Abstract
The oral cavity is a unique environment in which antimicrobial peptides play a key role in maintaining health and may have future therapeutic applications. Present evidence suggests that α-defensins, β-defensins, LL-37, histatin, and other antimicrobial peptides and proteins have distinct but overlapping roles in maintaining oral health and preventing bacterial, fungal, and viral adherence and infection. The expression of the inducible hBD-2 in normal oral epithelium, in contrast to other epithelia, and the apparent differential signaling in response to commensal and pathogenic organisms, provides new insights into innate immunity in this body site. Commensal bacteria are excellent inducers of hBD-2 in oral epithelial cells, suggesting that the commensal bacterial community acts in a manner to benefit the overall innate immune readiness of oral epithelia. This may have major significance for understanding host defense in the complex oral environment.
The oral environment
The oral cavity is a unique environment. Oral mucosa is a critical protective interface between external and internal environments and must serve as a barrier to the myriad microbial species present in this warm, moist environment. The oral cavity is the only area of the body in which hard tissues break through the epithelial surface. The periodontal epithelium surrounding the tooth is specialized to form an attachment and seal around each tooth. This unique function imparts special challenges to the tissue and leads to certain vulnerabilities associated with periodontal disease, especially in view of the continual exposure to the bacterial biofilm (dental plaque) that forms on the tooth surface at the junction of the soft tissue. Thus, this anatomical region is one where there is a significant risk of bacterially induced infection and inflammation.
Antimicrobial peptides are important contributors to maintaining the balance between health and disease in this complex environment. These include several salivary antimicrobial peptides, the β-defensins expressed in the epithelium, the α-defensins expressed in neutrophils, and the cathelicidin, LL-37, expressed in both epithelium and neutrophils. These peptides are part of the host innate immune response in this environment. Epithelia, polymorphonuclear leukocytes (neutrophils), and saliva all contribute to the maintaining the health of the oral cavity in overlapping but independent ways. This review will focus on the human oral cavity and include 1) the expression and function of antimicrobial peptides in the oral cavity in the context of innate immune responses, 2) regulation of β-defensins which has led to advances in our understanding of oral epithelial innate immunity, and 3) functional efficacy against oral microbes when it is known.
Epithelial antimcrobial peptides
Historically, the oral epithelium has been considered mainly as a passive covering that becomes damaged and ulcerated in disease. This view has changed dramatically and the epithelial compartment is now seen as providing both a physical barrier to infection and playing an active role in innate host defense (Dale, 2002; Darveau et al., 1997; Ganz, 2003; Tonetti, 1997). Epithelial cells are in constant contact with bacterial products from supra- and sub-gingival biofilms on the tooth surface as well as from bacteria attached to mucosal surfaces. These cells respond to bacteria in an interactive manner; they secrete IL-8 and other chemokines and cytokines to alert various cell types and attract neutrophils. They also produce natural antimicrobial peptides and proteins constitutively and inducibly in response to bacterial exposure. These antimicrobial peptides are part of the innate immune system, a complex set of responses that keeps microbial invaders in check and maintains the microbial ecology of the healthy mucosa (Weinberg et al., 1998). Thus, the epithelium functions to actively respond to the environment, participates in response to infection, in signaling further host responses, and in integrating innate and acquired immune responses.
The first antimicrobial peptide identified in oral epithelium was the β-defensin, lingual antimicrobial peptide (LAP), described in bovine tongue (Zasloff et al., 1995). We now know that several families of natural antibiotic peptides or proteins are expressed in oral epithelium. These include members of the β-defensin family of peptides, the protein calprotectin (also known as calgranulin), and the multifunctional peptide, adrenomedullin (Dale and Krisanaprakornkit, 2001; Devine, 2003; Ganz, 2003; Kapas et al., 2001a; Ross and Herzberg, 2001). These antimicrobials have broad specificity with activity against Gram-positive and Gramnegative bacteria (reviewed in Hancock, 1997), as well as against yeast and some viruses (Ganz, 2003; Lehrer and Ganz, 2002; Quinones-Mateu et al., 2003). They complement the antimicrobial factors of saliva, such as the histatins, lysozyme, and salivary immunoglobulins. Both constitutive and inducible antimicrobial peptides are expressed in gingival epithelium, suggesting that they have a normal surveillance function as well as a specific role in innate host defense in response to infection. The role of these natural antibiotics is only just beginning to be appreciated, with potential applications for enhanced natural expression or as new therapeutic agents. Their role may be especially important for the oral cavity in which there is constant exposure to microbial challenges (Dale and Krisanaprakornkit, 2001; Weinberg et al., 1998). Further, individual variation in expression may contribute to disease susceptibility. Overall expression of antimicrobial peptides in the oral cavity is summarized in Table 1.
Table 1.
Antimicrobial peptides expressed in the oral cavity.
| Antimicrobial peptide | Site of expression | Role/comments1 |
|---|---|---|
| HNP1–4 (α-Defensins) | Neutrophils (azurophilic granules)
Gingival sulcus Sites of inflammation |
Antibacterial, antifungal, and antiviral. Functional levels in GCF Expression defective in Morbus Kostmann syndrome (congenital neutropenia associated with periodontal disease) |
| LL-37 | Neutrophils
Gingival sulcus Saliva |
Primarily antibacterial.
Expression is defective in Morbus Kostmann syndrome. |
| β-Defensins
hBD-1 hBD-2 hBD-3 |
Suprabasal layers of stratified epithelium; hBD-1 and hBD-2 absent in junctional epithelium; hBD-3 mRNA is widely expressed but peptide localization is not known | Antibacterial, antifungal, and antiviral. Part of the protective barrier function of epithelium. Secreted, may be associated with cell or mucosal surface. Also found in salivary glands and saliva. |
| Histatin | Saliva (parotid and submandibular) | Antifungal. Histidine-rich group of peptides. Histatin 5 (24 amino acids) is most active. Antifungal action requires metabolic activity. |
| Adrenomedullin | Epithelium | Antibacterial, mitogenic, vasodilator, inducible peptide. |
See text for references.
β-Defensins in oral cells and tissue
Human β-defensins (hBDs) are expressed in all human epithelial tissues tested to date including those of the oral cavity (Dale and Krisanaprakornkit, 2001; Lehrer and Ganz, 2002). β-defensins are expressed in gingiva, tongue, salivary glands, and mucosa (Mathews et al., 1999). They are present in oral inflammatory conditions, oral carcinomas, and some cell lines derived from oral carcinomas (Abiko et al., 1999; Mizukawa et al., 2000). Oral keratinocytes have been a useful model for investigation of the regulation of β-defensin expression. The human peptide hBD-1 is constitutively expressed while hBD-2 and hBD-3 are upregulated in inflamed skin and other epithelia (Diamond et al., 1991; Harder et al., 1997a; Harder et al., 2001; Krisanaprakornkit et al., 2000; Krisanaprakornkit et al., 1998; Liu et al., 2002; Schonwetter et al., 1995; Singh et al., 1998; Stolzenberg et al., 1997; Valore et al., 1998; Zhao et al., 1996). These and similar peptides in other mammalian species are induced by inflammation and proinflammatory cytokines, and in some cell types, by bacterial lipopolysaccharide (LPS) (Diamond and Bevins, 1994; Diamond et al., 2000; Diamond et al., 1996; Russell et al., 1996). Oral epithelial tissues and oral epithelial cells derived from gingiva express hBD-1, -2, and -3 (Dunsche et al., 2002; Dunsche et al., 2001; Krisanaprakornkit et al., 2000; Krisanaprakornkit et al., 1998). Oral expression of other members of the large β-defensin family recently identified by Schutte and coworkers is not yet characterized (Schutte et al., 2002).
β-Defensins in oral tissue
Normal uninflamed gingival tissues express both hBD-1 and hBD-2. Both the constitutive and inducible β-defensins have been found in essentially all gingival biopsies tested, both healthy uninflamed and inflamed samples (Krisanaprakornkit et al., 2000; Krisanaprakornkit et al., 1998). Thus, normal non-inflamed oral epithelial tissue is activated to express hBD-2, in contrast to normal epidermis, trachea, and gut. This expression of hBD-2 is not accompanied by upregulation of markers of host innate immune response such as IL8. It seems to be part of the normal barrier function of gingival epithelia. The hypothesis is that this exposure represents a useful interaction between the commensal bacteria and the tissue, resulting in enhanced expression of hBD-2 and therefore providing an advantage in subsequent response to other potentially pathogenic organisms. This may be a general paradigm, since it has been shown that Staphylococcus epidermidis, the major commensal skin bacterium, also is an excellent inducer of hBD-2 in epidermal keratinocytes (Chung and Dale, 2004; Dinulos et al., 2003). The role of hBD-1 may be in preventing commensal bacteria from becoming opportunistic pathogens, while hBD-2 and hBD-3 and other inducible antimicrobials may be more effective against pathogens.
β-Defensins are differentially regulated in oral keratinocytes
HBD-1 is constitutively expressed in oral keratinocytes while hBD-2 is upregulated by bacterial and proinflammatory stimuli (Krisanaprakornkit et al., 2000; Mathews et al., 1999). The unstimulated level of hBD-2 is extremely low and upregulation of mRNA expression occurs rapidly, within 2–4 hr after stimulation with bacterial products or TNFα. Stimulants include TNFα, IL1β, Fusobacterium nucleatum (a commensal oral bacteria, not associated with periodontal disease), and Porphyromonas gingivalis, a periodontal pathogen. Nevertheless, bacterial LPS preparations from F. nucleatum, P. gingivalis and E.coli are relatively poor inducers of hBD-2 in oral keratincytes (Krisanaprakornkit et al., 2000). This is in contrast to regulation in tracheal epithelial cells in which hBD-2 is upregulated by E. coli LPS (Becker et al., 2000; Diamond et al., 2000). HBD-3 is upregulated by interferon-γ (Garcia et al., 2001), various bacteria (Harder et al., 2001), and by Actinobacillus actinomycetemcomitans in oral epithelial cells (Feucht et al., 2003).
