Abstract
Chronic feeding of methyl-donor (methionine, choline, folic acid, and vitamin B12) deficient diet induces hepatocellular carcinoma formation in rats. Previous studies have shown that promoter CpG islands in various cancer-related genes are aberrantly methylated in this model. Moreover, the global genome in methyl-donor-deficient diet fed rats contains a lesser amount of 5-methylcytosine than control livers. It is speculated that more than 90% of all 5-methylcytosines lie within the CpG islands of the transposons, including the long/short interspersed nucleotide elements (LINE and SINE). It is considered that the 5-methylcytosines in LINE-1 limit the ability of retrotransposons to be activated and transcribed; therefore, the extent of hypomethylation of LINE-1 could be a surrogate marker for aberrant methylation in other tumor-related genes as well as genome instability. Additionally, LINE-1 methylation status has been shown to be a good indicator of genome-wide methylation. In this study, we determined cytosine methylation status in the LINE-1 repetitive sequences of rats fed a choline-deficient (CD) diet for various durations and compared these with rats fed a choline-sufficient (CS) diet. The methylation status of LINE-1 was assessed by the combined bisulfite restriction analysis (COBRA) method, where the amount of bisulfite-modified and RsaI-cleaved DNA was quantified using gel electrophoresis. Progressive hypomethylation was observed in LINE-1 of CD livers as a function of feeding time; that is, the amount of cytosine in total cytosine (methylated and unmethylated) increased from 11.1% (1 week) to 19.3% (56 weeks), whereas in the control CS livers, it increased from 9.2% to 12.9%. Hypomethylation in tumor tissues was slightly higher (6%) than the nontumorous surrounding tissue. The present result also indicates that age is a factor influencing the extent of cytosine methylation.
INTRODUCTION
When rats are chronically fed a diet devoid of a methyl-donor source (choline, methionine, folic acid, and vitamin B12), they spontaneously develop hepatocellular carcinomas (HCCs) [1–5]. This is a unique carcinogenesis model in which no known carcinogen is involved. After the initial discovery [6] of this model, many questions were raised concerning its validity including the possibility of carcinogenic contaminants in the diet. Later, it was definitively demonstrated that diets lacking in methionine and choline and containing no detectable level of carcinogens acted as a complete carcinogen [7, 8]. In spite of extensive phenomenological studies, the mechanism by which dietary methyl-donor deficiency causes HCC formation is not understood, but it is suggested that various concurrent carcinogenic pathways may be involved [4]. Oxidative stress appears to play a major role in this model [4] because there is a significant increase in oxidized DNA (8-hydroxy-deoxyguanosine) levels from day 1 of feeding [4] and because antioxidant cofeeding inhibits cancer formation [9]. Nevertheless, no direct connection between oxidative stress and carcinogenesis has been elucidated.
Genome-wide demethylation of 5-methylcytosine has been regarded as a common epigenetic event in malignancies and may play a crucial role in carcinogenesis. In the rat methyl-donor deficiency models, promoter CpG islands in several cancer-related genes are known to be aberrantly methylated, as noted by changes in DNA 5-methylcytosine content [10]. Methyl donors including choline and methionine are required for S-adenosyl methionine (SAM) biosynthesis [11], and SAM is the substrate for DNA cytosine methyltransferase, the enzyme responsible for maintaining DNA cytosine methylation. Therefore, it is possible to speculate that the dysregulation of Dnmt activity is a cause of the genome-wide decrease of 5-methylcytosine and aberrant methylation of specific genes. However, the selective nature of the presence of aberrantly methylated genes suggests that other factors are also involved. More than 90% of all 5-methylcytosines lie within the CpG islands in the transposons, including long/short interspersed nucleotide elements (LINE and SINE). The presence of 5-methylcytosine in LINE-1 is considered to limit the ability of retrotransposons to be activated and transcribed; therefore, loss of 5-methylcytosine in LINE-1 could result in an increase in retrotransposon activity, leading to propagation of aberrant methylation to other genes [12, 13]. Genome-wide instability inevitably results from hypomethylation. LINE-1 methylation status has also been shown to be a good indicator of genome-wide methylation [14, 15]. In humans, LINE-1 hypomethylation was demonstrated in patients having various cancers [16–18]. In the present study, using the combined bisulfite restriction analysis (COBRA) method [14, 19], we evaluated the amount of cytosine/5-methylcytosine in the LINE-1 repetitive sequence in rats fed a choline-deficient (CD) diet for various times.
