Abstract
Heptameric pores formed in the endosomal membrane by the protective antigen moiety of anthrax toxin serve as portals for entry of the enzymatic moieties of the toxin into the cytosol. In the aqueous lumen of each pore is a “Phe clamp,” a heptad of narrowly apposed Phe residues (Phe-427), that catalyzes the unfolding and translocation of the enzymatic moieties across the membrane. Here, we provide evidence for a “loop swap” between neighboring protective antigen subunits, which is required for efficient translocation and is mediated by a salt bridge formed between the side chains of Lys-397 and Asp-426. We propose that the interaction between residues 397 and 426 creates a structural framework that positions Phe-427 within the pore lumen, forming a functional Phe clamp and, hence, a translocation-competent pore.
Keywords: intersubunit, translocation, planar lipid bilayer, lethal factor, protective antigen
Pathogenic bacteria have evolved various strategies for delivering selected enzymes into the cytosolic compartment of mammalian cells, generally with the goal of inhibiting the host’s immune system. One such strategy is illustrated by anthrax toxin, an ensemble of three proteins secreted by Bacillus anthracis. This toxin consists of two intracellularly acting enzymes, termed edema factor (EF) and lethal factor (LF), and a third protein, termed protective antigen (PA), which binds the enzymatic moieties and delivers them to the cytosol. EF, a Ca2+- and calmodulin-dependent adenylate cyclase, and LF, a Zn2+-dependent protease, access their respective intracellular targets after unfolding and translocating through pores generated in the endosomal membrane by the PA moiety (1, 2).
The series of events leading to the membrane crossing of EF and LF begins when PA (83 kDa) binds to a cell-surface receptor (3, 4) and is activated by furin-catalyzed cleavage, causing a 20-kDa segment to be removed from the N terminus (5, 6). The remaining receptor-bound fragment, PA63, then spontaneously self-assembles into a heptameric ring, termed the prepore (6). The prepore binds up to three copies of LF and/or EF with nanomolar affinities (7), yielding a series of toxic complexes on the cell surface. These complexes then are internalized into endosomes, where, in response to acidic pH, the prepore undergoes a conformational change that enables it to form a membrane-spanning pore (8). EF and LF then unfold (1, 9) and translocate through the PA63 pore into the cytosol (10), where they modify their respective intracellular targets.
The various roles of PA in the intoxication pathway underscore the need to elucidate the structural and functional features of this protein at each step. With the crystallographic structures of monomeric PA83 and the heptameric prepore solved (11–14), both alone and bound to a receptor, and with solid evidence regarding how EF and LF bind to the prepore (15, 16), we now have a good understanding of the self-assembly process. Remaining to be described are the detailed structure of the pore, the full panoply of conformational changes occurring in response to acidic pH, and how the pore functions in delivering EF and LF across the endosomal membrane. Extant data indicate that the pore formed by PA63, like that formed by the Staphylococcal α-toxin, is a heptameric mushroom-shaped structure, containing a globular cap connected to a membrane-spanning 14-strand β-barrel. Domain 2 (D2; residues 254–487) contains critical determinants for pore formation and translocation and is known to undergo large conformational changes during prepore-to-pore conversion. These changes include relocation of a loop and its flanking β-strands (i.e., residues 275–350) from the body of D2 to the base of the prepore and reorganization of this stretch to form the long, ≈100 Å, 14-stranded β-barrel stem that inserts into the endosomal membrane (17, 18). The remaining residues of D2 are believed to remain as a part of the globular cap, and some of them line the central channel of the pore. Domain 1′ forms the mouth of the pore and the binding sites for EF and LF. The flexible N terminus of bound EF or LF enters the pore, initiating translocation in an N- to C-terminal direction (2, 19).
Scanning and site-directed mutagenesis studies on PA63 (20–23) have revealed that many point mutations that disable PA-mediated toxicity cluster to two solvent-exposed loops of D2 within the lumen of the prepore (loop 395–408 and loop 421–431). Although many of these mutations inhibit conversion of the prepore to the pore (20), two of them, F427A and K397Q, allow pores to form, but block translocation of LF derivatives through the pores. This result suggests that the parent residues, K397 and F427, are involved in mediating protein translocation. Recently, we reported that the seven F427 residues of the heptamer (one from each subunit) converge as the prepore converts to the pore to create a structure that is required for translocation. We also presented evidence that this structure, termed the Phe clamp, facilitates unfolding of substrate proteins before or during translocation (2). Here, we address the role of K397.