β-Defensin expression is associated with differentiation
HBD-1 and hBD-2 mRNA and peptides are expressed as a function of differentiation in cultured oral keratinocytes. In keratinocytes in vitro, the peptides are detected only in cells that are expressing involucrin, an early marker of differentiation (Dale et al., 2001; Dale and Krisanaprakornkit, 2001). In normal gingival tissue mRNAs for both hBD-1 and hBD-2 are most strongly expressed in the spinous layer of the tissue, while the peptides are detected in the upper spinous, granular, and cornified layers. The strongest expression is at the gingival margin, adjacent to the region of plaque formation on the tooth surface and in inflamed sulcular epithelium (Dale et al., 2001). The tissue location is consistent with a role for these peptides in the epithelial antimicrobial barrier. HBD-1 and hBD-2 are not detected in junctional epithelium although this region is frequently the site of inflammation. However, the cells of the junctional epithelium are relatively undifferentiated in contrast to other regions of the oral cavity (Schroeder and Listgarten, 1997). The lack of expression in the junctional epithelium, the suprabasal localization in stratified epithelia, and the association with differentiation in vitro, all point to a dependence on normal differentiation for expression of β-defensins in stratified oral epithelia as well as in the epidermis (Liu et al., 2002).
Epithelial antimicrobials
Calprotectin
Calprotectin or calgranulin is a heterodimeric calcium- and zinc-binding protein, also referred to as S100A8 and S100A9. It is expressed in neutrophils, monocytes, macrophages, and mucosal keratinocytes and is involved in leukocyte trafficking and arachidonic acid metabolism (Nacken et al., 2003). It is upregulated in inflammatory conditions, including periodontal disease in which it is elevated in the gingival cervicular fluid (Kido et al., 1999). It is constitutively expressed in cells of stratified oral epithelia and in cultured gingival epithelial cells (Ross and Herzberg, 2001) and is highly responsive to stress in epidermis (Marionnet et al., 2003). The mechanism of antimicrobial action of calprotectin appears to be due to competition for zinc, a growth requirement for many microbial species (Brandtzaeg et al., 1995). Calprotectin expression confers protection from bacterial binding and invasion and may contribute to resistance of gingival cells to invasion by Porphyromonas gingivalis, a gram-negative periodontal pathogen (Nisapakultorn et al., 2001).
Adrenomedullin
Adrenomedullin is a multifunctional peptide that was initially characterized for its vasodilatory effects and subsequently has been recognized to have antibacterial function against both Gram positive and Gram negative bacteria from the oral cavity, skin, respiratory tract and gut (Allaker and Kapas, 2003). It does not display antifungal activity. It is constitutively expressed and secreted by oral epithelial cells; expression is also increased in response to live oral bacteria, IL1, and TNFα (Kapas et al., 2001a; Kapas et al., 2001b). Adrenomedullin consists of 52 amino acids with one intramolecular disulfide bond. The precursor is encoded by the ADM gene which maps to human chromosome 11. Although there are some functional parallels to the β-defensins, the adrenomedullin gene and protein structure differ from that of the β-defensins. Adrenomedullin has some homology with calcitonin gene-related peptide (Kitamura et al., 1993) and binds to the calcitonin receptor-like receptor (McLatchie et al., 1998) and is thought to have hormone-like functions in the control of circulation. Mice lacking expression of adrenomedullin die in mid-gestation due to cardiovascular abnormalities (Caron and Smithies, 2001).
Epithelial and neutrophil antimicrobial peptides function together in gingiva
Neutrophil antimicrobial peptides work together with the epithelium to provide a barrier to microbial colonization in the oral cavity, particularly in the region adjacent to the tooth surface. The junctional epithelium which forms the attachment of the soft tissue to the tooth surface is less differentiated and more permeable than the sulcular or oral surface epithelium. Neutrophils migrate through the junctional epithelium in response to a gradient of IL8 expressed in the junctional and sulcular epithelia (Tonetti et al., 1998). The importance of neutrophils in oral health is readily seen by the severity of problems in individuals with defects in neutrophil chemotaxis or function. Such defects are associated with severe periodontal disease occurring at a young age (see below).
LL-37 and α-defensins
The cathelicidin, LL-37, is expressed in epithelial cells especially following inflammatory stimulation, and both the protein and mRNA have been detected in human tongue and buccal mucosa (Frohm Nilsson et al., 1999) and saliva (Murakami et al., 2002). However, LL-37 detected in gingival epithelium by immunohistochemisty appeared to be the product of neutrophil migration through the tissue rather than the epithelial cells per se (Dale et al., 2001). In addition, the neutrophil α-defensins, HNP-1–3, are readily detected in the junctional epithelium (Dale et al., 2001). They can be detected in quantities consistent with their antimicrobial function in the gingival crevicular fluid that lies between the epithelium and the tooth surface (McKay et al., 1999).
Localized expression of antimicrobial peptides in gingiva
The α- and β-defensins and LL-37 are localized in different sites in the gingiva, suggesting that they may serve different roles in the several ecological niches of the periodontium. The β-defensin peptides, hBD-1 and hBD-2 are localized in the differentiated layers of the gingival epithelium in a pattern of expression consistent with function as a microbial barrier (Dale et al., 2001). Their expression is particularly strong at the gingival margin where the tissue can be expected to be in nearly continuous contact with supragingival plaque. While β-defensins are poorly expressed in the undifferentiated cells of junctional epithelium, the α-defensins and LL-37 are present in high amounts in neutrophils that migrate through the junctional epithelium to the gingival sulcus. Thus, junctional epithelium is protected by α-defensins and LL-37 released from neutrophils, whereas the differentiated stratified epithelia are protected by β-defensins (Dale et al., 2001; McKay et al., 1999). Taken together in the context of the oral cavity, the observations on tissue and gingival fluid suggest that the gingiva is protected by α- and β-defensins, LL-37, and salivary antimicrobial peptides as summarized in Fig. 1. This is quite analogous to the findings in intestine in which the crypts are protected by α-defensins and the epithelia of intestinal villae express β-defensins (Bevins et al., 1999). Both α- and β-defensins, as well as LL-37, signal other innate and acquired immune responses (see below, reviewed in Lehrer and Ganz, 2002) in addition to their antimicrobial properties.
Fig. 1.
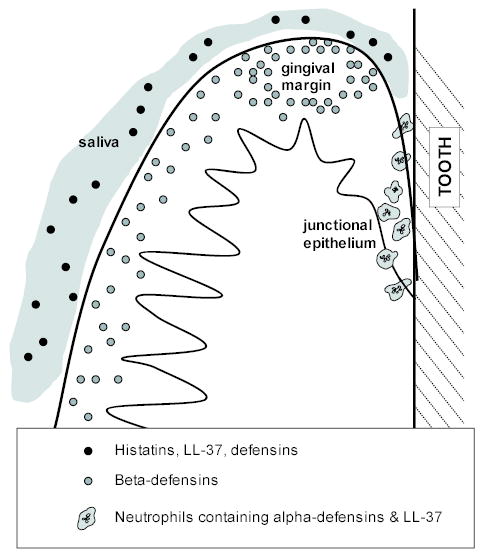
Localized expression of antimicrobial peptides in gingiva. Diagrammatic view of a cross-section of the gingiva illustrating the oral aspect of the tissue, the gingival margin at the crest of the gum tissue, and the junctional epithelium attaching to the tooth surface. The sulcular epithelium is adjacent to the space (sulcus) between the tissue and the tooth. Neutrophils, containing α-defensins and LL-37, migrate through the junctional epithelium into the gingival sulcus. β-Defensins are expressed in the gingival epithelium. The tissue is bathed in saliva containing histatins, LL-37 and α- and β-defensins.
The expression and regulation of antimicrobial peptides indicates heightened innate immune responses in oral epithelia
HBD-2 is expressed in both normal and inflamed oral epithelia
In contrast to most other epithelia, in which hBD-2 is expressed only in the presence of infection or inflammation (including skin, trachea, gut epithelium), this peptide is expressed in normal uninflamed gingival tissue (Dale et al., 2001). Present evidence suggests that the high level of hBD-2 expression is due to the exposure of the tissue to commensal, non-pathogenic bacteria (Krisanaprakornkit et al., 2000) and that the innate immune responses of normal oral epithelia are at a heightened state of readiness. This has been most clearly demonstrated in studies of β-defensin expression in oral epithelial cells in vitro. Normal oral gingival epithelial cells do not express hBD-2 in cell culture unless stimulated. Commensal and pathogenic bacteria, proinflammatory cytokines and phorbol ester, a general cell activator, are all effective stimulants. Cell wall preparations of the commensal organism, F. nucleatum, are particularly effective in up-regulating hBD-2 mRNA expression (Krisanaprakornkit et al., 2000).
Signaling pathways for hBD-2 in oral epithelial cells
HBD-2 up-regulation in response to oral commensal bacteria does not seem to utilize the NF-κB intracellular signaling cascade typically associated with recognition of bacterial products, but instead utilizes the JNK and p38 mitogen activated protein kinase (MAPK) pathways that are associated with cytokine and stress responses (Krisanaprakornkit et al., 2002). Intracellular calcium signaling is also involved (Krisanaprakornkit et al., 2003). Utilization of this signaling pattern has now been extended to show that commensal organisms from both oral and skin sites do not utilize the NF-κB pathway for hBD-2 up-regulation, in contrast to pathogens which use an NF-κB pathway (Chung and Dale, 2004). This differs from results in other tissues, for example, tracheal epithelium in which NF-κB signaling has been demonstrated (Diamond et al., 2000). This signaling pathway may represent an adaptation to the normal population of both Gram positive and Gram negative bacteria within the oral cavity. Similarly, colon epithelial cells upregulate hBD-2 expression poorly in response to LPS (O’Neil et al., 1999). The findings in oral epithelial cells are consistent with the hypothesis that oral epithelia are capable of distinguishing commensal and pathogenic organisms, that immune response signaling mechanisms in the presence of commensal bacteria differ from those in response to pathogens, and that the expression of hBD-2 is part of this scenario. Because commensal bacteria are excellent inducers of hBD-2 in oral epithelial cells, these findings suggest that the oral commensal bacterial community acts in a manner to benefit the overall innate immune readiness of oral epithelia. This may have major significance for understanding host defense in the complex oral environment. The receptors, molecular mechanisms and signaling pathways responsible for the distinction are not yet fully understood. Further, interpretation of various experimental approaches may be complicated by cross-talk between various receptor-mediated signaling pathways.