MATERIAL AND METHODS
Animals and diets
Rats were treated strictly following the animal use protocol approved by the Institutional Animal Care and Use Committee in the Oklahoma Medical Research Foundation. Weaned male Fisher 344 rats were obtained from Charles River (Indianapolis, IN, USA) and divided into 2 groups each containing 3–5 animals and fed a choline sufficient (CS) or a choline deficient (CD) diet (Dyets Inc Bethlehem, IN, USA). Feeding periods were 1, 4, 24, and 56 weeks. In these experiments, protein in the diet was substituted by a defined amino acid diet (CSAA or CDAA diet), because the CDAA diet has been shown to considerably accelerate carcinogenesis without altering cancer pathology as compared with the conventional CD (Lombardi's CD diet) [20].
Sequence of LINE-1
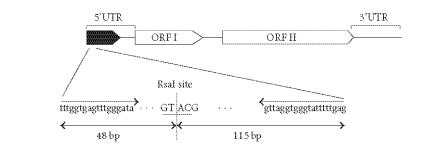
The LINE-1 sequence of interest in this study is illustrated in Figure 1. The consensus sequence revealed that LINE-1 elements have a 5' untranslated region (UTR) with internal promoter activity, two open reading frames (ORFs), a 3'UTR that ends in an AATAAA polyadenylation signal, and a poly A tail [21, 22].
Figure 1.
Genomic structure of LINE-1 in rats. Total length is 6 kb which is composed of three portions, 5' UTR, ORF I, ORF II, and 3'UTR. Bisulfite PCR was conducted in the 5'UTR sequence. The primer sequences are shown with arrows.
Combined bisulfite restriction analysis
COBRA is a simple method of CpG methylation analysis which utilizes the cleaving ability of the restriction enzyme RsaI specifically at bisulfite-modified CpG sites [14]. Genomic DNA from rat liver tissues was extracted using a Quiagen's genomic DNA extraction kit (Qiagen, Valencia, CA, USA). Bisulfite modification of genomic DNA was performed as follows: 3 μg of DNA, digested with the restriction enzyme EcoRI, was incubated with 0.3 N NaOH in a volume of 20 μL for 15 minutes, and then combined with a 120 μL portion of 3.6 M sodium bisulfite (Sigma, St Louis, MO, USA)/0.6 mM hydroquinone (Sigma) (adjusted to pH 5.0 with NaOH). The bisulfite reaction was performed by utilizing a thermocycler (Perkin Elmer 9600, Boston, MA, USA) with 15 cycles of 95°C for 30 seconds followed by 50°C for 15 minutes. The samples were desalted with a Wizard DNA Clean-Up System (Promega, Madison, WI, USA) and desulfonated with 5 minutes incubation in 0.3 N NaOH. Bisulfite modified DNA was PCR-amplified with custom-synthesized primers (Molecular Biology Resource Facility, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA). The primer sequences which correspond to the nucleotides in the regulatory region of LINE-1 sequence (GenBank: U87600) are as follows: L1bisF, 5'-TTT GGT GAG TTT GGG ATA-3'; L1bisR, 5'-CTC AAA AAT ACC CAC CTA AC-3'. The PCR conditions were 30 cycles of 94°C 30 seconds, 55°C 30 seconds, 72°C 30 seconds. Subsequently, the PCR product was digested with 10 unit of RsaI at 37°C, for 5 hours, isolated with ethanol-chloroform precipitation, and subjected to polyacrylamide gel electrophoresis. The gel was stained with ethidium bromide, and the band intensity in the fluorogram was analyzed with an imaging workstation (NucleoTech Corp, Hayward, CA, USA). The data for densitometric analysis is presented as mean ± SE Tests for statistical significance were evaluated using Student t test.
