31. San Diego: Academic; 2002. pp. 559–589. [Google Scholar]
- 33.Xu J., Gordon J. I. Bioinformatics. 2005;21:1265–1266. doi: 10.1093/bioinformatics/bti122. [DOI] [PubMed] [Google Scholar]
- 34.Passonneau J. V., Lowry O. H. Enzymatic Analysis: A Practical Guide. Totawa, NJ: Humana; 1993. [Google Scholar]
- 35.Moreau N. M., Goupry S. M., Antignac J. P., Monteau F. J., Le Bizec B. J., Champ M. M., Martin L. J., Dumon H. J. J. Chromatogr. B. Anal. Technol. Biomed. Life Sci. 2003;784:395–403. doi: 10.1016/s1570-0232(02)00827-9. [DOI] [PubMed] [Google Scholar]
- 36.Bernal-Mizrachi C., Weng S., Li B., Nolte L. A., Feng C., Coleman T., Holloszy J. O., Semenkovich C. F. Arterioscler. Thromb. Vasc. Biol. 2002;22:961–968. doi: 10.1161/01.atv.0000019404.65403.71. [DOI] [PubMed] [Google Scholar]
- 37.Marshall B. A., Tordjman K., Host H. H., Ensor N. J., Kwon G., Marshall C. A., Coleman T., McDaniel M. L., Semenkovich C. F. J. Biol. Chem. 1999;274:27426–27432. doi: 10.1074/jbc.274.39.27426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.