Epithelial receptors for hBD-2 regulation
Mammalian cells sense the presence of bacteria via Pattern Recognition Receptors, such as the Toll-like receptor or TLR family (Medzhitov, 2001). Lipoteichoic acid and peptidoglycans of Gram positive organisms and LPS of Gram negative bacteria signal via TLR2 and TLR4, respectively, activating NF-κB transcription factors and the JNK pathways that mediate up-regulation of multiple innate and inflammatory responses. TLR2 and -4 are expressed on oral epithelial cells and up-regulated with interferon gamma stimulation (Uehara et al., 2002b). The LPS binding factor, CD14, a cofactor in the association of bacterial LPS to TLR4, is poorly expressed on epithelial cells but is present in soluble form in plasma. HBD-2 regulation in tracheal epithelial cells occurs via CD14, and NF-κB-mediated signaling (Becker et al., 2000; Diamond et al., 2000), and/or via TLR2-mediated signaling (Hertz et al., 2003). However, there is not yet convincing evidence that this pathway is involved in hBD-2 regulation in oral epithelial cells.
A second family of recently described receptors is the proteinase-activated receptors or PARs (Coughlin, 2000; Mackie et al., 2002). PARs act as sensors of possible danger, have a role in inflammation, and may also have a central role in the recognition of, and response to, bacteria within the oral cavity (Mackie et al., 2002), including regulation of the inducible β-defensins. Because the three major Gram-negative pathogens associated with periodontal disease, P. gingivalis, Bacteroides forsythus (recently renamed Tannerella forsythensis) and Treponema denticola (Socransky et al., 1998) each have proteases as part of their virulence mechanisms (Curtis et al., 2001; Fenno et al., 2001; Saito et al., 1997); these receptors may aid in differentially stimulating epithelial cell responses to pathogenic vs. commensal oral bacteria. PARs are implicated in platelet activation and response to injury via thrombin and trypsin (Lourbakos et al., 2001; Uehara et al., 2002a). PARs are seven-transmembrane domain G-protein coupled receptors. PAR activation involves proteolytic cleavage of the extracellular domain, resulting in a new amino-terminus that acts as a tethered ligand that binds to one of the extracellular loops of the receptor (Coughlin and Camerer, 2003; Mackie et al., 2002). PAR-1, -3, and -4 are activated by thrombin; PAR-2 is activated by various trypsin-like enzymes, including mast cell tryptase, neutrophil proteinase 3, as well as P. gingivalis proteinases (Lourbakos et al., 1998). These receptors can also be activated by peptide agonists that mimic the proteolytically produced amino-terminus. A PAR-2 peptide agonist up-regulated hBD-2 mRNA in oral epithelial cells and P. gingivalis mutants lacking proteinase fail to upregulate hBD-2, suggesting a role for PAR-2 in regulation of hBD-2 expression (Chung et al., 2004).
Other functions of antimicrobial peptides in the oral cavity
There is a growing recognition that the functions of chemokines and antimicrobial peptides overlap (Cole et al., 2001; Durr and Peschel, 2002). Epithelial cells signal Langerhans cells, the antigen presenting cells within epithelia, via cytokines, chemokines, and β-defensins, to connect innate immune responses with acquired immunity (Banchereau and Steinman, 1998; Yang et al., 1999). HBD-1 and hBD-2 act as chemoattractants for dendritic cells and T-cells. They also act as ligands for the receptor CCR6, a G-protein coupled receptor located in immature dendritic cells. Activation of the receptor results in dendritic cell maturation (Yang et al., 1999). Interestingly, the natural ligand for CCR6 is the chemokine CCL20 which has been shown to have structural similarity to hBD-2 (Hoover et al., 2002) and has antimicrobial activity (Starner et al., 2003). HBD-2 has also been reported as a ligand for TLR4 in dendritic cells (Biragyn et al., 2002), however, this finding is somewhat controversial and could be due to the ability of cationic hBD-2 to efficiently bind and act as a carrier for LPS delivery to the TLR4 receptor. HBD-2 also has effects on mast cells resulting in histamine release and intracellular Ca++ mobilization in a G protein-phopholipase C dependent manner (Niyonsaba et al., 2001).
The antimicrobial peptide LL-37 is a chemoattractant for neutrophil, monocytes and T-cells (Chertov et al., 1996; Yang et al., 2000). It also stimulates mast cells and alters macrophage gene expression to upregulate chemokines and their receptors resulting in greater responsiveness to the environment (Scott et al., 2002; Scott et al., 2000). HBD-2 also up-regulates gene expression for numerous cytokine and chemokine receptors in oral epithelial cells in a manner reminiscent of LL-37 (Yin and Dale, manuscript in preparation). Thus, hBD-2 enhances epithelial cell responses and alters gene expression in oral epithelial cells as well as mast cells and dendritic cells.
α-Defensins also have multiple functions as was recently reviewed (Yang et al., 2002). They selectively attract naïve CD4+ T-cells and immature dendritic cells via a G-protein coupled receptor (Yang et al., 2000). α-Defensins also stimulate mast cell degranulation (Befus et al., 1999), regulate complement activation (Prohaszka et al., 1997; van den Berg et al., 1998), and enhance macrophage phagocytosis (Ichinose et al., 1996).
Together these observations support the view that antimicrobial peptides promote adaptive immunity against microorganisms by recruiting both naïve T-cells, memory T-cells and antigen-presenting dendritic cells to regions of infection and stimulate repair and clean-up of infected sites via effects on mast cells and macrophages. In the oral cavity, this is an important ongoing process, particularly in the region of the gingival sulcus.
Salivary antimicrobial peptides
Antimicrobial components of the saliva (excluding salivary IgA) are generally referred to as the non-immune factors, but can be considered as part of the innate immune system of gene encoded factors that aid in protection against oral microbial colonization and infection (reviewed by Nieuw Amerongen and Veerman, 2002). These include a number of proteins such as salivary peroxidase, lysozyme, lactoferrin, cystatins, SLPI, agglutinin and mucins as well as peptides of the histatin family. In addition, the cathelicidin, LL-37, and α- and β-defensins are expressed and secreted by salivary glands and/or ducts.
α- and β-Defensins
β-Defensins have been demonstrated in salivary duct cells, but not acinar cells by immunohistochemical means (Sahasrabudhe et al., 2000) and in salivary glands and saliva by protein and RT-PCR analyses (Bonass et al., 1999; Mathews et al., 1999; Sahasrabudhe et al., 2000). HBD-1 mRNA has been identified in all major and minor salivary gland samples, while hBD-2 mRNA has been identified in a portion of samples, possibly associated with inflammation (Bonass et al., 1999). The secretion of β-defensins in saliva as well as their secretion from epithelial cells (Diamond et al., 2001) may contribute to the protection of oral mucosal surfaces. Levels of β-defensins in saliva have not been quantified. The α-defensins, HNP1–3, have also been detected in saliva and are elevated in patients with oral inflammation (Mizukawa et al., 1999). Levels of HNP1–3 vary in healthy individuals ranging from undetectable to ~12 μg/ml (Goebel et al., 2000; Mizukawa et al., 1999). The presence of α-defensins in saliva is most likely derived from neutrophils and is a reflection of gingival or mucosal inflammation and loose or exfoliating teeth.
LL-37
Expression of the mouse cathelicidin CRAMP was detected in salivary acinar cells of submandibular and palatine minor glands (mRNA and protein), ducts cells (protein) and mucosa in both embryonic and adult mice. In the human, LL37 was detected in saliva (Murakami et al., 2002). The results suggest localized expression of different antimicrobial peptides in salivary glands and ducts, similar to that of the gingiva (see above).
Histatins
Histatins are a family of at least 12 histidine-rich, cationic peptides synthesized by parotid and submandibular salivary duct cells and present in saliva (reviewed by Devine, 2002). Multiple histatins are generated from proteolytic cleavage of histatin 1 (38 amino acids) and 3 (32 amino acids). Histatins 1, 3, and 5 comprise approx. 85% of the total histatin protein in saliva. Histatins exhibit antifungal properties in vitro with histatin 5 having the most potent fungicidal activity (Oppenheim et al., 1988). The HTN genes map to chromosome 4q13, and the histatins are expressed only in salivary glands (vanderSpek et al., 1989), in contrast to the more general expression pattern of other antimicrobial peptides.
Histatins and oral candida
Saliva plays a critical role in regulating Candida adhesion to mucosal surfaces (Ueta et al., 2000). A high frequency of oral candidiasis occurs in patients with reduced salivary flow, such as in patients with Sjögren’s syndrome and histatins appear to be a main player in control of oral Candida (Jainkittivong et al., 1998). The fungicidal action of histatins requires active mitochondria (Helmerhorst et al., 1999a). α-defensins and histatins share some aspects of their antifungal action which is associated with ATP transport and reaction with purinergic-like cell surface receptors (Edgerton et al., 2000). The relationship of histatins and α- and β-defensins with oral Candida is not clear, but these peptides may work synergistically.
Altered antimicrobial peptide expression in the oral cavity
The innate immune system is essential in preventing infection, and the relationship of antimicrobial peptides and an individual’s susceptibility to infection is an important area of investigation. A current approach to identification of protein function is to identify the physiologic consequences in null mice lacking expression of the gene under investigation. This approach has led to advances in the understanding of the role of LL-37 in systemic and skin infection by S. aureus (Nizet et al., 2001). However, because there are numerous defensins, it is not possible to unequivocally prove their importance by knock out technology in a mouse model system. An alternate approach was used by Wilson and coworkers (Wilson et al., 1999) in which they disrupted the gene for matrilysin, the enzyme required for activation of Paneth cell α-defensins in the intestine. A second approach is to take advantage of experiments of nature–via genetics and the occurrence of genetic disorders.
