Abstract
Occludin and claudin are the major integral membrane components of the mammalian tight junction. Although more than 11 distinct claudins have been identified, only 1 occludin transcript has been reported thus far. Therefore, we searched by reverse transcription–PCR for occludin-related sequences in Madin–Darby canine kidney (MDCK) mRNA and identified a transcript encoding an alternatively spliced form of occludin, designated occludin 1B. The occludin 1B transcript contained a 193-base pair insertion encoding a longer form of occludin with a unique N-terminal sequence of 56 amino acids. Analysis of the MDCK occludin gene revealed an exon containing the 193-base pair sequence between the exons encoding the original N terminus and the distal sequence, suggesting that occludin and occludin 1B arise from alternative splicing of one transcript. To assess the expression and distribution of occludin 1B, an antibody was raised against its unique N-terminal domain. Immunolabeling of occludin 1B in MDCK cells revealed a distribution indistinguishable from that of occludin. Furthermore, occludin 1B staining at cell-to-cell contacts was also found in cultured T84 human colon carcinoma cells and in frozen sections of mouse intestine. Immunoblots of various mouse tissues revealed broad coexpression of occludin 1B with occludin. The wide epithelial distribution and the conservation across species suggests a potentially important role for occludin 1B in the structure and function of the tight junction.
INTRODUCTION
Tight junctions, or zonulae occludentes, are specialized membrane domains that seal the intercellular spaces in epithelia and endothelia and thus contribute to the permeability barrier between lumenal and interstitial compartments (Diamond, 1977; Powell, 1981). Tight junctions not only separate distinct physiological compartments, but they also confer selectivity to the transepithelial flux of molecules and ions through the intercellular spaces between the epithelial cells, the so-called paracellular pathway (Schneeberger and Lynch, 1992; Gumbiner, 1993; Mitic and Anderson, 1998; Goodenough, 1999). In addition, tight junctions limit the diffusion of lipids between the apical and basolateral plasma membrane domains (van Meer and Simons, 1986).
Fundamental to these functions are tight junction–associated integral membrane proteins. To date, these include JAM (junctional-associated molecule), a type I membrane protein of the immunoglobulin gene superfamily (Martin-Padura et al., 1998), and two types of four-transmembrane domain proteins: occludin (Furuse et al., 1993; Ando-Akatsuka et al., 1996) and the claudins (Furuse et al., 1998a; Morita et al., 1999a; for review, see Tsukita and Furuse, 1999). JAM seems to be capable of homotypic interaction via its extracellular domain, thus producing cell-to-cell adhesion, and was implicated in the process of monocyte infiltration through an endothelial monolayer (Martin-Padura et al., 1998). Occludin and the members of the claudin family exhibit two short hydrophobic extracellular loops that are potentially involved in homotypic or heterotypic interactions between adjacent cells and hence in both permeability barrier and selectivity functions.
Transfection of claudins into fibroblasts lacking tight junctions is sufficient to cause the assembly of freeze-fracture fibrils similar to those found in tight junctions (Furuse et al., 1998b) and to increase cell-to-cell adhesion (Kubota et al., 1999). Although the roles of claudins in junctional permeability are not fully studied (Tsukita and Furuse, 1999), it has been shown that a mutation in paracellin-1, a member of the claudin family, causes profound Mg2+ wasting (Simon et al., 1999) and that the specific removal of claudin 4 from tight junctional strands of Madin–Darby canine kidney (MDCK) cells decreases drastically the transepithelial resistance (Sonoda et al., 1999). These results strongly support a role for claudins in the architecture and function of tight junctions (Wong and Goodenough, 1999).
The role of occludin in the physiology of the tight junctions is unknown. Although occludin is distributed throughout most tight junctional fibrils, as visualized by freeze-fracture immunocytochemistry (Fujimoto, 1995), transfection experiments in nonepithelial cells have shown that occludin alone forms only short, discontinuous fibrils (Furuse et al., 1998b). This result demonstrates that occludin is a component of the junctional fibrils but is accompanied by other proteins, notably the claudins. Occludin has been implicated in cell adhesivity (Van Itallie and Anderson, 1997) and in the barrier function of epithelial tight junctions (Balda et al., 1996a; McCarthy et al., 1996; Chen et al., 1997; Wong and Gumbiner, 1997; Lacaz-Vieira et al., 1999). However, other studies have shown that in the absence of claudins, occludin alone was unable to confer cell adhesivity (Kubota et al., 1999). Also, epithelia formed from embryonic stem cells lacking occludin maintain a permeability barrier (Saitou et al., 1998).
The existence of multiple forms of claudins conferring particular properties to tight junctions from specific locations (Morita et al., 1999a,b,c; Simon et al., 1999) raises the possibility that additional forms of occludin may also exist. Here we have cloned a cDNA from MDCK epithelial cells encoding an occludin splice variant. This variant, designated occludin 1B, arises by the insertion of a novel exon that contains an alternative start. The resultant transcript encodes a longer form of occludin with a unique N-terminal sequence of 56 amino acids. Occludin 1B colocalized with occludin at the tight junctions in canine (MDCK) and human (T84) epithelial cells in culture and in mouse intestinal epithelium in situ. Occludin 1B is expressed along with occludin in canine and human epithelial cell lines, as well as in several organs from mouse. This broad distribution and the conservation of occludin 1B across species suggests a potentially important role for this isoform in the structure and function of the tight junction.