RNA extraction and Semiquantitative RT-PCR
Total RNA was extracted from liver tissues of CS- or CD-diet fed rats with Qiagen RNeasy kit (Qiagen, Crawley, UK). After RNA samples were treated with DNase I (Invitrogen, Carlsbad, CA, USA), RT-PCR was performed with SuperScript III One-Step RT-PCR System (Invitrogen, Carlsbad, CA, USA). Common GenBank accession number of ORF I and ORF II sequences in LINE-1 is DQ100473. PCR was performed under the following conditions: annealing temperatures were 60°C for ORF I and 55 °C for ORF II, cycle number was 23, and primer sequences were ORF I forward; 5′-AAG AAA CAC CTC CCG TCA CA-3′, ORF I reverse; 5′-CCT CCT TAT GTT GGG CTT TAC C-3′, ORF II forward; 5′-CCC ACT CTC TCC CTA CTT A-3′, and ORF II reverse; 5′-TAT AGA GGA AGG CAA CTG AT-3′. The expression of the glyceraldehydes phosphate dehydrogenase gene (GAPDH) was used to normalize the transcript band intensity.
RESULTS
LINE-1 COBRA in artificially methylated DNA
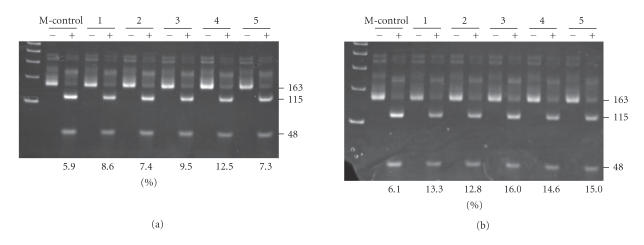
To confirm the accuracy of COBRA, control genomic DNA obtained from rats fed a regular diet was enzymatically methylated with SssI methylase (Sigma) in the presence of the substrate S-adenosylmethionine, and its LINE-1 promoter CpG island methylation was determined with COBRA. The results indicate that there was 5.9–6.1% undigested DNA (Figure 2, 163 kb band in the lanes marked with M-control +), suggesting that the amount of mutated DNA plus SssI methylation resistant cytosine is within this level.
Figure 2.
Typical COBRA for LINE-1 cytosine methylation status in the livers of rats fed CD or CS diet for 4 weeks. Numbers shown at the bottom of each lane indicate the percentage of unmethylated cytosine (115 bp band + 48 bp band) versus total cytosine (163 bp band). Panel A: CS livers (#1–#5) after 4-week feeding; Panel B: CD livers (#1–#5) after 4-week feeding. Isolated genomic DNA was treated with sodium bisulfite and PCR-amplified with LINE-1 primers and divided into two portions. The one portion was digested with RsaI (designated with +), the other was not digested (designated with −), and then both were run on the gel and stained with ethidium bromide. The band at 163 bp was identified as the LINE-1 sequence and those at 115 bp and 48 bp were identified as RsaI-digested LINE-1. The numbers on the top of each pair of lanes indicate the rat identification numbers. Lanes designated with M-control are COBRA treated samples obtained from SssI-hypermethylated rat DNA.