Neutrophil defects and defects in expression of α-defensins and LL-37
It has been known for many years that periodontal disease in prepubertal children is associated with defects in neutrophil function (Page et al., 1985). With a general neutrophil defect, for example in the chemotactic response, or in cyclic neutropenia, periodontal disease may occur in association with increased occurrence of otitis media and other repeated infections (reviewed by Hart and Kornman, 2000). Recently a genetic form of periodontal disease (PD) in young people, Morbus Kostmann syndrome, was shown to have a deficiency in the α-defensins, HNP1–3, and a near absence of LL-37 (Putsep et al., 2002). This led to the suggestion that LL-37 may be particularly important in its effects vs. the Gram-negative, Actinobacillus actinomycetemcomitans, an organism associated with rapidly progressive PD especially in young people.
Another early onset inherited form of PD is the Papillon-Lefevre syndrome in which the genetic defect is in the gene for cathepsin C, a lysosomal protease (Hart et al., 1999; Toomes et al., 1999). Periodontal disease and palmar/plantar hyperkeratosis occur in this disorder but without an associated high frequency of other types of infections. Apparently a specific effect of cathepsin C is important in each of these body sites, however, the key substrate(s) for cathepsin C in neutrophils and epidermis is(are) not known. To date, altered processing of antimicrobial peptides has not been examined in this disorder, but is among the possible candidate substrates.
Genetics and the defensins
HBD-1 polymorphism and oral candida carriage
Occurrence of genetic polymorphisms offers another means to ask questions about the role of antimicrobial peptides in oral health and disease. Multiple single nucleotide polymorphisms (SNPs) have been identified in the DEFB1 and DEFB2 (now renamed DEFB4) genes, encoding hBD-1 and hBD-2, respectively (Dork and Stuhrmann, 1998; Jurevic et al., 2002; Vatta et al., 2000). Such SNPs may alter the expression or function of defensins and could lead to altered susceptibility to infection. The SNPs, especially those that had a reasonably high frequency, occur mainly in promoter and untranslated regions of these genes. These SNPs could potentially alter the amount of the peptide expressed, but not the peptide itself. Individuals with Type 1 diabetes are susceptible to oral Candida infection and have marginal immunosuppression, therefore, these minor differences in β-defensin genes may have significant effects in this population. The diabetic population had nearly 3-fold higher average oral Candida carriage than a non-diabetic group. However, in both populations, a SNP in DEFB1 (-44 C to G) was found to be associated with protection from oral Candida carriage. In other words, people who had the SNP had very low levels of oral Candida. The difference in relative risk for high Candida carriage was 25 (p<0.01) for the diabetic population and 8.5 for the nondiabetic group (Jurevic et al., 2003). The most obvious interpretation of this finding is that this SNP alters hBD-1 expression. Initial results using a molecular approach to address this possibility strongly suggest that the SNP results in increased hBD-1 protein expression, and this may explain the basis of the apparent protection from oral Candida carriage. Alternatively, the SNP may be linked to a genetic change in another functional gene in this chromosomal region. Nevertheless, the results suggest that individuals who are immunosuppressed, and thus more susceptible to opportunistic infections, may have greater reliance on innate immune defenses than healthy individuals who have multiple ways to fight infection.
Defensin gene duplications
Hollox and co-workers (Hollox et al., 2003) recently demonstrated variable copy number indicative of gene duplication within the β-defensin cluster on Chr. 8p23. They showed that individuals can have from 2 to 12 copies of the region of DEFB4, DEF103, and DEFB104 encoding hBD-2, hBD-3, and hBD-4, respectively, per diploid genome. This repeat region does not include the genes encoding the α-defensins or hBD-1. Inheritance of genes encoding the α-defensins can also vary, but less extensively (Mars et al., 1995). The frequency of low vs. high copy number and the effect on peptide expression is not yet known, but may be an important factor in oral antimicrobial function.
Efficacy of antimicrobial peptides in the oral cavity
The oral cavity provides an inviting warm, moist environment conducive to colonization by many types of microorganisms. Thus the normal oral flora is extremely complex, requiring multiple types of defenses in order to prevent infection. These defenses include antimicrobial peptides with different, but overlapping, ranges of microorganisms against which they are effective. The oral cavity also provides an environment that allows optimal effectiveness of these peptides. Activity of most of the oral antimicrobial peptides is inhibited by ions/salt at physiologic levels (Goldman et al., 1997; Nagaoka et al., 2000; Turner et al., 1998); however, in the oral environment, they function at the surface away from high salt concentrations and interfering substances in the blood. Table 2 provides data from numerous sources on the activity of these peptides against various, mainly pathogenic, oral bacteria and fungi (see table for references for this section). The antimicrobial function of these peptides has been obtained through a myriad of methods, making comparisons and interpretations difficult. It should also be recognized that very few methods test for bactericidal (lethal) as opposed to bacteriostatic activity.
Table 2.
Activity of antimicrobial peptides against oral microbes*
| Organism | hBD1 | hBD2 | hBD3 | HNP-1 | HNP-2/-3 | LL-37 | Histatin | Adrenomedulin |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli1,3,4,5,11,20,21,23 | 40 (≥3); 5 (0.5) | 10 (1); 0.6 (0.5) | 5 (≥3); 0.1 (0.5) | 50 (≥3) | 1.0; 10 (≥3) | 0.4 | ||
| Streptococcus mutans1,6,8,10,12 | 4.1–7.8b | 2.6–5 b; 2 (≥3) | NS | 12.5 | ||||
| S. sanguis3,6,8,10 | 8–25 b | 7.6–21.3 b; 16 (≥3) | >100 (≥3) | NS | ||||
| S. salvarius6 | NS | |||||||
| S. sobrinus10 | 16 (≥3) | |||||||
| Actinomyces naeslundii1,6,8 | 8–14 b | 4.1–7.2 b | NS | 12.5 | ||||
| Fusobacterium nucleatum3,6,8,14 | 6.3->250 b | 4.5->250 b | >500 | >100 (≥3) | NS | |||
| Veillonella parvula6 | (0.3) | |||||||
| Prevotella intermedia6 | NS | |||||||
| Porphyromonas gingivalis1,3,8,10,12 | 34.6->250b | 5.7->250 b; 100 (≥3) | >100 (≥3) | 7.75x10–4 | ||||
| Actinobacillus actinomycetemcomitans3,8,10,12,13,15 | >250 | 9.6->250 b; 2.5 (≥3) | NS (1) | NS (1) | 10 (2); >100 (≥3) | |||
| Lactobacillus acidophilus10 | 8 (≥3) | |||||||
| Capnocytophaga sputigena13,19 | 200 (1); 500 (2) | 500 (1) | 7.5 (2) | |||||
| C. gingivalis13,19 | 200 (1) | 500 (1) | 9.1 (2) | |||||
| C. ochracea13,19 | 10 (1); 500 (2) | 10 (1) | 11.0 (2) | |||||
| Candida albicans1,2,3,7,8,9,17,18,20 | >500 | 25 (1); 125; 4.6–59 b | 25;2.8–7.1b | >250 (≥3); 96% (52 ug/ml) LOV | >250 (≥3); >100 (≥3) | 97% (14 ug/ml) LOV; >500 [24 hr]; 2.4 (0.5); 50 uM (1.5); 100 uM (2) | NS | |
| resist. C. albicans9,22 | 125 | 25 | 125 [24 hr] | |||||
| C. glabrata7,8,9,15,22 | 23->250b; >1000 | 34->250; 50 | 62.5 [24 hr]; 29 (0.5); 42–87% LOVa | |||||
| resist. C. glabrata7,9 | >1000 | 200 | 1.0 (0.5) | |||||
| C. krusei3,7,8,9,15,22 | 12->250b; >1000 | 2–13.7 b; 200 | >100 (≥3) | 500 [24 hr]; 1.2 (0.5); 80–95% LOVa | ||||
| C. neoformans7,22 | 31.3 [24 hr]; 0.7 (0.5) | |||||||
| C. pseudotropicalis7 | 1.0 (0.5) | |||||||
| C. parapsilosis7,8,9,15 | 9.3–17.8b; >1000 | 1.4–12.4 b; 50–100 | 2.0e; 72–98% LOVa | |||||
| C. tropicalis3,8,9,15 | >500 | 3.9–13.1b; 125 | 3.3–14.4 b; 6.25 | >100 (≥3) | 90–98% LOVa | |||
| C. guilleirmondii15 | 90–98% LOVa | |||||||
| Aspergillus fumigatus22 | >500 [24 hr] | |||||||
| HIV15,16,24 | NO | YES | YES | YES | YES |
Values in MIC (μg/ml) unless otherwise noted.
(#) = # log reduction
[24 hr] = read at 24 hr
LOV = loss of viability
NS = no significant activity
range due to different strains (50 μM)
MIC range due to strain differences
Jurevic, 2004
Miyasaki et al., 1990
Zang et al., 2002
Peptide activity
LL-37
LL-37 is a cathelicidin with the conserved proregion which keeps it inactive until proteases cleave this portion after the protein is secreted (Zanetti et al., 2000). Against oral microbes, LL-37 has the greatest activity against A. actinomycetemcomitans and Capnocytophaga spp. A. actinomycetemcomitans is an important pathogen associated with rapidly progressive forms of periodontal disease which often affect younger individuals. Capnocytophaga spp. have been implicated in juvenile gingivitis and periodontitis and can cause sepsis in immunocompromised patients (Gomez-Garces et al., 1994).
α-Defensins
α-Defensins are relatively ineffective against most of the oral microbes tested, with the exceptions of Capnocytophaga ochracea and C. albicans. However, synergy between HNP-1 and LL-37 has been demonstrated against E. coli and Staphylococcus aureus even at physiological salt concentrations (150 mM) (Nagaoka et al., 2000). HNP-1 requires target microbes to be metabolically active (Edgerton, et al., 2000). HNP-1–3 also show inhibitory effects on HIV infectivity.
