Abstract
We previously developed a deterministic record linkage algorithm demonstrating sensitivities approaching 90% while maintaining 100% specificity.1 Substantially better performance has been reported using probabilistic linkage techniques; however, such methods often incorporate human review into the process. To avoid human review, we employed an estimator function using the Expectation Maximization (EM) algorithm to establish a single true-link threshold. We compared the unsupervised probabilistic results against the manually reviewed gold-standard for two hospital registries, as well against our previous deterministic results. At an estimated specificity of 99.95%, actual specificities were 99.43% and 99.42% for registries A and B, respectively. At an estimated sensitivity of 99.95%, actual sensitivities were 99.19% and 98.99% for registries A and B, respectively. The EM algorithm estimated linkage parameters with acceptable accuracy, and was an improvement over the deterministic algorithm. Such a methodology may be used where record linkage is required, but human intervention is not possible or practical.
INTRODUCTION
Increasingly health care information is distributed across many independent databases and systems, both within and among organizations as separate islands with different patient identifiers.2 This is the case for data collected within an institution where there may be multiple identifiers, or for data collected about the same patient at different health care institutions, different pharmacy systems, different payers, and so on. This situation interferes with the aggregation of information about individuals across such databases as needed for public health reporting, clinical research, outcomes management, and drug toxicity reporting. Aggregation is important not only to determine a patient’s health care status, but also for population based studies. Record linkage is the process of combining information about an individual, family, or entity residing in one or more databases.
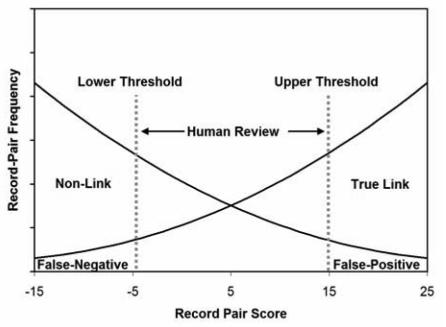
Several kinds of linkage algorithms exist.3 We previously developed an exact-agreement deterministic linkage algorithm and reported on its performance, which yielded sensitivities approaching 90% while maintaining 100% specificity.1 On the surface these results compare poorly with the success of probabilistic linkage methods, which can approach greater than 95% for both sensitivity and specificity.4 However these comparisons can be misleading because they reflect the success of the algorithm assisted by a human, not the algorithm alone. Probabilistic linkage software will declare a link for record-pairs with high match likelihood scores and will declare a non-link if the score is very low, but requires a human operator to evaluate the record-pair when the computed likelihood is within an indeterminate middle range.5–7 (Figure 1)
Figure 1. Typical two-threshold scheme for probabilistic scores using human review.
Record pairs between the upper and lower thresholds are manually reviewed for true- or false-link status.
We hypothesize that the Felligi-Sunter (FS) probabilistic linkage method will perform better than our exact-agreement deterministic method because the FS method produces match likelihood scores that are tailored to the unique characteristics of the specific records being linked. That being the case, we wanted to evaluate a probabilistic method without human intervention, so that we can make the right methodology choice when we cannot afford the high cost of a human operator, or because privacy concerns dominate. While the computational methods described in this research are well-known, there are no known reports describing the unsupervised performance of such methods using hospital registry data.
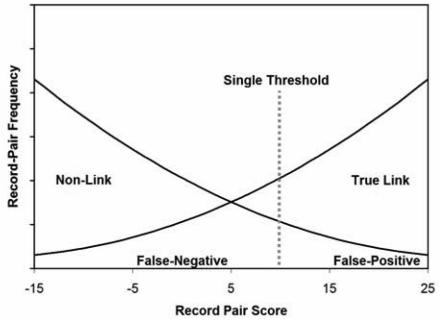
Probabilistic linkage algorithms generate a match likelihood score for each comparison. We can remove the human operator by picking a single threshold above which we declare a link and below which a non-link. (Figure 2) In this report we describe the performance of a probabilistic linkage algorithm implemented without human intervention, assess its performance, and compare it to our deterministic method.
Figure 2. Single probabilistic score threshold without human review.
No scores are tagged for human review.