PA homologues are found in toxins produced by certain Bacillus and Clostridium species, and a sequence alignment of these homologues revealed an interesting pattern relevant to K397. We observed that, whereas the crucial Phe-clamp residue, F427, was universally conserved, K397 was conserved only in the Bacillus homologues. At the equivalent position in all of the Clostridial homologues was Gln, which was surprising given that K397Q was known to be a translocation-blocking mutation in PA (21). This finding raised the question of how the Clostridial K397Q-bearing homologues translocate their enzymatic moieties to the cytosol. A residue adjacent to Phe-427, namely Asp-426, was found to display sequence covariance with K397; in the Clostridial homologues, Gln was present at both the 426-equivalent and the 397-equivalent positions. This initial observation led us to perform a mutational analysis to examine whether Asp-426 in PA functionally complements K397 and, thus, whether the corresponding Gln residues at these positions in the Clostridium homologues functionally complement each other. Our results indicate that this hypothesis is correct and, further, that this complementation reflects an intersubunit, rather than an intrasubunit, interaction within the pore. We propose that the 397–426 interaction positions F427 to form a fully functional Phe clamp, thereby enabling the translocation function of the pore.
Results
Sequence Variations in PA Homologues Suggest a Functional Interaction.
The known homologues of PA from Bacillus and Clostridium species were aligned to investigate sequence conservation patterns among residues in D2 that might be involved in translocation (Fig. 1a). As expected, most of the residues previously identified as being critical for proper PA function, such as D425 and F427, were absolutely conserved among the nine homologues (2, 20–23). K397, on the other hand, displayed a genus-specific conservation pattern: Whereas all Bacillus homologues contained Lys at position 397, the Clostridium homologues contained Gln. This observation was noteworthy because it had been shown earlier (21) that, like F427A pores, those with the K397Q mutation were unable to translocate LF derivatives (21). Because the Clostridium homologues must translocate their substrates in a manner analogous to PA, we hypothesized that compensatory mutation(s) were present elsewhere in the Clostridium members that could “rescue” this translocation-blocking mutation.
Fig. 1.
Conservation and covariance within pore lining loops. (a) An alignment of the PA homologues by using clustalx (24). Abbreviations are defined in Supporting Text, which is published as supporting information on the PNAS web site. Shown is the alignment between residues T390 and Y436 according to protective antigen. Shown above are the secondary structural features including the loop regions, residues 395–408 (i.e., the 397 loop) and residues 421–431 (i.e., the 426 loop). Residues with an asterisk denote residues that were identified from mutagenesis studies as being important for mediating toxicity to CHO-K1 cells (20, 21). F427 (‡; colored red) is the translocation active site residue required for PA-mediated toxicity (2, 21, 22). Highlighted in blue, red, and gray are residues that showed an identical conservation pattern and, thus, are referred to as correlated mutations. (b) Side-view of a single subunit of PA taken from the PA83-CMG2 costructure (PDB ID code 1T6B) (12), showing the position of the 397 and 426 loops, in blue and red, respectively, and the side chains of K397, D426, and F427. (c) Cutaway side view surface rendering of the PA prepore heptamer (1TZO) showing loop positions relative to the putative 14-stranded β-barrel. (d) Normalized planar lipid bilayer translocation records of LFN through PA63 channels. After PA63 channel formation reached steady state, 10 nM LFN was added to the cis chamber (at ΔΨ = 10 mV; symmetric pH 5.5 conditions), resulting in a rapid decrease in conductance. Once conductance block reached a steady state, the cis chamber was perfused, and translocation was initiated by adding a fixed amount of KOH to the trans compartment to bring the final pH to 7.2. (e) Cellular translocation assay. PA83 was titrated, mixed with a fixed concentration of LFN-DTA (100 pM), added to CHO-K1 cells, and incubated overnight. Cell death is quantitated by measuring the ability of cells to take up and cleave WST-1.