LINE-1 COBRA in CD livers
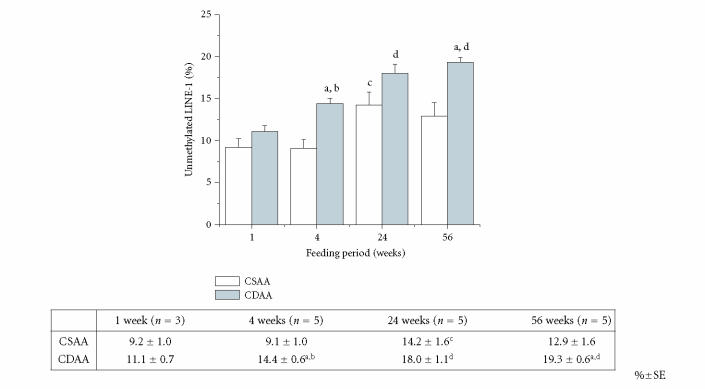
LINE-1 methylation was analyzed with COBRA in CD and CS livers after various feeding periods, including 1, 4, 24, and 56 weeks. Figure 2 illustrates typical COBRA fluorograms obtained from livers of rats fed either CD or CS diet for 4 weeks. There was a clear tendency for the CD diet to promote hypomethylation in LINE-1 during the entire feeding period (1 week to 56 weeks). The amount of unmethylated cytosine in LINE-1 ranged from 9.2% at 1 week to 12.9% at 56 weeks in CS livers, while in the CD livers, it ranged from 11.1% at 1 week to 19.3% at 56 weeks (Figure 3). There was a statistical significant difference between CS and CD livers at 4 and 56 weeks of feeding (Figure 3).
Figure 3.
Percentage of unmethylated cytosine versus total (methylated plus unmethylated) cytosine in LINE-1 sequences in the livers of CSAA- and CDAA-diet fed rats. Actual data are shown in the table under the graph. Symbols: (a) significantly different from CSAA at the same feeding period; (b) significantly different from CDAA at 1 week; (c) significantly different from CSAA at 1 and 4 weeks; (d) significantly different from CDAA at 1 and 4 weeks. %±SE; standard error
LINE-1 hypomethylation in tumor and nontumor tissues
After 24 weeks of feeding the CD diet, rats began to have tumor nodules which were histologically identified as adenomas (19), and at 56 weeks, most of these tumor nodules had developed into HCC. COBRA of LINE-1 DNA for tumor and nontumor tissues showed that there is a tendency for the DNA in tumor tissues to be more hypomethylated than nontumor tissues, however, a statistically significant difference was obtained only in the 56-week fed animals (Figure 4).
Figure 4.
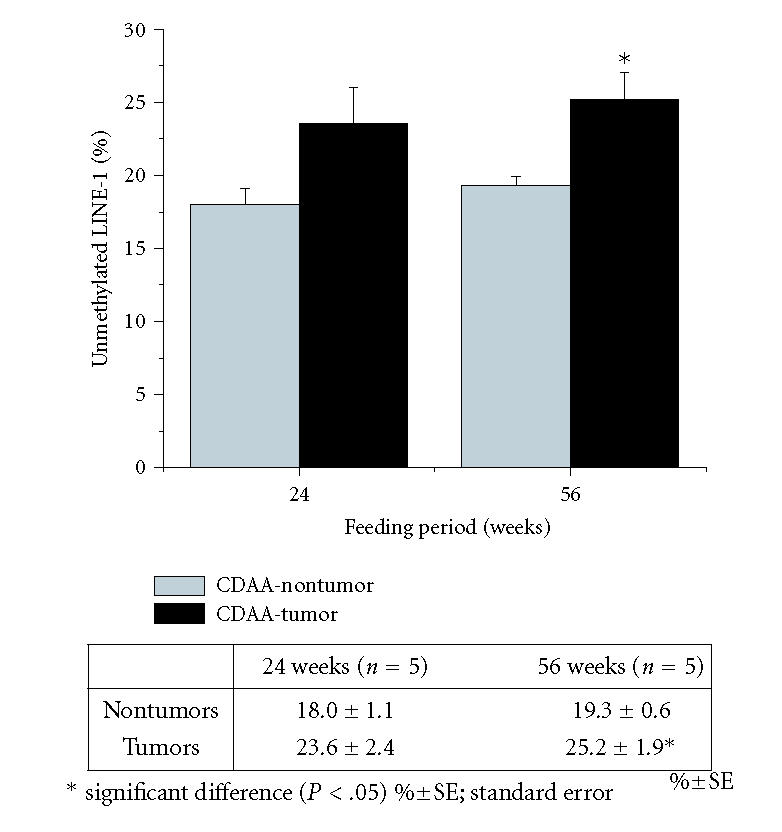
Percentage of unmethylated cytosine in total (methylated plus unmethylated) cytosine in LINE-1 sequences in tumor or nontumor liver tissue in CD-diet fed rats. Actual data are shown in the table under the graph. Statistically significant differences (*) are seen between nontumor and tumor at 56 weeks.