MATERIALS AND METHODS
DNA Cloning and Analysis
cDNAs containing the complete coding regions for occludin and occludin 1B were obtained by reverse transcription–PCR (RT-PCR). Total RNA was extracted from MDCK cells (total RNA extraction kit, Qiagen, Chatsworth, CA), and poly(A)+ RNA was purified on poly(dT) oligotex beads (Qiagen). RT was carried out on ∼2 μg of poly(A)+ RNA with the use of the First-Strand Synthesis Retroscript Kit according to the manufacturer's directions (Ambion, Austin, TX). PCR used 50% of the RT reaction as a template, with sense and antisense primers corresponding to the regions around canine occludin start and stop codons, respectively (P1 and P2). PCR was carried out with the use of Amplitaq DNA polymerase (Perkin Elmer-Cetus, Foster City, CA) in a volume of 30 μl with the following cycling parameters: 25 cycles at 95°C for 3 min, 60°C for 30 s, and 72°C for 1.5 min. PCR products were subcloned into pCR2.1 (Invitrogen, Carlsbad, CA) and sequenced on both strands. To eliminate the possibility of a PCR artifact, the presence of occludin 1B mRNA was independently verified by RT-PCR with the use of primers within the cDNA segment encoding for the unique 56-amino acid sequence of occludin 1B (P3 and P4) paired with primers shared by both occludins (P1 and P5). In addition, occludin 1B was identified in multiple independent RT-PCR reactions.
To confirm the authenticity of the novel transcript found by RT-PCR, genomic DNA was extracted from MDCK cells and amplified with the following primers: P6 (sense) at base 30 in U49221, and P7 (antisense) at base 474 in U49221. A prominent band of ∼3 kilobases was subcloned into the pCR2.1 (Invitrogen) and sequenced.
Exogenous Expression of Occludin 1B in MDCK Cells
The cDNA encoding for the full-length occludin 1B was amplified (sense primer P8 and antisense primer P9) and subcloned in the vector pCRII-TOPO (Invitrogen, San Diego, CA). The mammalian expression vector was prepared as follows. The vector pcDNA 3.1(−)/Myc-His A (Invitrogen), carrying the DNA sequence for myc and polyhistidine tags downstream from the multiple cloning site, was linearized with BamHI. A cDNA segment encoding six tandem repeats of the myc tag was excised from the vector CS2-MT (Roth et al., 1991) with the use of BamHI/BglII digestion and was ligated in frame with the existing myc sequence. This resulting vector was digested with NotI and BamHI, and the cDNA encoding for occludin 1B was inserted in frame, upstream of the myc tags. The accuracy of the obtained construct was verified by sequencing. The construct was transfected in MDCK cells with the use of lipofectamine plus (Life Technologies, Grand Island, NY). Transfectants were selected in medium containing 800 μg/ml (effective concentration) G418 (Life Technologies).
Primer Sequences
The primer sequences were as follows: P1, GACACCCGGGATGTCATCGAGGCCTTTTGAGAGTCCACCT; P2, GGCATCGTCGACCTATGTTTTCTGTCTATCATAGTCTCC; P3, GAGGCTGCCAGGAATTGGACATGA; P4, GCTGAGAAATGTGTAAAATCCCCATGG; P5, GGCGATGCACATCACAATGAC; P6, ATCCTGCTCGTCCTGAAGAT; P7, ATGGACAGAATCCGAATTACTC; P8, GGATCCCCGCCGCCAGCCATGACCCTGGGCGGG; P9, CAGATCTTGTTTTCTGTCTATCATAGTCTCC; P10, GCATGCATGACCCTGGGCGGG; and P11, TCAGCAGCTGAGAAATGTGTA.
Production of Antisera Specific for Occludin 1B
A SphI/HindIII-linkered cDNA segment encoding the unique 56 N-terminal amino acids of occludin 1B was prepared by PCR with the use of full-length occludin 1B as a template (P10 sense and P11 antisense) and was subcloned into the bacterial expression vector pQE30 (Qiagen), placing the occludin 1B sequence in frame downstream from a hexahistidine tag. The tagged occludin 1B peptide was expressed in BL21 by induction with 2 mM isopropylthio-β-galactoside for 5 h. Cells were collected and extracted for 3 h at room temperature in buffered 6 M guanidinium chloride, pH 8.0, and the tagged peptide was purified on nickel-nitrilotriacetic acid agarose (Qiagen), according to the protocol recommended by the manufacturer. The tagged peptide was further purified by SDS-PAGE and used to immunize two guinea pigs.