Using a simple covariance algorithm we found that two sites, positions 426 and 431, both of which are on the same loop as F427, varied in an identical, genus-specific fashion as K397. Thus, as Lys varied to Gln at position 397 in Clostridium homologues, Asp varied to Gln at position 426 and Pro varied to Leu at position 431 (Fig. 1a). Correlated mutations in multiple sequence alignments often point to residues that are linked functionally and/or are in close proximity in a particular conformation within the protein (25). In the prepore crystal structure, K397 is seen to reside on a large disordered loop, herein referred to as the K397 loop, which lies directly above the loop containing D426, F427, and P431, referred to as the D426 loop (14) (Fig. 1 b and c). All of these residues cluster together, at least in the prepore, suggesting that they may be linked structurally and/or functionally.
Functional Complementation of Mutations at Positions 397 and 426.
To address whether the covariance between residues 397 and 426 reflects a functional interaction between these residues, we compared the ability of channels formed by wild-type PA and various mutants to translocate LF derivatives across planar lipid bilayers (26). (Wild-type PA, with K at position 397 and D at position 426, is abbreviated below as K/D, and various mutants are designated in a similar manner; e.g., the K397Q mutant is Q/D.) At symmetric pH 5.5, with the membrane potential held at modest levels (ΔΨ < +20 mV, cis-positive; ref. 27), LFN enters PA63 channels and blocks conductance but does not proceed through to the trans side unless the trans pH is raised. As shown in Fig. 1d, increasing the trans pH to 7.2 caused LFN to translocate through wild-type K/D PA channels with a half time of 13 ± 1 seconds. In Q/D channels, translocation was virtually nil, but adding a second mutation, D426Q, yielding the Clostridial Q/Q sequence, restored translocation activity to a level (t1/2 = 16 ± 2 seconds) close to that of wild-type channels. Thus Q at position 426 complemented Q at position 397 in this assay.
The same PA mutants also were tested for the ability to mediate translocation of LFN-DTA, a fusion between LFN and the catalytic chain of diphtheria toxin (DTA), into CHO-K1 cells. The DTA moiety catalyzes the ADP ribosylation of elongation factor-2, which inactivates the elongation factor, inhibiting protein synthesis and ultimately causing cell death (ref. 26; Fig. 1e). As in the planar bilayer experiments, wild-type K/D pores and Q/Q pores were able to mediate toxicity, whereas Q/D pores were inactive. We also made a triple mutant, K397Q/D426Q/P431L, to address the role of the remaining covariant 431 position but found its translocation activity to be identical to Q/Q both in bilayers and in cells (data not shown). Thus, we focused on the interaction between residues 397 and 426.
K397 Interacts with D426 in a Neighboring Subunit.
Given the relatively narrow confines of the pore lumen, we considered the possibility that, instead of interacting within a given subunit, position 397 might associate with position 426 from a neighboring subunit. Such an intermolecular interaction might, among other things, reposition the K397 and D426 loops and, thus, the Phe clamp more prominently within the pore lumen via an iris-like closing mechanism. One potential way of verifying an intersubunit interaction model would be to show that mixtures of certain nonfunctional mutants are able to complement each other to produce functional heteroheptamers. To this end, we first characterized a panel of single and double mutants at positions 397 and 426 and found that three of the mutants, K/K, Q/D, and Q/E, were completely inactive (<1/10,000 as active as wild-type PA) in mediating the toxicity of LFN-DTA in CHO-K1 cells (Fig. 2 and Table 1). We then combined these mutants in a pairwise fashion at a 1:1 ratio and tested the mixtures for the ability to mediate toxicity of LFN-DTA in CHO-K1 cells and for activity in mediating translocation in planar bilayers. Whereas the individual K/K and Q/D homoheptamers virtually were devoid of activity in transporting LFN-DTA into cells or in translocating LF constructs across planar bilayers, the heteroheptamers formed from the mixture of Q/D and K/K showed a greatly enhanced level of activity, approximately one-tenth that of wild-type PA, in both assays (Fig. 3a). Prepores formed from an equimolar mixture of K/K and the Q/E mutant gave similar results, whereas the Q/D + Q/E mixture gave no activity in either assay (data not shown).