LINE-1 transcript expression
The expression of LINE-1 (ORF1 and ORF2) gene transcript was assessed with semiquantitative RT-PCR. Densitometry-analysis indicated (in arbitrary unit) (1) for ORF I CS(8 weeks →16 weeks): 1.60 ± 0.42 → 1.35 ± 0.49, and CD (8 weeks → 16 weeks): 1.40±0.42 → 1.90±0.28, and (2) for ORF II CS(8 weeks → 16 weeks): 1.45±0.21 → 1.35±0.07, and CD (8 weeks → 16 weeks): 0.75±0.21 → 0.70±0.14. The comparison of the numbers of 8 weeks and 16 weeks in each group indicates that there was no increase in LINE-1 transcript expression. However, these results may have marginal statistical significance because of the small number of samples (N = 2 for each group).
DISCUSSION
Chronic CD diet is hepatocarcinogenic in male rats and global hypomethylation has been shown to exist from early feeding times [10, 23]. In many cancers, global hypomethylation as well as hypo- or hypermethylation in specific genes are widely accepted epigenetic changes [24]; however, which gene or DNA region responsible for aberrant methylation, especially in the methyl-deficient diet models, is not clear. Using a rat CD model, we applied the COBRA method to analyze LINE-1 methylation. COBRA requires small amount of DNA samples and was previously employed to analyze human LINE-1 hypomethylation [18].
We showed that the LINE-1 promoter was hypomethylated in the livers of rats fed a CD diet from as early as 4-weeks' feeding (14.4%) as compared to CS livers of the same feeding period (9.1%), and that hypomethylation increased as a function of feeding period up to 19.3% at 56 weeks (Figure 3). It is not unexpected that there was a significant increase in unmethylated cytosine (or a decrease in 5-methylcytosine) in LINE-1 of control CS livers as a function of feeding period (12.9% at 56 weeks) because it has been shown that aging is a major cause of genome-wide hypomethylation in mice and rats [25, 26]. For example, using Southern blotting, Mays-Hoopes et al estimated that there is an 8% decrease in LINE-1's 5-methylcyotsine content in the livers of 27-month old mice [26]. Furthermore, it is possible that the age-dependent increase of mutation in LINE-1 could provide false signals of hypomethylation in this assay. In humans, LINE-1 hypomethylation was detected only in HCC but not in nontumor liver cirrhosis [16]. Also, a recent report has indicated that in some cancers, such as lymphoma, renal cell carcinoma, and papillary carcinoma of the thyroid, the LINE-1 hypomethylation level is not significantly different from normal tissues [18]. The authors suggested that human cancers may be classified into two groups, a low (0–3.4%) LINE-1 hypomethylation group and a moderately high (6.8–9.5%) group [18]. We speculate that HCC in this model may mimic the situation in the low group. The same study also showed a linear correlation of methylation levels analyzed by COBRA with levels determined by using semiquantitative conventional Southern blotting hybridization analysis [18].