Immunoprecipitation
MDCK cells (strain II) were grown in 100-mm dishes in high-glucose DMEM containing 10% FBS and 50 U/ml penicillin/streptomycin (Life Technologies, Rockville, MD). Cells were plated at 50% confluence in culture medium containing 100 μCi/ml Express (ICN Biomedicals, Irvine, CA) and allowed to grow until they reached 90% confluence. At this point, labeled cells were rinsed three times with cold PBS containing 1.8 mM CaCl2 and 1 mM MgCl2 (PBS++) and extracted in IP buffer (10 mM Tris, pH 7.4, 50 mM NaCl, 4 mM EDTA, with the following protease inhibitors: 1 mM PMSF and 10 μg/ml each leupeptin, pepstatin, chymostatin, and aprotinin) containing 1% SDS. The extract was boiled for 3 min, triturated by passing 15 times through a 21-gauge needle, and cleared by spinning for 20 min at 12,000 rpm at 4°C. This extract was diluted 10 times with IP buffer containing 1% NP-40 instead of SDS and cleared by incubation for 1 h with a 50 μl/ml 50% suspension of protein A–agarose (Zymed Laboratories, San Francisco, CA). Sepharose beads were pelleted, and the supernatant was incubated overnight with immune serum, preimmune serum, or no antibody. Immunoprecipitates were collected on protein A–Sepharose, washed 5 times with IP buffer, eluted from beads by boiling in sample buffer, and analyzed by SDS-PAGE and autoradiography.
Dephosphorylation of Occludin 1B
MDCK cells were grown in 100-mm dishes to 80% confluence. They were rinsed with PBS++ and extracted in IP buffer containing phosphatase inhibitors (1 mM sodium vanadate and 25 mM NaF), protease inhibitors, and 1% SDS. Extracts were immunoprecipitated as described above. Immunoprecipitates were washed five times with 1% NP-40 in IP buffer and four times with alkaline phosphatase buffer (50 mM Tris, pH 8.3, 50 mM NaCl, 1 mM MgCl2, 2 mM 2-mercaptoethanol, 1 mM PMSF) and incubated for 1 h at room temperature with 20 U/ml calf intestinal alkaline phosphatase (Boehringer Mannheim, Indianapolis, IN) or in phosphatase buffer alone. Dephosphorylation was terminated by washing beads three times with 1 ml of 1% NP-40 in IP buffer. Immunoprecipitated proteins were eluted by boiling in electrophoresis sample buffer, separated by SDS-PAGE, and immunoblotted with a mouse monoclonal anti-occludin antibody (Zymed).
Immunofluorescence
MDCK or T84 cells were grown on glass coverslips. Cells were rinsed three times with PBS++. They were then fixed for 30 min with 4% paraformaldehyde in PBS++ or for 2 min in methanol, rinsed with PBS, and permeabilized for 10 min with 0.2% Triton X-100 in PBS (T-PBS). Specimens were blocked for 30 min with 5% normal goat serum in T-PBS and incubated for 1 h with the antibody diluted in T-PBS. For double immunolabeling, cells were incubated for 1 h with a 1:100 dilution of crude antiserum to occludin 1B and a 1:200 dilution of affinity-purified rabbit anti-occludin antibody (Zymed). Coverslips were rinsed with T-PBS and incubated for 30 min with a 1:200 dilution of Cy3-conjugated goat anti-guinea pig antibody (Chemicon, Temecula, CA) and a 1:100 dilution of FITC-conjugated goat anti-rabbit antibody (Jackson Immunoresearch Laboratories, West Grove, PA). Specimens were rinsed with T-PBS and mounted with Vectashield (Vector Laboratories, Burlingame, CA). For immunolocalization in mouse duodenum, fresh tissue was placed in Tissue-Tek (Miles, Elkhart, IN) and snap frozen in liquid nitrogen. Seven-micrometer cryosections were air dried, fixed for 2 min with methanol, dried, and extracted with T-PBS for 10 min. Sections were blocked with 5% normal goat serum for 30 min and double labeled by overnight incubation with a mixture of anti-occludin and anti-occludin 1B antibodies, each diluted 1:50 in T-PBS. After rinsing three times for 5 min with T-PBS, sections were incubated with Cy3-conjugated goat anti-guinea pig antibody (Chemicon) and FITC-conjugated goat anti-rabbit antibody (Jackson Immunoresearch), each diluted 1:100. Sections were rinsed with T-PBS, mounted, and viewed with a Zeiss (Thornwood, NY) photomicroscope. Exogenously expressed occludin 1B in MDCK cells was visualized with mouse monoclonal anti-myc antibody 9E10 (Calbiochem, La Jolla, CA).
Western Blotting
MDCK or T84 cells grown in 100-mm dishes were rinsed three times with PBS++ and extracted in 400 μl of hot (90°C) sample buffer containing protease inhibitors. The extract was boiled immediately for 3 min, triturated by passing 15 times through a 21-gauge needle, and cleared by spinning for 20 min at 12,000 rpm at 4°C. Samples from mouse tissues were finely minced with a razor blade, placed in SDS sample buffer containing protease inhibitors (0.4 μg/ml), and homogenized with a Teflon pestle fitted to a microfuge tube. The homogenate was boiled for 3 min and cleared by spinning for 20 min at 12,000 rpm. Protein concentration in detergent extracts was determined by Dot Metric (Geno Technology, Maplewood, MO) with the use of pancreatic RNase A (Sigma, St. Louis, MO) as a standard. Samples were separated by SDS-PAGE and transferred to an Immobilon membrane. The membrane was blocked for 1 h with 6% nonfat milk and then incubated for 1 h with the relevant antibodies (1:2000 rabbit polyclonal anti-occludin, 1:1000 mouse monoclonal anti-occludin, and 1:500 guinea pig anti-occludin 1B). Alkaline phosphatase–conjugated species-specific secondary antibodies (anti-rabbit, Kirkegard and Perry, Gaithersburg, MD; anti-mouse and anti-guinea pig, Jackson Immunoresearch Laboratories) were used at the dilutions recommended by the manufacturers.