β-Defensins
β-Defensins show both anti-fungal and antibacterial action. HBD-3 is consistently more active against both bacteria and fungi, with hBD-2 next and hBD-1 last in activity. The study by Joly and coworkers compared the effectiveness of hBD-2 and hBD-3 against oral microorganisms. HBD-2 and hBD-3 show considerable variability against multiple strains of the same species, but overall, aerobes are more susceptible than anaerobes. These peptides also show strain-specific activity toward various Candida spp. (Joly et al., 2004). Interestingly, Wu and coworkers showed that hBD-3 activity against E. coli was unaffected by disulfide bonding which was previously thought to be critical to antimicrobial function (Wu et al., 2003). Along with the α-defensins, hBD-2 and -3 inhibit HIV replication. HBD-2 has anti-fungal activity against C. albicans and C. tropicalis, while hBD-3 is active against other Candida spp. tested.
Histatins
Histatins are primarily anti-Candida agents. They have little effect on oral bacteria or on the fungus Aspergillus fumigatus. Histatin-5 is the most effective histatin and is active against C. albicans as well as other Candida spp. Histatins have activity in the concentration range found in saliva. Johnson, Yeh and Dodds (Johnson et al., 2000) measured histatin concentrations in saliva at 31 μg/ml in parotid secretions and 62 μg/ml in submandibular/submaxillary secretions. They also found that these concentrations decline by one half and one third, respectively, between the ages of 45–75. Synergistic effects have been demonstrated between histatin-5 and amphotericin B against Candida spp., including an amphotericin B-resistant strain, Cryptococcus neoformans and A. fumigatus (van’t Hof et al., 2000).
Antimicrobial peptides as therapeutic agents in the oral cavity
Antimicrobial peptides are under investigation for control of oral infections. One such drug is Iseganan, an analog of protegrin-1, a porcine cathelicidin, which has broad spectrum bactericidal activity. Iseganan has recently completed Phase III clinical trials for prevention of ulcerative oral mucositis. It showed encouraging results in reducing the occurrence of oral mucositis and associated clinical problems such as mouth pain, throat pain and difficulty swallowing (Giles et al., 2003). A second investigational peptide is the antifungal, histatin-5. The yeast Candida is a commensal organism on skin and mucosal surfaces but is a growing problem in causing opportunistic infections. Frequency of mucosal and invasive fungal infections has increased in association with increased numbers of immunocompromised patients due to HIV, chemotherapy, and the use of immunosuppressive drugs. In addition, there has been an increase in Candida strains that are resistant to the main therapeutic drugs in current use. This has led to an increased interest in new antifungals, primarily antifungal peptides. Histatin-5 and several variants are under active investigation for this purpose since their mechanism of action differs from the main antifungal drugs in current use and they are effective against azole-resistant Candida strains (Situ and Bobek, 2000; Tsai and Bobek, 1998).
Conclusions
The oral cavity is a unique environment in which antimicrobial peptides play a key role in maintaining health. Present evidence suggests that α-defensins, β-defensins, LL-37, histatin, and other antimicrobial peptides and proteins have distinct but overlapping roles in maintaining oral health. The expression of the inducible hBD-2 in normal oral epithelium, and the apparent differential signaling in response to commensal and pathogenic organisms, provides new insights into innate immunity in this body site. Commensal bacteria are excellent inducers of hBD-2 in oral epithelial cells, suggesting that the commensal bacterial community acts in a manner to benefit the overall innate immune readiness of oral epithelia. This may have major significance for understanding host defense in the complex oral environment.
Future trends
Future studies will emphasize approaches to learn more about the role of antimicrobial peptides in oral health and susceptibility to disease and infection. Because periodontal disease is largely a situation in which the inflammatory process has gone awry, it is critical to understand the role of innate immunity in the recognition of, response to, and effectiveness against oral microbes. This includes more detailed information about both α- and β-defensins, LL-37, and other oral antimicrobial peptides and proteins. These peptides very likely have specific functions in keeping the level of commensal bacteria in check as well as being effective against pathogens. The involvement of antimicrobial peptides in other mucosal disorders is also important to understand. In oral squamous cell carcinoma and lichen planus, β-defensin expression may reveal clues on disease progression and prognosis. Recent evidence of the involvement of β-defensins in the defense against HIV infection and transmission in the oral environment is fascinating (Quinones-Mateu et al., 2003). There is much more to learn about this effect and how it may be useful therapeutically. Further, studies of expression and function of the newly described members of the β-defensin family (Schutte et al., 2002) may provide critical new pieces of information relating to oral health.
The initial genetic studies suggesting a protective role of an hBD-1 SNP need extension and confirmation. This finding opens a whole new area of investigation. Does the hBD-1 SNP affect Candida carriage directly via hBD-1 or indirectly via a linked gene? Can this SNP be used as part of a diagnostic susceptibility profile in situations in which Candida infection could become life threatening, such as transplant recipients? Are defensin SNPs correlated with other oral infections? Because defensins are secreted and may function synergistically with salivary antimicrobials they may protect against dental caries, or conversely contribute to genetic susceptibility to dental decay. Similar questions can be asked of the role of LL-37 in oral health and disease. Genetic investigations could be extended to include molecular mechanisms and defects in processing of antimicrobial peptides in neutrophils, such as in Morbus Kostmann syndrome and possibly in Papillon-Lefevre syndrome in view of the profound effects of neutrophil antimicrobials on periodontal health.
Another major area of future study is the regulation by and functional efficacy against oral commensal vs. pathogenic organisms. Present evidence suggests that innate immune responses of oral epithelia are primed by commensal bacteria and this occurs via pathways other than those utilized in response to pathogens. This ability of epithelial cells to distinguish commensal and pathogens may represent a general phenomonen since epithelia from different body sites co-exist with different populations of commensal bacteria. Study of the regulation of β-defensins is a window opening our understanding of this important aspect of innate immunity of oral epithelia as well as epithelia in other body sites. It will also be important to understand the molecular mechanism by which the defensins and LL-37 exert their multiple functions in promoting wound healing and linking innate immunity to acquired immunity.
Finally, the development of new peptide antimicrobial agents for therapeutic use in the oral environment is an important area for future investigation and testing. Peptide antimicrobial agents may augment the innate defenses of individuals at high risk for oral infection because of compromised immune status.
Acknowledgments
Support for the work of this laboratory on the role of β-defensins in the oral cavity is from USPHS NIDCR Grants DE P60 013061 and R01 DE 13573 (to B.A.D.). Dr. Fredericks is supported by NIDCR T32 DE07023. We thank Dr. W. O. Chung, and Ms. Janet Kimball for discussions and critical reading of the manuscript.
Footnotes
Added note: Activity of antimicrobial peptides against some addition oral bacterial species and strains was recently reported by Ouhara et al. (2005) J. Antimicro. Chemother. Doi:10.1093/jac/dki103.
References
- Abiko Y, Mitamura J, Nishimura M, Muramatsu T, Inoue T, Shimono M, Kaku T. Pattern of expression of beta-defensins in oral squamous cell carcinoma. Cancer Lett. 1999;143:37–43. doi: 10.1016/s0304-3835(99)00171-8. [DOI] [PubMed] [Google Scholar]
- Allaker RP, Kapas S. Adrenomedullin and mucosal defence: interaction between host and microorganism. Regul Pept. 2003;112:147–152. doi: 10.1016/s0167-0115(03)00033-8. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Becker MN, Diamond G, Verghese MW, Randell SH. CD14-dependent lipopolysaccharide-induced beta-defensin-2 expression in human tracheobronchial epithelium. J Biol Chem. 2000;275:29731–29736. doi: 10.1074/jbc.M000184200. [DOI] [PubMed] [Google Scholar]
- Befus AD, Mowat C, Gilchrist M, Hu J, Solomon S, Bateman A. Neutrophil defensins induce histamine secretion from mast cells: mechanisms of action. J Immunol. 1999;163:947–953. [PubMed] [Google Scholar]
- Bevins CL, Martin-Porter E, Ganz T. Defensins and innate host defence of the gastrointestinal tract. Gut. 1999;45:911–915. doi: 10.1136/gut.45.6.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, Shirakawa AK, Farber JM, Segal DM, Oppenheim JJ, Kwak LW. Toll-like receptor 4-dependent activation of dendritic cells by beta-defensin 2. Science. 2002;298:1025–1029. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- Bonass WA, High AS, Owen PJ, Devine DA. Expression of beta-defensin genes by human salivary glands. Oral Microbiol Immunol. 1999;14:371–374. doi: 10.1034/j.1399-302x.1999.140607.x. [DOI] [PubMed] [Google Scholar]