Fig. 2.
Typical record of a planar bilayer experiment showing changes in K+ current in response to PA63 pore formation, LFN-induced block, and pH-induced LFN translocation at a membrane potential of +10 mV (cis-positive).
Table 1.
Pore formation, LFN block, and translocation across planar lipid bilayers and into cells for a panel of 397/426 mutant constructs
| PA 397/426 | Pore formation | LFN block, % | Translocation t1/2sec | Toxicity EC50, fold decrease |
|---|---|---|---|---|
| K/D | + | 95 | 13 | — |
| Q/Q | + | 95 | 16 | 6.0 |
| Q/D | + | 98 | >1,000 | >10,000 |
| Q/E | + | 98 | >1,000 | >10,000 |
| K/K | + | — | — | >10,000 |
| Q/K | + | 35 | 48 | 85 |
| K/N | + | 30 | 27 | 2.7 |
| Q/N | + | 90 | 38 | 17 |
| K/E | + | 93 | 12 | 3.1 |
| K/Q | + | 57 | 17 | 3.4 |
| N/N | + | — | — | >10,000 |
| E/E | + | — | — | >10,000 |
Positive (+) pore formation was determined by the ability of a particular mutant to form both SDS-resistant heptamers on SDS/PAGE and K+-conducting channels in planar lipid bilayers. The degree of LFN block was quantitated after perfusing the 1-ml cis chamber twice with 3 ml of pH 5.5 buffer. Translocation across planar lipid bilayers was initiated by increasing the pH of the trans compartment to 7–7.2 with a fixed amount of KOH with a holding potential, Δψ = 10 mV. The fold decrease in toxicity to cells was calculated by taking the ratio of the mutant EC50 to the WT EC50 (1.3 × 10−13 M). Mutants that did not result in an decreases in cell viability within the concentrations used for these assays were deemed as >10,000-fold defective.
Fig. 3.
Complementation indicates an intersubunit interaction model. (a) Planar lipid bilayer and cellular translocation assays. Both single mutants K397Q and D426K are translocation-defective. ∗, D426K translocation could not be measured, because there was no block by LFN observed. To create mixed heptamers, K397Q and D426K mutants (PA83) were combined in equal quantities, nicked with trypsin, and purified over a Q-Sepharose column (see Materials and Methods). In planar lipid bilayers, mixed heptamers were able to bind and translocate LFN with a translocation half-time of 63 ± 3 seconds (n = 3). In the LFN-DTA cellular translocation assay, both single mutants were at least six orders of magnitude defective, but the mixture was approximately one order of magnitude defective. (b) Model illustrating intersubunit interactions. In wild-type heptamers, K397 interacts with D426 from a neighboring subunit. In K/K and Q/D homoheptamers, these interactions are disrupted. In heteroheptamers, translocation K–D and Q–K interactions are restored.
These results support an intersubunit model for the 397–426 interaction (Fig. 3b) because, in the intrasubunit interaction scenario, an admixture of two defective mutants still should be defective. Thus, with the K/K + Q/D mutant pair, K397 on one subunit interacts with D426 on a neighbor (we designate this interaction as K–D), and Q397 on one subunit would interact with K426 on a neighbor (Q–K) (Fig. 3b). The K–D interaction should be productive, because it reflects that predicted by the intersubunit model in wild-type PA pores. As for the Q–K interaction, we see in Fig. 2b that the Q/K homoheptamer retains significant, although lower than wild-type, levels of activity in both the cell-based and planar bilayer assays. It is not surprising that the level of activity shown by the K/K + Q/D mixture is considerably lower than that of wild-type PA, for three reasons: the low activity of the Q–K interaction; the range of productive intersubunit contacts within the heterogeneous population of heptamers; and the fact that, because of the odd number of subunits, not all neighbor-to-neighbor interactions could be productive within any single heptamer.