RESULTS
Cloning of the Canine Occludin 1B cDNA
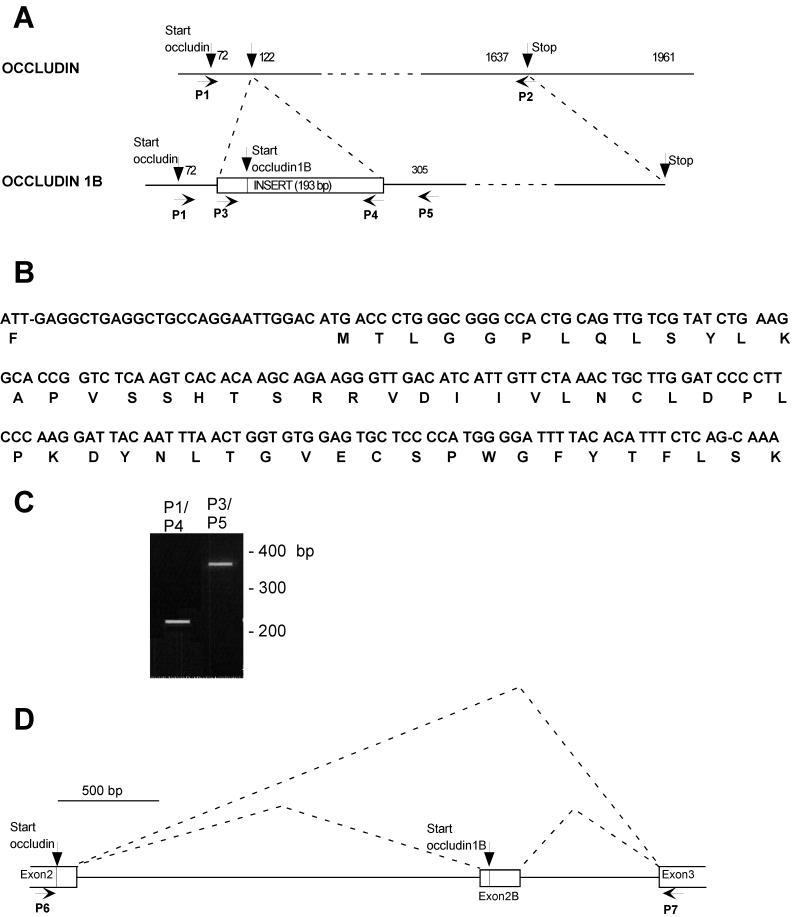
Two cDNA products were obtained from RT-PCR with the use of poly(A)+ RNA from MDCK cells as a template. One of them corresponded to the previously cloned dog occludin (Ando-Akatsuka et al., 1996); the other contained a 193-base pair (bp) insert after nucleotide 121 of canine occludin cDNA (Figure 1A). At position 27 of the insert, there was a start codon and an ORF of 56 amino acids (Figure 1B) that was continuous with that of occludin at lysine 18. In this insert, the reading frame of occludin was interrupted by a stop codon. As a result, the second cDNA encoded an occludin variant with a total size of 560 amino acids. We will refer to this variant as occludin 1B. To eliminate the possibility of a PCR artifact, the presence of occludin 1B mRNA was independently verified by RT-PCR with the use of additional primer pairs (Figure 1A, P1/P4 and P3/P5). In each pair, one of the primers corresponded to sequence common to both occludins and the other corresponded to sequence unique to occludin 1B. The PCR products obtained had the expected sizes and confirmed the nucleotide sequence of the insert and its contiguity at both ends with occludin cDNA (Figure 1C).
Figure 1.
(A) Scheme of occludin and occludin 1B cDNA. Occludin 1B cDNA is aligned relative to the cDNA of occludin, and the positions of the primers used for PCR amplification (P1–P5) are indicated. The 193-bp exon specific for occludin 1B is inserted in the cDNA of occludin and is shown as a box. This exon contains the start for occludin 1B, as indicated, and encodes 56 amino acids. P1 and P2 indicate the positions of the primers used to clone the transcript for occludin 1B. (B) Nucleotide sequence of the exon specific for occludin 1B and the deduced amino acid sequence. (C) RT-PCR verification of expression of occludin 1B in MDCK cells as a transcript containing an insert contiguous at both ends with occludin mRNA. The products obtained with the primer pairs P1/P4 and P3/P5 (shown in Figure 1A) have the expected size and confirm the contiguity between occludin mRNA and the sequence shown in Figure 1B. Similar data were obtained with three separatemRNA preparations from MDCK cells. (D) Scheme of the structure of the canine occludin gene between exons 2 and 3. P6 and P7 indicate the positions of the primers used for the mapping of this portion of the gene. P6 is at the start of occludin (analogous to exon 2 in the mouse occludin gene), and P7 is on the previously designated exon 3 in the mouse gene. Downstream from exon 3, occludin and occludin 1B cDNAs are identical. The alternative start used for occludin 1B is contained in a novel exon (exon 2B) ∼700 bp upstream from exon 3.