Florl et al reported that in human urothelial cancer, there were coordinate changes of LINE-1 and HERV-K DNA methylation, suggesting that hypomethylation affects a variety of retroelements to similar extents [12]. Thus, LINE-1 hypomethylation is thought to be one of the important surrogate markers of global hypomethylation [14, 15]. In the present study, LINE-1 hypomethylation in CD livers increased in a time-dependent fashion from 11.1% at 1 week to 19.3% at 56 weeks. The cytosine (demethylated 5-methylcytosine) level in LINE-1 in this experiment was much lower than that of the global genome as determined by using an HpaII/MspI-based cytosine extension assay [23], which resulted in cytosine content increasing from 46% at 9 weeks up to 54% at 36 weeks. However, another study using HPLC analysis indicated that genome-wide unmethylated cytosine increased from 6% at 8 weeks to 11–14% at 22 weeks [10]. The cause of discrepancies in the three methods is unknown.
LINE-1 promoter methylation is thought to play an important role in transcriptional activation of retrotransposons [27]. Active retrotransposition can cause the movement of LINE-1 to anywhere within one chromosome and, as such, could disrupt tumor suppressor genes and/or activate oncogenes [28, 29]. Indeed, human colon cancer has been shown to be associated with APC retrotransposon activity [28]. In addition, aberrant LINE-1 methylation may cause specific gene modification as well as genomic instability [13, 30]. In rat methyl-deficiency models, CpG island aberrant methylation has been seen in several oncogenes and tumor-suppressor genes, such as alpha-fetoprotein (AFP) [31], c-Ha-ras [32], c-Ki-ras [32], c-fos [32], c-myc [33], Dnmt [34], glutathione S-transferase pi (GSTP) [35], p16 [36], and protein tyrosine phosphatase receptor O gene (PTPRO) [37]. Although LINE-1 hypomethylation should be followed by the increase in LINE-1 transcripts, our RT-PCR assessment did not show the increasing tendency. Previously, the similar discrepancy was reported in human liver cancer tissues [38].
In conclusion, the degree of hypomethylation in promoter CpG islands in LINE-1 repetitive sequences in the livers of rats fed a CD diet progresses as a function of feeding period. The level of LINE-1 hypomethylation was similar to that found in previous HPLC analysis for genome-wide hypomethylation. These results suggest that genome-wide hypomethylation occurs because of the choline deficiency diet and that LINE-1 methylation status is a good indicator of such. Moreover, because LINE-1 hypomethylation can activate its retrotransposon activity, it may be the root cause of aberrant methylation in several cancer-related genes in this model. This notion is yet to be proven.
ACKNOWLEDGMENT
Support from the National Cancer Institute, National Institutes of Health (R01 CA82506) is gratefully acknowledged.
References
- 1.Poirier LA. The role of methionine in carcinogenesis in vivo. Advances in Experimental Medicine and Biology. 1986;206:269–282. doi: 10.1007/978-1-4613-1835-4_20. [DOI] [PubMed] [Google Scholar]
- 2.Perera MI, Betschart JM, Virji MA, Katyal SL, Shinozuka H. Free radical injury and liver tumor promotion. Toxicologic Pathology. 1987;15(1):51–59. doi: 10.1177/019262338701500106. [DOI] [PubMed] [Google Scholar]
- 3.Zeisel SH, da Costa KA, Albright CD, Shin OH. Choline and hepatocarcinogenesis in the rat. Advances in Experimental Medicine and Biology. 1995;375:65–74. doi: 10.1007/978-1-4899-0949-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Nakae D. Endogenous liver carcinogenesis in the rat. Pathology International. 1999;49(12):1028–1042. doi: 10.1046/j.1440-1827.1999.00990.x. [DOI] [PubMed] [Google Scholar]