- Brandtzaeg P, Gabrielsen TO, Dale I, Muller F, Steinbakk M, Fagerhol MK. The leucocyte protein L1 (calprotectin): a putative nonspecific defence factor at epithelial surfaces. Adv Exp Med Biol. 1995;371A:201–206. doi: 10.1007/978-1-4615-1941-6_41. [DOI] [PubMed] [Google Scholar]
- Caron KM, Smithies O. Extreme hydrops fetalis and cardiovascular abnormalities in mice lacking a functional Adrenomedullin gene. Proc Natl Acad Sci USA. 2001;98:615–619. doi: 10.1073/pnas.021548898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertov O, Michiel DF, Xu L, Wang JM, Tani K, Murphy WJ, Longo DL, Taub DD, Oppenheim JJ. Identification of defensin-1, defensin-2, and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J Biol Chem. 1996;271:2935–2940. doi: 10.1074/jbc.271.6.2935. [DOI] [PubMed] [Google Scholar]
- Chung WO, Dale BA. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect Immun. 2004;72:352–358. doi: 10.1128/IAI.72.1.352-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WO, Hansen SR, Rao D, Dale BA. Protease-activated receptor signaling increases epithelial antimicrobial peptide expression. J Immunol. 2004;173:1565–1570. doi: 10.4049/jimmunol.173.8.5165. [DOI] [PubMed] [Google Scholar]
- Cole AM, Ganz T, Liese AM, Burdick MD, Liu L, Strieter RM. Cutting edge: IFN-inducible ELR-CXC chemokines display defensin-like antimicrobial activity. J Immunol. 2001;167:623–627. doi: 10.4049/jimmunol.167.2.623. [DOI] [PubMed] [Google Scholar]
- Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- Coughlin SR, Camerer E. PARticipation in inflammation. J Clin Invest. 2003;111:25–27. doi: 10.1172/JCI17564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MA, Aduse-Opoku J, Rangarajan M. Cysteine proteases of Porphyromonas gingivalis. Crit Rev Oral Biol Med. 2001;12:192–216. doi: 10.1177/10454411010120030101. [DOI] [PubMed] [Google Scholar]
- Dale BA. Periodontal epithelium: a newly recognized role in health and disease. Periodontology 2000. 2002;30:70–78. doi: 10.1034/j.1600-0757.2002.03007.x. [DOI] [PubMed] [Google Scholar]
- Dale BA, Kimball JR, Krisanaprakornkit S, Roberts F, Robinovitch M, O’Neal R, Valore EV, Ganz T, Anderson GM, Weinberg A. Localized antimicrobial peptide expression in human gingiva. J Periodontal Res. 2001;36:285–294. doi: 10.1034/j.1600-0765.2001.360503.x. [DOI] [PubMed] [Google Scholar]
- Dale BA, Krisanaprakornkit S. Defensin antimicrobial peptides in the oral cavity. J Oral Pathol Med. 2001;30:321–327. doi: 10.1034/j.1600-0714.2001.300601.x. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Tanner A, Page RC. The microbial challenge in periodontitis. Periodontology 2000. 1997;14:12–32. doi: 10.1111/j.1600-0757.1997.tb00190.x. [DOI] [PubMed] [Google Scholar]
- Devine DA. Antimicrobial peptides in defence of the oral and respiratory tracts. Mol Immunol. 2003;40:431–443. doi: 10.1016/s0161-5890(03)00162-7. [DOI] [PubMed] [Google Scholar]
- Diamond DL, Kimball JR, Krisanaprakornkit S, Ganz T, Dale BA. Detection of beta-defensins secreted by human oral epithelial cells. J Immunol Methods. 2001;256:65–76. doi: 10.1016/s0022-1759(01)00442-2. [DOI] [PubMed] [Google Scholar]
- Diamond G, Bevins CL. Endotoxin upregulates expression of an antimicrobial peptide gene in mammalian airway epithelial cells. Chest. 1994;105:51S–52S. doi: 10.1378/chest.105.3_supplement.51s. [DOI] [PubMed] [Google Scholar]
- Diamond G, Kaiser V, Rhodes J, Russell JP, Bevins CL. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect Immun. 2000;68:113–119. doi: 10.1128/iai.68.1.113-119.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond G, Russell JP, Bevins CL. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc Natl Acad Sci USA. 1996;93:5156–5160. doi: 10.1073/pnas.93.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond G, Zasloff M, Eck H, Brasseur M, Maloy WL, Bevins CL. Tracheal antimicrobial peptide, a cysteine rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc Natl Acad Sci USA. 1991;88:3952–3956. doi: 10.1073/pnas.88.9.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinulos JG, Mentele L, Fredericks LP, Dale BA, Darmstadt GL. Keratinocyte expression of human β-defensin-2 following bacterial infection: role in cutaneous host defense. Clin Diag Lab Immunol. 2003;10:161–166. doi: 10.1128/CDLI.10.1.161-166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dork T, Stuhrmann M. Polymorphisms of the human beta defensin 1 gene. Mol Cell Probes. 1998;12:171–173. doi: 10.1006/mcpr.1998.0165. [DOI] [PubMed] [Google Scholar]
- Dunsche A, Acil Y, Dommisch H, Siebert R, Schroder JM, Jepsen S. The novel human betadefensin-3 is widely expressed in oral tissues. Eur J Oral Sci. 2002;110:121–124. doi: 10.1034/j.1600-0722.2002.11186.x. [DOI] [PubMed] [Google Scholar]
- Dunsche A, Acil Y, Siebert R, Harder J, Schroder JM, Jepsen S. Expression profile of human defensins and antimicrobial proteins in oral tissues. J Oral Pathol Med. 2001;30:154–158. doi: 10.1034/j.1600-0714.2001.300305.x. [DOI] [PubMed] [Google Scholar]
- Durr M, Peschel A. Chemokines meet defensins: the merging concepts of chemoattractants and antimicrobial peptides in host defense. Infect Immun. 2002;70:6515–6517. doi: 10.1128/IAI.70.12.6515-6517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgerton M, Koshlukova SE, Araujo MW, Patel RC, Dong J, Bruenn JA. Salivary histatin 5 and human neutrophil defensin 1 kill Candida albicans via shared pathways. Antimicrob Agents Chemother. 2000;44:3310–3316. doi: 10.1128/aac.44.12.3310-3316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Lee SY, Bayer CH, Ning Y. The opdB locus encodes the trypsin-like peptidase activity of Treponema denticola. Infect Immun. 2001;69:6193–6200. doi: 10.1128/IAI.69.10.6193-6200.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohm Nilsson M, Sandstedt B, Sorensen O, Weber G, Borregaard N, Stahle-Backdahl M. The human cationic antimicrobial protein (hCAP18), a peptide antibiotic, is widely expressed in human squamous epithelia and colocalizes with interleukin-6. Infect Immun. 1999;67:2561–2566. doi: 10.1128/iai.67.5.2561-2566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- Garcia JR, Jaumann F, Schulz S, Krause A, Rodriguez-Jimenez J, Forssmann U, Adermann K, Kluver E, Vogelmeier C, Becker D, et al. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 2001;306:257–264. doi: 10.1007/s004410100433. [DOI] [PubMed] [Google Scholar]
- Giles FJ, Miller CB, Hurd DD, Wingard JR, Fleming TR, Sonis ST, Bradford WZ, Pulliam JG, Anaissie EJ, Beveridge RA, et al. A phase III, randomized, double-blind, placebo-controlled, multinational trial of iseganan for the prevention of oral mucositis in patients receiving stomatotoxic chemotherapy (PROMPT-CT trial) Leuk Lymphoma. 2003;44:1165–1172. doi: 10.1080/1042819031000079159. [DOI] [PubMed] [Google Scholar]
- Goebel C, Mackay LG, Vickers ER, Mather LE. Determination of defensin HNP-1, HNP-2, and HNP-3 in human saliva by using LC/MS. Peptides. 2000;21:757–765. doi: 10.1016/s0196-9781(00)00205-9. [DOI] [PubMed] [Google Scholar]
- Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta defensin 1 is a salt sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- Gomez-Garces JL, Alos JI, Sanchez J, Cogollos R. Bacteremia by multidrug-resistant Capnocytophaga sputigena. J Clin Microbiol. 1994;32:1067–1069. doi: 10.1128/jcm.32.4.1067-1069.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmiller JM, Vargas KG, Srikantha R, Schomberg LL, Weistroffer PL, McCray PB, Jr, Tack BF. Susceptibilities of oral bacteria and yeast to mammalian cathelicidins. Antimicrob Agents Chemother. 2001;45:3216–3219. doi: 10.1128/AAC.45.11.3216-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock REW. Peptide antibiotics. Lancet. 1997;349:418–422. doi: 10.1016/S0140-6736(97)80051-7. [DOI] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. 1997a;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human betadefensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–5713. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- Hart TC, Hart PS, Bowden DW, Michalec MD, Callison SA, Walker SJ, Zhang Y, Firatli E. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. J Med Genet. 1999;36:881–887. [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Kornman KS. Genetic factors in the pathogenesis of periodontitis. Periodontology. 2000;14:202–215. doi: 10.1111/j.1600-0757.1997.tb00198.x. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Breeuwer P, van’t Hof W, Walgreen-Weterings E, Oomen LC, Veerman EC, Amerongen AV, Abee T. The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J Biol Chem. 1999a;274:7286–7291. doi: 10.1074/jbc.274.11.7286. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Hodgson R, van ‘t Hof W, Veerman EC, Allison C, Nieuw Amerongen AV. The effects of histatin-derived basic antimicrobial peptides on oral biofilms. J Dent Res. 1999b;78:1245–1250. doi: 10.1177/00220345990780060801. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Reijnders IM, van’t Hof W, Simoons-Smit I, Veerman EC, Amerongen AV. Amphotericin B- and fluconazole-resistant Candida spp. Aspergillus fumigatus, and other newly emerging pathogenic fungi are susceptible to basic antifungal peptides. Antimicrob Agents Chemother. 1999c;43:702–704. doi: 10.1128/aac.43.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz CJ, Wu Q, Porter EM, Zhang YJ, Weismuller KH, Godowski PJ, Ganz T, Randell SH, Modlin RL. Activation of toll-like receptor 2 on human tracheobronchial epithelial cells induces the antimicrobial peptide human beta defensin-2. J Immunol. 2003;171:6820–6826. doi: 10.4049/jimmunol.171.12.6820. [DOI] [PubMed] [Google Scholar]