Although the canine occludin gene has not been characterized, the mouse occludin gene contains at least five exons (Saitou et al., 1998). Interestingly, exon 2 encodes the first 17 amino acids, whereas exon 3 encodes residues 18–243. Thus, the location of the 193-bp insertion in MDCK occludin 1B corresponds precisely to a well-characterized splice site in the mouse occludin gene. This observation supports both the notion of alternative splicing and the possibility of a conserved occludin gene structure. To determine if an additional exon was present in the canine gene, MDCK genomic DNA was amplified with the use of primers spanning the putative splice site (P6 and P7). An ∼3-kilobase fragment was obtained and sequenced (our unpublished results). The predicted 193-bp exon (referred to as exon 2B) was found 700 bp upstream from exon 3, indicating that occludin and occludin 1B are encoded by splicing variations of transcripts from the same gene (Figure 1D).
Biochemical Properties of Occludin 1B
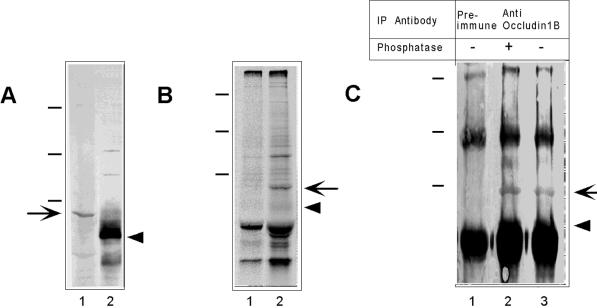
To determine if occludin 1B was present in MDCK cells, we raised a polyclonal antibody to the 56-amino acid peptide unique to occludin 1B. This antiserum immunoblotted a protein of ∼70 kDa (Figure 2A, lane 1, arrow) distinct from that of occludin (∼65 kDa; Figure 2A, lane 2, arrowhead). The specificity of the anti-occludin 1B antibody for the ∼70-kDa band was confirmed by immunoprecipitation of metabolically labeled detergent extracts of MDCK cells. As shown in Figure 2B, anti-occludin 1B precipitated one band (lane 2, arrow) not visible in the preimmune control (lane 1). This band had a mobility in SDS-PAGE corresponding to that of occludin 1B seen in the immunoblot in Figure 2A, lane 1.
Figure 2.
Biochemical characterization of occludin 1B. (A) Immunoblot of extracts of MDCK cells with polyclonal anti-occludin 1B (lane 1) and anti-occludin (lane 2). No band was detected by the preimmune serum (not shown). The arrow and arrowhead indicate occludin 1B (∼70 kDa) and occludin (∼65 kDa), respectively. (B) SDS-PAGE and autoradiography of immunoprecipitates of extracts of MDCK cells. Cells metabolically labeled with [35S]methionine were immunoprecipitated with preimmune (lane 1) or anti-occludin 1B (lane 2) sera. The arrow points to the occludin 1B band. The arrowhead indicates the expected position of occludin. (C) Anti-occludin 1B immunoprecipitates were treated with (+) or without (−) alkaline phosphatase and then displayed by SDS-PAGE followed by Western blotting with anti-occludin. Anti-occludin 1B immunoprecipitated a specific band (∼70 kDa; arrow, lanes 2 and 3) not seen in the preimmune immunoprecipitate (lane 1) that was recognized by anti-occludin. Phosphatase treatment did not shift the position of this band. The expected position of occludin is indicated by the arrowhead. Molecular mass standards (dashes, from top to bottom) are 209, 125, and 78 kDa.
Because the antibody to occludin was raised against a domain common to both occludins, it was expected that it would also recognize occludin 1B. Indeed, this antibody stained a diffuse region that overlapped with the position of migration of occludin 1B (Figure 2A, lane 2). However, because much of this staining is known to arise from the multiple bands attributable to extensive phosphorylation of occludin (Sakakibara et al., 1997), specific staining of occludin 1B could not be determined in these blots. To demonstrate cross-reactivity of anti-occludin with occludin 1B, anti-occludin 1B immunoprecipitates were displayed by SDS-PAGE followed by Western blotting with anti-occludin. As can be seen in Figure 2C, occludin 1B could be readily detected with anti-occludin in these immunoprecipitates (lane 2, arrow). Unlike that of occludin (Sakakibara et al., 1997), phosphatase treatment did not change detectably the mobility of occludin 1B (lane 2), although the same treatment of occludin immunoprecipitates removed the upper bands associated with its phosphorylated forms (our unpublished results). These immunoblotting studies with anti-occludin indicated that the protein levels of occludin 1B were much lower than those of occludin (Figure 2A, lane 2).