- 5.Ghoshal AK, Farber E. Liver biochemical pathology of cho-line deficiency and of methyl group deficiency: a new orientation and assessment. Histology and Histopathology. 1995;10(2):457–462. [PubMed] [Google Scholar]
- 6.Copeland DH, Salmon WD. The occurrence of neoplasms in the liver, lungs and other tissues of rats as a result of prolonged choline deficiency. The American Journal of Pathology. 1946;22:1059–1076. [PubMed] [Google Scholar]
- 7.Mikol YB, Hoover KL, Creasia D, Poirier LA. Hepatocarcinogenesis in rats fed methyl-deficient, amino acid-defined diets. Carcinogenesis. 1983;4(12):1619–1629. doi: 10.1093/carcin/4.12.1619. [DOI] [PubMed] [Google Scholar]
- 8.Ghoshal AK, Farber E. The induction of liver cancer by dietary deficiency of choline and methionine without added carcinogens. Carcinogenesis. 1984;5(10):1367–1370. doi: 10.1093/carcin/5.10.1367. [DOI] [PubMed] [Google Scholar]
- 9.Nakae D, Kotake Y, Kishida H, et al. Inhibition by phenyl N-tert-butyl nitrone of early phase carcinogenesis in the livers of rats fed a choline-deficient, L-amino acid-defined diet. Cancer Research. 1998;58(20):4548–4551. [PubMed] [Google Scholar]
- 10.Wilson MJ, Shivapurkar N, Poirier LA. Hypomethylation of hepatic nuclear DNA in rats fed with a carcinogenic methyl-deficient diet. The Biochemical Journal. 1984;218(3):987–990. doi: 10.1042/bj2180987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shivapurkar N, Poirier LA. Tissue levels of S-adenosylmethionine and S-adenosylhomocysteine in rats fed methyl-deficient, amino acid-defined diets for one to five weeks. Carcinogenesis. 1983;4(8):1051–1057. doi: 10.1093/carcin/4.8.1051. [DOI] [PubMed] [Google Scholar]
- 12.Florl AR, Löwer R, Schmitz-Dräger BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. British Journal of Cancer. 1999;80(9):1312–1321. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz WA, Elo JP, Florl AR, et al. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes, Chromosomes and Cancer. 2002;35(1):58–65. doi: 10.1002/gcc.10092. [DOI] [PubMed] [Google Scholar]
- 14.Yang AS, Estécio MRH, Doshi K, Kondo Y, Tajara EH, Issa J-PJ. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Research. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz WA. L1 retrotransposons in human cancers. to appear in Journal of Biomedicine & Biotechnology. [DOI] [PMC free article] [PubMed]
- 16.Takai D, Yagi Y, Habib N, Sugimura T, Ushijima T. Hypomethylation of LINE1 retrotransposon in human hepatocellular carcinomas, but not in surrounding liver cirrhosis. Japanese Journal of Clinical Oncology. 2000;30(7):306–309. doi: 10.1093/jjco/hyd079. [DOI] [PubMed] [Google Scholar]
- 17.Kaneda A, Tsukamoto T, Takamura-Enya T, et al. Frequent hypomethylation in multiple promoter CpG islands is associated with global hypomethylation, but not with frequent promoter hypermethylation. Cancer Science. 2004;95(1):58–64. doi: 10.1111/j.1349-7006.2004.tb03171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalitchagorn K, Shuangshoti S, Hourpai N, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23(54):8841–8846. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 19.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Research. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakae D, Yoshiji H, Mizumoto Y, et al. High incidence of hepatocellular carcinomas induced by a choline deficient L-amino acid defined diet in rats. Cancer Research. 1992;52(18):5042–5045. [PubMed] [Google Scholar]