The intersubunit model also can explain the fact that the K/K + Q/E mutant pair showed approximately the same level of functional complementation as the K/K + Q/D pair; note in Table 1 that the K/E mutant is only slightly lower in activity than wild-type PA (K/D). The fact that the Q/D + Q/E pair did not show complementation also is consistent, because both of the predicted intersubunit interactions, Q–D and Q–E, should not be functional, as implied by the inactivity of the individual Q/D and Q/E mutants.
The 397–426 Interaction Positions F427 Within the Pore.
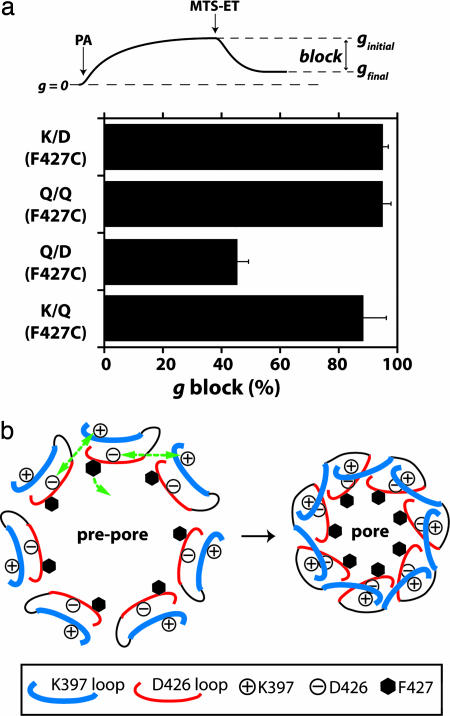
Direct, intersubunit interaction of K397 with D426 would necessarily generate a structural network surrounding the lumen of the pore and, because F427 is an immediate neighbor of D426, this network can be conceived to position this crucial Phe for optimal functionality within the lumen. Previously, it was shown that adding the cationic, Cys-reactive probe [2-(trimethylammonium) ethyl] methane-thiosulfonate (MTS-ET) (Fig. 4) to pores formed by F427C PA almost completely blocked conductance, confirming that this residue was not only accessible but was prominently positioned within the pore (2, 27). Here, we used the same approach to detect differences in location of F427C in the K/D, Q/D, and Q/Q backgrounds. Once a steady-state conductance was reached (ginitial), MTS-ET was added to the trans compartment, resulting in a decrease in conductance to a new steady-state level (gfinal) (Fig. 4 Upper). For the K/D, K/Q, and Q/Q pores, all of which are translocation-competent, the conductance decrease was >90% (Fig. 4). In contrast, in the nontranslocating Q/D mutant background, the conductance block was just 43 ± 3%, suggesting that the Phe clamp was less prominently situated in the lumen. In all cases, the rate of MTS-ET-mediated conductance block was similar, arguing against differences in either accessibility to reagent or reactivity with reagent (data not shown).
Fig. 4.
The 397–426 interaction determines prominence of F427 within lumen. (a Upper) Overview of MTS-ET experiments. Once PA63 channels, containing the F427C mutation have reached steady state, ginitial, MTS-ET is added to the trans compartment to a final concentration of 20 μg/ml. Channel conductance falls to a new steady state, gfinal, and the percent block (g block) is calculated by using (ginitial – gfinal)/ginitial. (a Lower) The percent reduction of macroscopic conductance for K/D, Q/D, and Q/Q pores with F427C (n = 3–6). (b) Intersubunit loop interactions cause a pore constriction that brings F427 more prominently into the pore.
Discussion
Over the past 10 years, it has become evident that unstructured luminal loops often play a critical role in substrate translocation through membrane channels. Perhaps the most well characterized example of how pore loops impact substrate transport comes from ion channels, where in many cases, there exist loops that protrude into the ion-conduction pathway and help catalyze the selective diffusion of ions across membranes (reviewed in ref. 28). Pore loops also have been shown to be involved in the transport of sugars through the maltoporin channel (29) and proteins through the ClpA chaperone (30). In the current work, we show that the K397 and D426 loops interact within the PA pore to generate a site that catalyzes the transport of EF and LF through the pore.