Immunolocalization and Tissue Distribution of Occludin 1B in Epithelial Cells
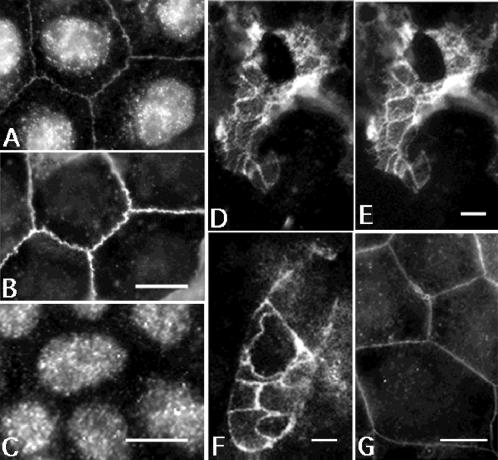
Occludin 1B colocalized with occludin by immunocytochemistry. Figure 3A shows that the antiserum to occludin 1B labeled the cell-to-cell contacts of MDCK cells at a position coincident with anti-occludin staining (Figure 3B). The nuclear staining that appears in Figure 3A was also observed in specimens incubated with the preimmune serum and therefore is nonspecific (Figure 3C). Occludin 1B colocalized with occludin at the cell contacts between epithelial cells from mouse duodenum (Figure 3, D and E). In addition, occludin 1B was evident in T84 human colon carcinoma cells (Figure 3F). Finally, epitope-tagged occludin 1B correctly targeted to cell-to-cell contacts when exogenously expressed in MDCK cells (Figure 3G). Together, these data suggested that occludin 1B is a component of the tight junction in multiple epithelial cell types.
Figure 3.
Indirect immunofluorescence with anti-occludin 1B (A, D, and F), preimmune occludin 1B (C), anti-occludin (B and E), and anti-myc (G). Specimens shown are MDCK cells (A–C and G), T84 cells (F), and mouse duodenum (D and E). (A and B) Double labeling of occludin 1B (A) and occludin (B) in MDCK cells. Note colocalization of the immune staining of both occludins at the cell-to-cell contacts. The nuclear labeling observed in A is nonspecific, because it is also detected by the preimmune serum (C). (D and E) Tangential section through the apical area of mouse intestinal epithelial cells double labeled with antiserum to occludin 1B (D) and with affinity-purified polyclonal anti-occludin (E). The staining for occludin 1B and occludin colocalized at the junctional complexes between intestinal epithelial cells. (F) T84 cells labeled with anti-occludin 1B. The staining is at the cell-to-cell contacts, most likely at the tight junction. (G) MDCK cells expressing myc-tagged full-length occludin 1B are labeled with anti-myc antibody. Transfected occludin 1B is targeted to the cell-to-cell contacts. Bars, 10 μm for A–C, F, and G; 5 μm for D and E.
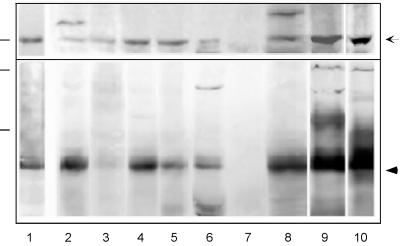
Several murine tissues (intestine, brain, heart, kidney, liver, pancreas, spleen, testis), and epithelial cell lines (MDCK and human T84) were analyzed by immunoblotting (Figure 4). Equal amounts of protein from each sample were probed separately with antibodies specific for each occludin. Figure 4 indicates that occludin 1B (top) showed a distribution among tissues that paralleled that of occludin. Both occludins were absent from spleen but were detected in all other tissues examined. Their presence in heart and brain suggested that tight junctions between some endothelial cells contained both occludins. In the brain and testis, additional immunoreactive bands (∼80–90 kDa) were detected by the antiserum to occludin 1B. The relationship of this band to occludin 1B was unclear.
Figure 4.
Tissue expression of occludin 1B and occludin. Mouse organ homogenates were subjected to SDS-PAGE and immunoblotting with the use of anti-occludin 1B (top) or anti-occludin (bottom). The same analysis was done for extracts of MDCK cells and T84 human colon carcinoma cells. Protein loading for extracts from tissues and cell lines was 30 μg/lane, except for duodenum, for which the loading was 75 μg/lane. Molecular mass standards (dashes at left) are 78 kDa for the top panel and 125 and 78 kDa for the bottom panel. Occludin 1B and occludin are indicated by the arrow and the arrowhead, respectively. The lanes contain extracts from the following sources: mouse duodenum (1), brain (2), heart (3), kidney (4), liver (5), pancreas (6), spleen (7), testis (8), MDCK cells (9), and T84 cells (10).
DISCUSSION
We have cloned a cDNA from MDCK cells encoding a novel occludin isoform, termed occludin 1B. According to the deduced sequence, occludin 1B has a unique stretch of 56 amino acids at the N terminus that replace the first 17 amino acids of occludin. Occludin and occludin 1B are identical from Lys-18 in occludin (Lys-57 in occludin 1B) to the C terminus and represent alternatively spliced transcripts of the single occludin gene. As revealed by immunofluorescence, occludin 1B colocalized at cell-to-cell contacts with occludin, a specific marker for the tight junction. In addition, exogenously expressed full-length, myc-tagged occludin 1B was recruited to apical cell-to-cell contacts in MDCK cells. Together with the finding of occludin 1B in several mammalian species, these results suggest that occludin 1B may be a characteristic component of all tight junctions. The novel sequences in MDCK occludin 1B were encoded in a 193-bp exon not identified in the initial characterization of the murine gene (Saitou et al., 1998). Strikingly, the location of the 193-bp insert in MDCK cDNA at the second base of codon 17 coincides precisely with the reported splice junction of exons 2 and 3 in the mouse gene (Saitou et al., 1998). Thus, it is likely that the murine and canine occludin genes have a similar structure, at least in this region.