- 21.Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annual Review of Genetics. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 22.Deininger PL, Batzer MA. Mammalian retroelements. Genome Research. 2002;12(10):1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- 23.Pogribny IP, James SJ, Jernigan S, Pogribna M. Genomic hypomethylation is specific for preneoplastic liver in folate/methyl deficient rats and does not occur in non-target tissues. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2004;548(1-2):53–59. doi: 10.1016/j.mrfmmm.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature Reviews. Genetics. 2002;3(6):415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 25.Dunn BK. Hypomethylation: one side of a larger picture. Annals of the New York Academy of Sciences. 2003;983:28–42. doi: 10.1111/j.1749-6632.2003.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 26.Mays-Hoopes L, Chao W, Butcher HC, Huang RC. Decreased methylation of the major mouse long interspersed repeated DNA during aging and in myeloma cells. Developmental Genetics. 1986;7(2):65–73. doi: 10.1002/dvg.1020070202. [DOI] [PubMed] [Google Scholar]
- 27.Steinhoff C, Schulz WA. Transcriptional regulation of the human LINE-1 retrotransposon L1.2B. Molecular Genetics and Genomics. 2003;270(5):394–402. doi: 10.1007/s00438-003-0931-2. [DOI] [PubMed] [Google Scholar]
- 28.Miki Y, Nishisho I, Horii A, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Research. 1992;52(3):643–645. [PubMed] [Google Scholar]
- 29.Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature. 1988;333(6168):87–90. doi: 10.1038/333087a0. [DOI] [PubMed] [Google Scholar]
- 30.Rockwood LD, Felix K, Janz S. Elevated presence of retrotransposons at sites of DNA double strand break repair in mouse models of metabolic oxidative stress and MYC-induced lymphoma. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2004;548(1-2):117–125. doi: 10.1016/j.mrfmmm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Locker J, Hutt S, Lombardi B. alpha-Fetoprotein gene methylation and hepatocarcinogenesis in rats fed a choline-devoid diet. Carcinogenesis. 1987;8(2):241–246. doi: 10.1093/carcin/8.2.241. [DOI] [PubMed] [Google Scholar]
- 32.Zapisek WF, Cronin GM, Lyn-Cook BD, Poirier LA. The onset of oncogene hypomethylation in the livers of rats fed methyl-deficient, amino acid-defined diets. Carcinogenesis. 1992;13(10):1869–1872. doi: 10.1093/carcin/13.10.1869. [DOI] [PubMed] [Google Scholar]
- 33.Tsujiuchi T, Tsutsumi M, Sasaki Y, Takahama M, Konishi Y. Hypomethylation of CpG sites and c-myc gene overexpression in hepatocellular carcinomas, but not hyperplastic nodules, induced by a choline-deficient L-amino acid-defined diet in rats. Japanese Journal of Cancer Research. 1999;90(9):909–913. doi: 10.1111/j.1349-7006.1999.tb00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopatina NG, Vanyushin BF, Cronin GM, Poirier LA. Elevated expression and altered pattern of activity of DNA methyltransferase in liver tumors of rats fed methyl-deficient diets. Carcinogenesis. 1998;19(10):1777–1781. doi: 10.1093/carcin/19.10.1777. [DOI] [PubMed] [Google Scholar]
- 35.Steinmetz KL, Pogribny IP, James SJ, Pitot HC. Hypomethylation of the rat glutathione S-transferase pi (GSTP) promoter region isolated from methyl-deficient livers and GSTP-positive liver neoplasms. Carcinogenesis. 1998;19(8):1487–1494. doi: 10.1093/carcin/19.8.1487. [DOI] [PubMed] [Google Scholar]
- 36.Pogribny IP, James SJ. De novo methylation of the p16INK4A gene in early preneoplastic liver and tumors induced by folate/methyl deficiency in rats. Cancer Letters. 2002;187(1-2):69–75. doi: 10.1016/s0304-3835(02)00408-1. [DOI] [PubMed] [Google Scholar]
- 37.Motiwala T, Ghoshal K, Das A, et al. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene. 2003;22(41):6319–6331. doi: 10.1038/sj.onc.1206750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin C-H, Hsieh S-Y, Sheen I-S, et al. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Research. 2001;61(10):4238–4243. [PubMed] [Google Scholar]