We showed recently that the seven Phe-427 residues of PA converge during pore formation to create a narrow, hydrophobic constriction in the translocation pathway that helps the unraveling substrates to further unfold and translocate across the membrane (2). What was not addressed explicitly was how the seven highly hydrophobic F427 residues, which are on the flexible loop within the prepore, are held within the water-filled channel of the pore. On the basis of the results presented here, we propose a model where the intersubunit interaction between the 397 and 426 loops creates a structural scaffold, which positions F427 within the pore such that it can interact optimally with substrate polypeptide (Fig. 4b). One can draw an analogy between this scaffold and the extensive bonding network that holds the pore selectivity filter of KcsA at its optimum diameter for potassium selectivity (31). However, the Phe clamp must be much less specific, because it must accommodate the various side-chain sizes and chemistries presented by an unfolded polypeptide. It has been proposed that the Phe clamp serves as a site for hydrophobic side chains of the translocating polypeptide to interact transiently, thereby reducing the energy barrier for unfolding. In addition, or alternatively, the clamp may serve as a seal against the passage of protons, thereby preserving the transmembrane proton gradient as the primary energy source for translocation (32).
With respect to the specific interaction that brings the loops into contact, the most straightforward way for K397 of one subunit to interact with D426 of a neighboring subunit would be via a salt bridge between their respective, side-chain −NH3+ and COO− groups. In an analogous way, the side-chain −CONH2 groups of the Gln residues at the corresponding positions of the Clostridium PA homologues could interact via H bonds. Although the magnitude of each individual weak-bonding interaction may be modest (i.e., ≈1–10 kcal/mol), we posit that the multiplicity of these interactions within the pore lumen provides sufficient energy to overcome the entropic penalty associated with stabilizing unstructured loops. We do not yet have data that would allow us to determine whether K397 interacts with D426 on the subunit to its left (facing inward) or to its right. In addition, it is difficult to estimate how many productive 397–426 contacts within a heptameric pore are needed for it to be fully functional. Not every 397–426 contact in the heterogeneous population of pores formed from a mixture of inactive PA mutants, such as K/K + Q/D, can be fully productive, suggesting some level of dominance of the K–D interaction.
Interaction of the charged side chains of K397 and D426 in PA via salt bridges would have the effect of neutralizing a ring of seven positive charges and ring of seven negative charges, respectively, within the channel. This effect suggests another potential role for the loop network within the pore, namely preventing excessive pore–substrate interactions. A ring of exposed charges, either positive or negative, in the pore might interact strongly with exposed positively or negatively charged side chains of the passing polypeptide and stall protein flux through the pore. Thus, salt bridges between the K397 and D426 side chains could create a surface less prone to interaction with the passing polypeptide (see Fig. 5, which is published as supporting information on the PNAS web site). An analysis of this model and a more detailed discussion of the results in Table 1 are included in Supporting Text.
Finally, this work implies that the presence of a ring of seven Phe residues within the PA pore is necessary for protein translocation but is not sufficient, given the need for a structural framework to position these residues properly. Thus, any future attempts at engineering other nontranslocating, pore-forming toxins, such as α-hemolysin (33), into polypeptide translocases by simply adding a hydrophobic polypeptide clamp must consider carefully the geometric constraints imposed by pore loops as outlined here.
Materials and Methods
Proteins.
PA83 and LFN were purified from Escherichia coli as described in ref. 34. PA was activated by incubation with trypsin at a final trypsin:PA ratio of 1:1,000 (wt:wt) for 30 min at room temperature. Adding a 10-fold molar excess of soybean trypsin inhibitor stopped the reaction. PA63 heptamer was prepared as described in ref. 35. Amino-terminal His-6 tags were removed from LFN by treatment with bovine thrombin (10 units/mg protein) for 4 h at room temperature in 20 mM Tris·HCl, pH 8.5/150 mM NaCl.
Planar Lipid Bilayer Experiments.