A possible significance to multiple occludin expression would be to provide different functional properties to tight junctions in different locations. The claudins may be a useful comparison in this regard, because they are a multigene family in which each member has a distinctive tissue distribution (Tsukita and Furuse, 1999) and may confer specific properties of transjunctional permeability (Simon et al., 1999; Wong and Goodenough, 1999). There is no compelling evidence as yet for the existence of multiple occludin genes (Saitou et al., 1997), but multiple occludins could arise by alternative splicing. This notion is supported by the cloning of occludin 1B and may be extended to other isoforms, because the antibody to the 56-amino acid peptide detected additional immunoreactive bands (∼80–90 kDa; Figure 4) in mouse brain and testis. The unique N terminus of occludin 1B lacks the polyproline sequence RPFESPPPYRP contained within the 17 N-terminal amino acids specific for occludin. The amino acid sequence around this proline cluster fits the reported consensus sequence recognized by SH3 domains (XPXXPPP*XP, where * indicates any hydrophobic amino acid; Ren et al., 1993) and suggests that the N terminus of occludin, but not that of occludin 1B, may bind proteins containing SH3 domains. However, for occludin, the function of the N-terminal domain and the potential binding partners to this region have not been studied experimentally. For occludin 1B, database searches did not reveal any sequences homologous to the 56-amino acid sequence at the N terminus.
Some properties of occludin 1B are similar to those of occludin, whereas others are distinct. For example, both proteins were targeted with equal efficiency to junctional complexes, because double immunolabeling revealed the distribution of myc-tagged occludin 1B to be perfectly superimposable on that of occludin. Thus, replacement of the first 17 N-terminal amino acids in occludin with the novel 56-amino acid sequence specific for occludin 1B did not alter its intracellular trafficking. Because occludin 1B shares an identical cytoplasmic C-terminal domain with occludin, it is expected that the ZO family of proteins known to bind the C terminus of occludin (Furuse et al., 1994; Fanning et al., 1998) would also bind occludin 1B. Therefore, the cytoplasmic C terminus provides at least one of the targeting/anchoring cues necessary for the recruitment of occludin 1B to the tight junction (Mitic et al., 1999). The functions performed by the first N-terminal amino acids in occludin and occludin 1B remain to be determined. On the other hand, the patterns of occludin 1B phosphorylation appear to differ from those of occludin. Previous studies have shown that occludin phosphorylation regulates tight junction assembly and function (Cordenonsi et al., 1997; Sakakibara et al., 1997; Wong, 1997). Although no specific kinases have been directly implicated, several serine/threonine kinases are localized at tight junctions (Stuart and Nigam, 1995; Balda et al., 1996b; Izumi et al., 1998). Unlike occludin, occludin 1B did not undergo detectable shifts in mobility under dephosphorylating conditions. However, because of the lower level of expression of occludin 1B, phosphorylated forms of occludin 1B may be difficult to detect or may comigrate with unphosphorylated forms on SDS gels.
The existence of occludin 1B may explain a discrepancy in the literature regarding the relationship between transepithelial resistance and paracellular flux. Extracellular addition of a peptide corresponding to the second extracellular loop of occludin results in a decrease in the transepithelial resistance and an increase in the paracellular flux (Wong and Gumbiner, 1997). In contrast, overexpression of occludin in cultured epithelial cells results in an increase in the transepithelial resistance and a concomitant increase in the paracellular flux, rather than the expected reduction (Balda et al., 1996b; McCarthy et al., 1996). One possible explanation for this discrepancy is that extracellular application of a peptide will affect both occludin and occludin 1B, as well as other putative occludin isoforms that share this sequence, whereas the overexpression of occludin alone may alter the normal ratio of the two occludins or other tight junction proteins, resulting in a different form of junctional perturbation.
ACKNOWLEDGMENTS
We are grateful to the members of the Goodenough/Paul laboratory for their help with different phases of this project. This work was supported by National Research Service Award postdoctoral fellowship DK09411 to Z.M. and by National Institutes of Health grants GM18974 and GM37751 to D.A.G. and D.L.P.