- Hollox EJ, Armour JA, Barber JC. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover DM, Boulegue C, Yang D, Oppenheim JJ, Tucker K, Lu W, Lubkowski J. The structure of human macrophage inflammatory protein-3alpha/CCL20. Linking antimicrobial and CC chemokine receptor-6-binding activities with human beta-defensins. J Biol Chem. 2002;277:37647–37654. doi: 10.1074/jbc.M203907200. [DOI] [PubMed] [Google Scholar]
- Ichinose M, Asai M, Imai K, Sawada M. Enhancement of phagocytosis by corticostatin I (CSI) in cultured mouse peritoneal macrophages. Immunopharmacology. 1996;35:103–109. doi: 10.1016/s0162-3109(96)00118-x. [DOI] [PubMed] [Google Scholar]
- Jainkittivong A, Johnson DA, Yeh CK. The relationship between salivary histatin levels and oral yeast carriage. Oral Microbiol Immunol. 1998;13:181–187. doi: 10.1111/j.1399-302x.1998.tb00730.x. [DOI] [PubMed] [Google Scholar]
- Johnson DA, Yeh CK, Dodds MW. Effect of donor age on the concentrations of histatins in human parotid and submandibular/sublingual saliva. Arch Oral Biol. 2000;45:731–740. doi: 10.1016/s0003-9969(00)00047-9. [DOI] [PubMed] [Google Scholar]
- Joly S, Maze D, McCray PB, Jr, Guthmiller JM. Human β-defensin 2 and 3 demonstrate strain-selective activity against oral microorganisms. J Clin Microbiol. 2004;42:1024–1029. doi: 10.1128/JCM.42.3.1024-1029.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurevic RJ, Bai M, Chadwick RB, White TC, Dale BA. Single nucleotide polymorphisms (SNPs) in human β-defensin-1: High throughput SNP assays and association with Candida carriage in type 1 diabetics and nondiabetic controls. J Clin Microbiol. 2003;41:90–96. doi: 10.1128/JCM.41.1.90-96.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurevic RJ, Chrisman P, Mancl L, Livingston R, Dale BA. Single nucleotide polymorphisms and haplotype analysis in β-defensin genes in different ethnic populations. Genet Test. 2002;6:261–269. doi: 10.1089/10906570260471787. [DOI] [PubMed] [Google Scholar]
- Kapas S, Bansal A, Bhargava V, Maher R, Malli D, Hagi-Pavli E, Allaker RP. Adrenomedullin expression in pathogen-challenged oral epithelial cells. Peptides. 2001a;22:1485–1489. doi: 10.1016/s0196-9781(01)00470-3. [DOI] [PubMed] [Google Scholar]
- Kapas S, Tenchini ML, Farthing PM. Regulation of adrenomedullin secretion in cultured human skin and oral keratinocytes. J Invest Dermatol. 2001b;117:353–359. doi: 10.1046/j.0022-202x.2001.01426.x. [DOI] [PubMed] [Google Scholar]
- Kido J, Nakamura T, Kido R, Ohishi K, Yamauchi N, Kataoka M, Nagata T. Calprotectin in gingival crevicular fluid correlates with clinical and biochemical markers of periodontal disease. J Clin Periodontol. 1999;26:653–657. doi: 10.1034/j.1600-051x.1999.261004.x. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Sakata J, Kangawa K, Kojima M, Matsuo H, Eto T. Cloning and characterization of cDNA encoding a precursor for human adrenomedullin. Biochem Biophys Res Comm. 1993;194:720–725. doi: 10.1006/bbrc.1993.1881. [DOI] [PubMed] [Google Scholar]
- Krisanaprakornkit K, Kimball JR, Weinberg A, Darveau RP, Bainbridge BW, Dale BA. Inducible expression of human β-defensin-2 (hBD-2) by Fusobacterium nucleatumin oral epithelial cells: Multiple signaling pathways and the role of commensal bacteria in innate immunity and the epithelial barrier. Infect Immun. 2000;68:2907–2915. doi: 10.1128/iai.68.5.2907-2915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisanaprakornkit S, Jotikasthira D, Dale BA. Intracellular calcium in signaling human β-defensin-2 expression in oral epithelial cells. J Dent Res. 2003;82:877–882. doi: 10.1177/154405910308201106. [DOI] [PubMed] [Google Scholar]
- Krisanaprakornkit S, Kimball JR, Dale BA. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J Immunol. 2002;168:316–324. doi: 10.4049/jimmunol.168.1.316. [DOI] [PubMed] [Google Scholar]
- Krisanaprakornkit S, Weinberg A, Perez CN, Dale BA. Expression of the peptide antibiotic human beta defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect Immun. 1998;66:4222–4228. doi: 10.1128/iai.66.9.4222-4228.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- Liu AY, Dstoumieux D, Wong AV, Parak CH, Valore EV, Liu L, Ganz T. Human β-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J Invest Dermatol. 2002;118:275–281. doi: 10.1046/j.0022-202x.2001.01651.x. [DOI] [PubMed] [Google Scholar]
- Lourbakos A, Chinni C, Thompson P, Potempa J, Travis J, Mackie EJ, Pike RN. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 1998;435:45–48. doi: 10.1016/s0014-5793(98)01036-9. [DOI] [PubMed] [Google Scholar]
- Lourbakos A, Potempa J, Travis J, D’Andrea MR, Andrade-Gordon P, Santulli R, Mackie EJ, Pike RN. Arginine-specific protease from Porphyromonas gingivalis activates protease-activated receptors on human oral epithelial cells and induces interleukin-6 secretion. Infect Immun. 2001;69:5121–5130. doi: 10.1128/IAI.69.8.5121-5130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie EJ, Pagel CN, Smith R, de Niese MR, Song SJ, Pike RN. Proteaseactivated receptors: a means of converting extracellular proteolysis into intracellular signals. IUBMB Life. 2002;53:277–281. doi: 10.1080/15216540213469. [DOI] [PubMed] [Google Scholar]
- Maisetta G, Batoni G, Esin S, Luperini F, Pardini M, Bottai D, Florio W, Giuca MR, Gabriele M, Campa M. Activity of human beta-defensin 3 alone or combined with other antimicrobial agents against oral bacteria. Antimicrob Agents Chemother. 2003;47:3349–3351. doi: 10.1128/AAC.47.10.3349-3351.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Nagaraj R. Antibacterial activities and conformations of synthetic alpha-defensin HNP-1 and analogs with one, two and three disulfide bridges. J Pept Res. 2002;59:95–104. doi: 10.1034/j.1399-3011.2002.01945.x. [DOI] [PubMed] [Google Scholar]
- Marionnet C, Bernerd F, Dumas A, Verrecchia F, Mollier K, Compan D, Bernard B, Lahfa M, Leclaire J, Medaisko C, et al. Modulation of gene expression induced in human epidermis by environmental stress in vivo. J Invest Dermatol. 2003;121:1447–1458. doi: 10.1111/j.1523-1747.2003.12629.x. [DOI] [PubMed] [Google Scholar]
- Mars WM, Patmasiriwat P, Maity T, Huff V, Weil MM, Saunders GF. Inheritance of unequal numbers of the genes encoding the human neutrophil defensins HP-1 and HP-3. J Biol Chem. 1995;270:30371–30376. doi: 10.1074/jbc.270.51.30371. [DOI] [PubMed] [Google Scholar]
- Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, Tack BF, McCray PB., Jr Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–2745. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay MS, Olson E, Hesla MA, Panyutich A, Ganz T, Perkins S, Rossomando EF. Immunomagnetic recovery of human neutrophil defensins from the human gingival crevice. Oral Microbiol Immunol. 1999;14:190–193. doi: 10.1034/j.1399-302x.1999.140308.x. [DOI] [PubMed] [Google Scholar]
- McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, Thompson N, Solari R, Lee MG, Foord SM. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Mineshiba F, Takashiba S, Mineshiba J, Matsuura K, Kokeguchi S, Murayama Y. Antibacterial activity of synthetic human B defensin-2 against periodontal bacteria. J Int Acad Periodontol. 2003;5:35–40. [PubMed] [Google Scholar]
- Miyasaki KT, Lehrer RI. Beta-sheet antibiotic peptides as potential dental therapeutics. Int J Antimicrob Agents. 1998;9:269–280. doi: 10.1016/s0924-8579(98)00006-5. [DOI] [PubMed] [Google Scholar]
- Mizukawa N, Sawaki K, Yamachika E, Fukunaga J, Ueno T, Takagi S, Sugahara T. Presence of human beta-defensin-2 in oral squamous cell carcinoma. Anticancer Res. 2000;20:2005–2007. [PubMed] [Google Scholar]
- Mizukawa N, Sugiyama K, Ueno T, Mishima K, Takagi S, Sugahara T. Levels of human defensin-1, an antimicrobial peptide, in saliva of patients with oral inflammation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 1999;87:539–543. doi: 10.1016/s1079-2104(99)70130-7. [DOI] [PubMed] [Google Scholar]
- Murakami M, Ohtake T, Dorschner RA, Gallo RL. Cathelicidin antimicrobial peptides are expressed in salivary glands and saliva. J Dent Res. 2002;81:845–850. doi: 10.1177/154405910208101210. [DOI] [PubMed] [Google Scholar]
- Nacken W, Roth J, Sorg C, Kerkhoff C. S100A9/S100A8: Myeloid representatives of the S100 protein family as prominent players in innate immunity. Microsc Res Tech. 2003;60:569–580. doi: 10.1002/jemt.10299. [DOI] [PubMed] [Google Scholar]
- Nagaoka I, Hirota S, Yomogida S, Ohwada A, Hirata M. Synergistic actions of antibacterial neutrophil defensins and cathelicidins. Inflamm Res. 2000;49:73–79. doi: 10.1007/s000110050561. [DOI] [PubMed] [Google Scholar]
- Nieuw Amerongen AV, Veerman EC. Saliva--the defender of the oral cavity. Oral Dis. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- Nikawa H, Jin C, Fukushima H, Makihira S, Hamada T. Antifungal activity of histatin-5 against non-albicans Candida species. Oral Microbiol Immunol. 2001;16:250–252. doi: 10.1034/j.1399-302x.2001.160409.x. [DOI] [PubMed] [Google Scholar]
- Nisapakultorn K, Ross KF, Herzberg MC. Calprotectin expression in vitro by oral epithelial cells confers resistance to infection by Porphyromonas gingivalis. Infect Immun. 2001;69:4242–4247. doi: 10.1128/IAI.69.7.4242-4247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niyonsaba F, Someya A, Hirata M, Ogawa H, Nagaoka I. Evaluation of the effects of peptide antibiotics human beta-defensins-1/-2 and LL-37 on histamine release and prostaglandin D(2) production from mast cells. Eur J Immunol. 2001;31:1066–1075. doi: 10.1002/1521-4141(200104)31:4<1066::aid-immu1066>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- O’Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, Ganz T, Kagnoff MF. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- Oppenheim FG, Xu T, McMillian FM, Levitz SM, Diamond RD, Offner GD, Troxler RF. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J Biol Chem. 1988;263:7472–7477. [PubMed] [Google Scholar]