Membranes were made by painting diphytanoyl phosphatidylcholine (Avanti Polar Lipids) in decane across a 200-μm aperture in a Delrin cup by using the brush technique (36). Both cis and trans compartments contained 1 ml of solutions containing universal bilayer buffer (32) (100 mM KCl; 10 mM each of Mes, oxalic acid, and phosphoric acid; and 1 mM EDTA; pH 5.5). Translocation was initiated by adding appropriate amounts of 2 M KOH to the trans compartment to raise the pH to 7.2. Each compartment was stirred continuously throughout the experiment with a small stir bar. Agar salt bridges linked Ag/AgCl electrodes in 3 M KCl. The current was amplified through a BC-525C integrating bilayer clamp amplifier (Warner Instruments, Hamden, CT), filtered at a frequency of 0.1 kHz by a low-pass eight-pole Bessel filter and computer-displayed through an analog/digital converter (Instrutech, Mineola, NY) with axograph 4.0 (Axon Instruments, Union City, CA).
Cellular Translocation Assay.
LFN-DTA translocation assays on cells were performed as described in ref. 21. PA-dependent translocation of LFN-DTA was determined by measuring the CHO-K1 viability by using the WST-1 reagent after overnight treatment with varying concentrations of PA and fixed LFN-DTA (1 nM).
Substitution of Cysteine with MTS-ET.
MTS-ET (Toronto Research Chemicals, Downsview, ON, Canada) was prepared fresh at 10 mg/ml before each experiment. With the membrane voltage clamped at +20 mV, the nPA-induced conductance was allowed to plateau over several minutes before addition of MTS-ET to the trans compartment (final concentration of 20 μg/ml).
Supplementary Material
Acknowledgments
We thank Alan Finkelstein and Bryan A. Krantz for helpful discussions and suggestions; Henry C. Lin for help with covariance analysis; Stephen J. Juris for helpful discussions, reagents, and assay development; Ruth Pimental for help with protein expression and purification; and Antperro Partridge for critical reading of this manuscript. This work was supported by National Institutes of Health Grant AI022021.
Abbreviations
- D2
domain 2
- DTA
diphtheria toxin
- EF
edema factor
- LF
lethal factor
- MTS-ET
[2-(trimethylammonium) ethyl] methane-thiosulfonate
- PA
protective antigen.
Footnotes
Conflict of interest statement: R.J.C. holds equity in PharmAthene, Inc., and is a consultant for CombinatoRx, Inc.
References
- 1.Wesche J., Elliott J. L., Falnes P. O., Olsnes S., Collier R. J. Biochemistry. 1998;37:15737–15746. doi: 10.1021/bi981436i. [DOI] [PubMed] [Google Scholar]
- 2.Krantz B. A., Melnyk R. A., Zhang S., Juris S. J., Lacy D. B., Wu Z., Finkelstein A., Collier R. J. Science. 2005;309:777–781. doi: 10.1126/science.1113380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradley K. A., Mogridge J., Mourez M., Collier R. J., Young J. A. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 4.Scobie H. M., Rainey G. J. A., Bradley K. A., Young J. A. T. Proc. Natl. Acad. Sci. USA. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Molloy S. S., Bresnahan P. A., Leppla S. H., Klimpel K. R., Thomas G. J. Biol. Chem. 1992;267:16396–16402. [PubMed] [Google Scholar]
- 6.Milne J. C., Furlong D., Hanna P. C., Wall J. S., Collier R. J. J. Biol. Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- 7.Mogridge J., Cunningham K., Collier R. J. Biochemistry. 2002;41:1079–1082. doi: 10.1021/bi015860m. [DOI] [PubMed] [Google Scholar]
- 8.Blaustein R. O., Koehler T. M., Collier R. J., Finkelstein A. Proc. Natl. Acad. Sci. USA. 1989;86:2209–2213. doi: 10.1073/pnas.86.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krantz B. A., Trivedi A. D., Cunningham K., Christensen K. A., Collier R. J. J. Mol. Biol. 2004;344:739–756. doi: 10.1016/j.jmb.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S., Udho E., Wu Z., Collier R. J., Finkelstein A. Biophys. J. 2004;87:3842–3849. doi: 10.1529/biophysj.104.050864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petosa C., Collier R. J., Klimpel K. R., Leppla S. H., Liddington R. C. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 12.Santelli E., Bankston L. A., Leppla S. H., Liddington R. C. Nature. 2004;430:905–908. doi: 10.1038/nature02763. [DOI] [PubMed] [Google Scholar]