REFERENCES
- Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Furuse M, Tsukita S. Interspecies diversity of the occludin sequence: cDNA cloning of human, mouse, dog, and rat-kangaroo homologues. J Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Anderson JM, Matter K. The SH3 domain of the tight junction protein ZO-1 binds to a serine protein kinase that phosphorylates a region C-terminal to this domain. FEBS Lett. 1996a;399:326–332. doi: 10.1016/s0014-5793(96)01352-x. [DOI] [PubMed] [Google Scholar]
- Balda MS, Whitney JA, Flores C, Gonzalez S, Cereijido M, Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J Cell Biol. 1996b;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-H, Merzdorf C, Paul DL, Goodenough DA. COOH terminus of occludin is required for tight junction barrier function in early Xenopus embryos. J Cell Biol. 1997;138:891–899. doi: 10.1083/jcb.138.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordenonsi M, Mazzon E, De Rigo L, Baraldo S, Meggio F, Citi S. Occludin dephosphorylation in early development of Xenopus laevis. J Cell Sci. 1997;110:3131–3139. doi: 10.1242/jcs.110.24.3131. [DOI] [PubMed] [Google Scholar]
- Diamond J. The epithelial junction: bridge, gate, and fence. Physiologist. 1977;20:10–18. [PubMed] [Google Scholar]
- Fanning AS, Jameson BJ, Jesaitis LA, Anderson JM. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J Biol Chem. 1998;273:29745–29753. doi: 10.1074/jbc.273.45.29745. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins: application to the immunogold labeling in intercellular junctional complexes. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998a;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Itoh M, Hirase T, Nagafuchi A, Yonemura S, Tsukita S, Tsukita S. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol. 1994;127:1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol. 1998b;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough DA. Plugging the leaks. Proc Natl Acad Sci USA. 1999;96:319–321. doi: 10.1073/pnas.96.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Breaking through the tight junction barrier. J Cell Biol. 1993;123:1631–1633. doi: 10.1083/jcb.123.6.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y, Hirose T, Tamai Y, Hirai S, Nagashima Y, Fujimoto T, Tabuse Y, Kemphues KJ, Ohno S. An atypical PKC directly associates and colocalizes at the epithelial tight junction with ASIP, a mammalian homologue of Caenorhabditis elegans polarity protein PAR-3. J Cell Biol. 1998;143:95–106. doi: 10.1083/jcb.143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota K, Furuse M, Sasaki H, Sonoda N, Fujita K, Nagafuchi A, Tsukita S. Ca2+-independent cell-adhesion activity of claudins, a family of integral membrane proteins localized at tight junctions. Curr Biol. 1999;9:1035–1038. doi: 10.1016/s0960-9822(99)80452-7. [DOI] [PubMed] [Google Scholar]
- Lacaz-Vieira F, Jaeger MMM, Farshori P, Kachar B. Small synthetic peptides homologous to segments of the first external loop of occludin impair tight junction resealing. J Membr Biol. 1999;168:289–297. doi: 10.1007/s002329900518. [DOI] [PubMed] [Google Scholar]
- Martin-Padura I, et al. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and regulates monocytes transmigration. J Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KM, Skare IB, Stankewich MC, Furuse M, Tsukita S, Rogers RA, Lynch RD, Schneeberger EE. Occludin is a functional component of the tight junction. J Cell Sci. 1996;109:2287–2298. doi: 10.1242/jcs.109.9.2287. [DOI] [PubMed] [Google Scholar]
- Mitic LL, Anderson JM. Molecular architecture of tight junctions. Annu Rev Physiol. 1998;60:121–142. doi: 10.1146/annurev.physiol.60.1.121. [DOI] [PubMed] [Google Scholar]
- Mitic LL, Schneeberger EE, Fanning AS, Anderson JM. Connexin-occludin chimeras containing the ZO-binding domain of occludin localize at MDCK tight junctions and NRK cell contacts. J Cell Biol. 1999;146:683–693. doi: 10.1083/jcb.146.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999a;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin-11/OSP-based tight junctions in myelinated sheaths of oligodendrocytes and Sertoli cells in testes. J Cell Biol. 1999b;145:579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Furuse M, Tsukita S. Endothelial claudins: Claudin-5/TMVCF constitutes tight junction strands in endothelial cells. J Cell Biol. 1999c;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DW. Barrier function of epithelia. Am J Physiol. 1981;241:G275–G288. doi: 10.1152/ajpgi.1981.241.4.G275. [DOI] [PubMed] [Google Scholar]
- Ren R, Mayer BJ, Chichetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Roth MB, Zahler AM, Stolk JA. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Ando-Akatsuka Y, Itoh M, Furuse M, Inazawa J, Fujimoto K, Tsukita S. Mammalian occludin in epithelial cells: its expression and subcellular distribution. Eur J Cell Biol. 1997;73:222–231. [PubMed] [Google Scholar]
- Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, Takano H, Noda T, Tsukita S. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. J Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara A, Furuse M, Saitou M, Tsukita S. Possible involvement of phosphorylation of occludin in tight junction formation. J Cell Biol. 1997;137:1393–1401. doi: 10.1083/jcb.137.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. Structure, function, and regulation of cellular tight junctions. Am J Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- Simon DB, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J Cell Biol. 1999;147:195–204. doi: 10.1083/jcb.147.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RO, Nigam SK. Regulated assembly of tight junctions by protein kinase C. Proc Natl Acad Sci USA. 1995;92:6072–6076. doi: 10.1073/pnas.92.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Furuse M. Occludin and claudins in tight-junction strands: leading or supporting players? Trends Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- Van Itallie CM, Anderson JM. Occludin confers adhesiveness when expressed in fibroblasts. J Cell Sci. 1997;110:1113–1121. doi: 10.1242/jcs.110.9.1113. [DOI] [PubMed] [Google Scholar]
- van Meer G, Simons K. The function of tight junctions in maintaining differences in lipid composition between the apical and the basolateral cell surface domains of MDCK cells. EMBO J. 1986;5:1455–1464. doi: 10.1002/j.1460-2075.1986.tb04382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V. Phosphorylation of occludin correlates with occludin localization and function at the tight junction. Am J Physiol. 1997;273:C1859–C1867. doi: 10.1152/ajpcell.1997.273.6.C1859. [DOI] [PubMed] [Google Scholar]
- Wong V, Goodenough DA. Paracellular channels! Science. 1999;285:62. doi: 10.1126/science.285.5424.62. [DOI] [PubMed] [Google Scholar]
- Wong V, Gumbiner BM. A synthetic peptide corresponding to the extracellular domain of occludin perturbs the tight junction permeability barrier. J Cell Biol. 1997;136:399–409. doi: 10.1083/jcb.136.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]