- Page RC, Sims TJ, Geissler F, Altman LC, Baab DA. Defective neutrophil and monocyte motility in patients with early onset periodontitis. Infect Immun. 1985;47:169–175. doi: 10.1128/iai.47.1.169-175.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohaszka Z, Nemet K, Csermely P, Hudecz F, Mezo G, Fust G. Defensins purified from human granulocytes bind C1q and activate the classical complement pathway like the transmembrane glycoprotein gp41 of HIV-1. Mol Immunol. 1997;34:809–816. doi: 10.1016/s0161-5890(97)00097-7. [DOI] [PubMed] [Google Scholar]
- Putsep K, Carlsson G, Boman HG, Andersson M. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet. 2002;360:1144–1149. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- Quinones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, Marotta ML, Mirza M, Jiang B, Kiser P, et al. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. Aids. 2003;17:F39–48. doi: 10.1097/00002030-200311070-00001. [DOI] [PubMed] [Google Scholar]
- Raj PA, Edgerton M, Levine MJ. Salivary histatin 5: dependence of sequence, chain length, and helical conformation for candidacidal activity. J Biol Chem. 1990;265:3898–3905. [PubMed] [Google Scholar]
- Ross KF, Herzberg MC. Calprotectin expression by gingival epithelial cells. Infect Immun. 2001;69:3248–3254. doi: 10.1128/IAI.69.5.3248-3254.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JP, Diamond G, Tarver AP, Scanlin TF, Bevins CL. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor-a. Infect Immun. 1996;64:1565–1568. doi: 10.1128/iai.64.5.1565-1568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabudhe KS, Kimball JR, Morton T, Weinberg A, Dale BA. Expression of the antimicrobial peptide, human b-defensin 1, in duct cells of minor salivary glands and detection in saliva. J Dent Res. 2000;79:1669–1674. doi: 10.1177/00220345000790090601. [DOI] [PubMed] [Google Scholar]
- Saito T, Ishihara K, Kato T, Okuda K. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect Immun. 1997;65:4888–4891. doi: 10.1128/iai.65.11.4888-4891.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonwetter BS, Stolzenberg ED, Zasloff MA. Epithelial antibiotics induced at sites of inflammation. Science. 1995;267:1645–1648. doi: 10.1126/science.7886453. [DOI] [PubMed] [Google Scholar]
- Schroder JM, Harder J. Human betadefensin-2. Int J Biochem Cell Biol. 1999;31:645–651. doi: 10.1016/s1357-2725(99)00013-8. [DOI] [PubMed] [Google Scholar]
- Schroeder HE, Listgarten MA. The gingival tissues: the architecture of periodontal protection. Periodontology 2000. 1997;13:91–120. doi: 10.1111/j.1600-0757.1997.tb00097.x. [DOI] [PubMed] [Google Scholar]
- Schutte BC, Mitros JP, Bartlett JA, Walters JD, Jia HP, Welsh MJ, Casavant TL, McCray PB., Jr Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA. 2002;99:2129–2133. doi: 10.1073/pnas.042692699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MG, Davidson DJ, Gold MR, Bowdish D, Hancock RE. The human antimicrobial peptide LL-37 is a multifunctional modulator of innate immune responses. J Immunol. 2002;169:3883–3891. doi: 10.4049/jimmunol.169.7.3883. [DOI] [PubMed] [Google Scholar]
- Scott MG, Rosenberger CM, Gold MR, Finlay BB, Hancock RE. An alpha-helical cationic antimicrobial peptide selectively modulates macrophage responses to lipopolysaccharide and directly alters macrophage gene expression. J Immunol. 2000;165:3358–3365. doi: 10.4049/jimmunol.165.6.3358. [DOI] [PubMed] [Google Scholar]
- Singh PK, Jia HP, Wiles K, Hesselberth J, Liu L, Conway BA, Greenberg EP, Valore EV, Welsh MJ, Ganz T, et al. Production of beta-defensins by human airway epithelia [published erratum appears in Proc. Natl Acad Sci USA 1999 Mar 2;96(5):2569] Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Situ H, Bobek LA. In vitro assessment of antifungal therapeutic potential of salivary histatin-5, two variants of histatin-5, and salivary mucin (MUC7) domain 1. Antimicrob Agents Chemother. 2000;44:1485–1493. doi: 10.1128/aac.44.6.1485-1493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Starner TD, Barker CK, Jia HP, Kang Y, McCray PB., Jr CCL20 is an inducible product of human airway epithelia with innate immune properties. Am J Respir Cell Mol Biol. 2003;29:627–633. doi: 10.1165/rcmb.2002-0272OC. [DOI] [PubMed] [Google Scholar]
- Stolzenberg ED, Anderson GM, Ackermann MR, Whitlock RH, Zasloff M. Epithelial antibiotic induced in states of disease. Proc Natl Acad Sci USA. 1997;94:8686–8690. doi: 10.1073/pnas.94.16.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D, Miyasaki KT, Lehrer RI. Sensitivity of Actinobacillus actinomycetemcomitans and Capnocytophaga spp. to the bactericidal action of LL-37: a cathelicidin found in human leukocytes and epithelium. Oral Microbiol Immunol. 2000;15:226–231. doi: 10.1034/j.1399-302x.2000.150403.x. [DOI] [PubMed] [Google Scholar]
- Tonetti MS. Molecular factors associated with compartmentalization of gingival immune responses and transepithelial neutrophil migration. J Periodontal Res. 1997;32:104–109. doi: 10.1111/j.1600-0765.1997.tb01389.x. [DOI] [PubMed] [Google Scholar]
- Tonetti MS, Imboden MA, Lang NP. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J Periodontol. 1998;69:1139–1147. doi: 10.1902/jop.1998.69.10.1139. [DOI] [PubMed] [Google Scholar]
- Toomes C, James J, Wood AJ, Wu CL, McCormick D, Lench N, Hewitt C, Moynihan L, Roberts E, Woods CG, et al. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- Tsai H, Bobek LA. Human salivary histatins: promising anti-fungal therapeutic agents. Crit Rev Oral Biol Med. 1998;9:480–497. doi: 10.1177/10454411980090040601. [DOI] [PubMed] [Google Scholar]
- Turner J, Cho Y, Dinh NN, Waring AJ, Lehrer RI. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob Agents Chemother. 1998;42:2206–2214. doi: 10.1128/aac.42.9.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara A, Sugawara S, Muramoto K, Takada H. Activation of human oral epithelial cells by neutrophil proteinase 3 through protease-activated receptor-2. J Immunol. 2002a;169:4594–4603. doi: 10.4049/jimmunol.169.8.4594. [DOI] [PubMed] [Google Scholar]
- Uehara A, Sugawara S, Takada H. Priming of human oral epithelial cells by interferon-gamma to secrete cytokines in response to lipopolysaccharides, lipoteichoic acids and peptidoglycans. J Med Microbiol. 2002b;51:626–634. doi: 10.1099/0022-1317-51-8-626. [DOI] [PubMed] [Google Scholar]
- Ueta E, Tanida T, Doi S, Osaki T. Regulation of Candida albicans growth and adhesion by saliva. J Lab Clin Med. 2000;136:66–73. doi: 10.1067/mlc.2000.107304. [DOI] [PubMed] [Google Scholar]
- Valore EV, Park CH, Quayle AJ, Wiles KR, McCray PB, Jr, Ganz T. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–1642. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg RH, Faber-Krol MC, van Wetering S, Hiemstra PS, Daha MR. Inhibition of activation of the classical pathway of complement by human neutrophil defensins. Blood. 1998;92:3898–3903. [PubMed] [Google Scholar]
- vanderSpek JC, Wyandt HE, Skare JC, Milunsky A, Oppenheim FG, Troxler RF. Localization of the genes for histatins to human chromosome 4q13 and tissue distribution of the mRNAs. Am J Hum Genet. 1989;45:381–387. [PMC free article] [PubMed] [Google Scholar]
- van’t Hof W, Reijnders IM, Helmerhorst EJ, Walgreen-Weterings E, Simoons-Smit IM, Veerman EC, Amerongen AV. Synergistic effects of low doses of histatin 5 and its analogues on amphotericin B anti-mycotic activity. Antonie Van Leeuwenhoek. 2000;78:163–169. doi: 10.1023/a:1026572128004. [DOI] [PubMed] [Google Scholar]
- Vatta S, Boniotto M, Bevilacqua E, Belgrano A, Pirulli D, Crovella S, Amoroso A. Human beta defensin 1 gene: six new variants. Hum Mutat. 2000;15:582–583. doi: 10.1002/1098-1004(200006)15:6<582::AID-HUMU21>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Weinberg A, Krisanaprakornkit S, Dale BA. Epithelial antimicrobial peptides: review and significance for oral applications. Crit Rev Oral Biol Med. 1998;9:399–414. doi: 10.1177/10454411980090040201. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, Matrisian LM, Parks WC. Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science. 1999;286:113–117. doi: 10.1126/science.286.5437.113. [DOI] [PubMed] [Google Scholar]
- Wu Z, Hoover DM, Yang D, Boulegue C, Santamaria F, Oppenheim JJ, Lubkowski J, Lu W. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc Natl Acad Sci USA. 2003;100:8880–8885. doi: 10.1073/pnas.1533186100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Biragyn A, Kwak LW, Oppenheim JJ. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- Yang D, Chen Q, Chertov O, Oppenheim JJ. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J Leukoc Biol. 2000;68:9–14. [PubMed] [Google Scholar]
- Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, Anderson M, Schroder JM, Wang JM, Howard OM, Oppenheim JJ. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- Zanetti M, Gennaro R, Scocchi M, Skerlavaj B. Structure and biology of cathelicidins. Adv Exp Med Biol. 2000;479:203–218. doi: 10.1007/0-306-46831-X_17. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu W, He T, Yu J, Caffrey RE, Dalmasso EA, Fu S, Pham T, Mei J, Ho JJ, et al. Contribution of human alpha-defensin 1, 2, and 3 to the anti-HIV-1 activity of CD8 antiviral factor. Science. 2002;298:995–1000. doi: 10.1126/science.1076185. [DOI] [PubMed] [Google Scholar]
- Zhao C, Wang I, Lehrer RI. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 1996;396:319–322. doi: 10.1016/0014-5793(96)01123-4. [DOI] [PubMed] [Google Scholar]