- 13.Lacy D. B., Wigelsworth D. J., Scobie H. M., Young J. A. T., Collier R. J. Proc. Natl. Acad. Sci. USA. 2004;101:6367–6372. doi: 10.1073/pnas.0401506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacy D. B., Wigelsworth D. J., Melnyk R. A., Harrison S. C., Collier R. J. Proc. Natl. Acad. Sci. USA. 2004;101:13147–13151. doi: 10.1073/pnas.0405405101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melnyk R. A., Hewitt K. M., Lacy D. B., Lin H. C., Gessner C. R., Li S., Woods V. L., Jr., Collier R. J. J. Biol. Chem. 2006;281:1630–1635. doi: 10.1074/jbc.M511164200. [DOI] [PubMed] [Google Scholar]
- 16.Lacy D. B., Lin H. C., Melnyk R. A., Schueler-Furman O., Reither L., Cunningham K., Baker D., Collier R. J. Proc. Natl. Acad. Sci. USA. 2005;102:16409–16414. doi: 10.1073/pnas.0508259102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson E. L., Huynh P. D., Finkelstein A., Collier R. J. Biochemistry. 1998;37:3941–3948. doi: 10.1021/bi972657b. [DOI] [PubMed] [Google Scholar]
- 18.Nassi S., Collier R. J., Finkelstein A. Biochemistry. 2002;41:1445–1450. doi: 10.1021/bi0119518. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S., Finkelstein A., Collier R. J. Proc. Natl. Acad. Sci. USA. 2004;101:16756–16761. doi: 10.1073/pnas.0405754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mourez M., Yan M., Lacy D. B., Dillon L., Bentsen L., Marpoe A., Maurin C., Hotze E., Wigelsworth D., Pimental R.-A., et al. Proc. Natl. Acad. Sci. USA. 2003;100:13803–13808. doi: 10.1073/pnas.2436299100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sellman B. R., Nassi S., Collier R. J. J. Biol. Chem. 2001;276:8371–8376. doi: 10.1074/jbc.M008309200. [DOI] [PubMed] [Google Scholar]
- 22.Sellman B. R., Mourez M., Collier R. J. Science. 2001;292:695–697. doi: 10.1126/science.109563. [DOI] [PubMed] [Google Scholar]
- 23.Yan M., Collier R. J. Mol. Med. 2003;9:46–51. [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fodor A. A., Aldrich R. W. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- 26.Milne J. C., Blanke S. R., Hanna P. C., Collier R. J. Mol. Microbiol. 1995;15:661–666. doi: 10.1111/j.1365-2958.1995.tb02375.x. [DOI] [PubMed] [Google Scholar]
- 27.Akabas M. H., Kaufmann C., Cook T. A., Archdeacon P. J. Biol. Chem. 1994;269:14865–14868. [PubMed] [Google Scholar]
- 28.MacKinnon R. Neuron. 1995;14:889–892. doi: 10.1016/0896-6273(95)90327-5. [DOI] [PubMed] [Google Scholar]
- 29.Van Gelder P., Dumas F., Bartoldus I., Saint N., Prilipov A., Winterhalter M., Wang Y., Philippsen A., Rosenbusch J. P., Schirmer T. J. Bacteriol. 2002;184:2994–2999. doi: 10.1128/JB.184.11.2994-2999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnerwisch J., Fenton W. A., Furtak K. J., Farr G. W., Horwich A. L. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 31.Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 32.Krantz B. A., Finkelstein A., Collier R. J. J. Mol. Biol. 2006;355:968–979. doi: 10.1016/j.jmb.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 33.Song L., Hobaugh M. R., Shustak C., Cheley S., Bayley H., Gouaux J. E. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 34.Miller C. J., Elliott J. L., Collier R. J. Biochemistry. 1999;38:10432–10441. doi: 10.1021/bi990792d. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham K., Lacy D. B., Mogridge J., Collier R. J. Proc. Natl. Acad. Sci. USA. 2002;99:7049–7053. doi: 10.1073/pnas.062160399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller P., Rudin D. O., Tien H. T., Wescott W. C. Nature. 1962;194:979–980. doi: 10.1038/194979a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.