METHODS
We compared the performance of an unsupervised probabilistic technique to our earlier deterministic method using the same manually reviewed gold-standard data.1 We previously analyzed two separate 6,000 record pair files from two hospital registries linked to the Social Security Death Master File by Social Security Number. Each record pair was labeled as a link or non-link.
For the current study we generated match likelihood scores for each record-pair using the Felligi-Sunter model of record linkage, which sums the component weights of each identifier in the jth record pair:8
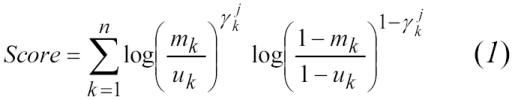 |
where, for the kth identifier in the jth record pair:
n = number of identifiers per record
γjk = observed agreement/disagreement value (1=agree, 0=disagree)
mk = estimated identifier agreement rate among true links
uk = estimated identifier agreement rate among false links
Because we don’t know which pairs are true links or false links, we have to estimate mk and uk from the data. Since commonly used identifiers demonstrate similar agreement rates across independent data sets,9, 10 a bootstrap method proposes using these standard agreement/disagreement rates for each identifier as initial estimates and iteratively recalculating the match weights.5, 9 However, such iterative recalculation requires manually reviewing indeterminate record pairs.
Recently investigators began using the expectation maximization (EM) algorithm to estimate the mk and uk parameters.11–13 The EM algorithm is a widely used probabilistic algorithm for obtaining maximum likelihood estimates of unknown parameters.14 Given an incomplete set of data D (in this dataset, the true link or false link status, and proportion of true links are missing) and a model for the incomplete data characterized by a parameter set θ (in this case mk and uk ), the fundamental goal of EM is to determine θ such that the probability P(D|θ) is maximized. To do so, an initial set of parameters (θ) are used to calculate an expected likelihood in the expectation step, providing estimates of missing data. In the maximization step, the derivative of the data log-likelihood is set to zero to update the estimates of the unknown parameters. We repeat the E and M steps until the parameter estimates converge.
The data log-likelihood for probabilistic record linkage is:12
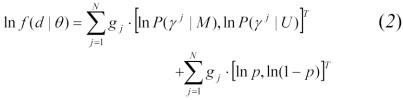 |
where:
d = observed, incomplete data
θ = parameter set (m1, m2, …, mn, u1, u2, …, un, p)
N = observed total number of record pairs
gj = (1,0) for matched pairs and (0,1) for unmatched pairs
γj = observed identifier agreement/disagreement vector for the jth record pair
p = proportion of truly matched record pairs
For the expectation step, the unknown values for gj are estimated using (gm(γj), gu(γj)) where:12
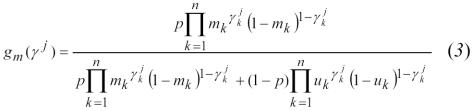 |
gu(γ j) is derived similarly.
For the maximization step, the partial derivatives for each of three maximization problems are set to zero, yielding equations for the unknown parameters:12
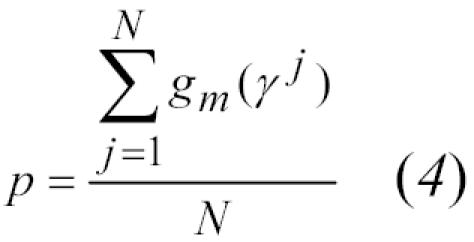 |
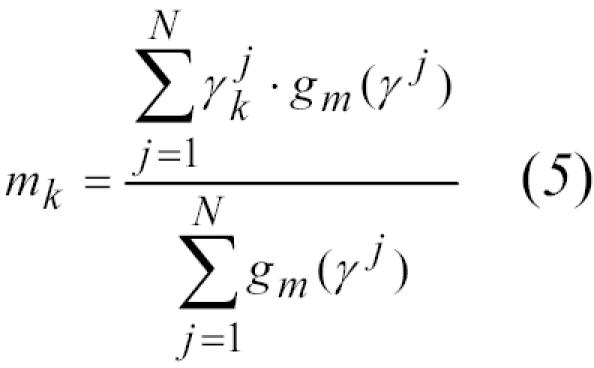 |
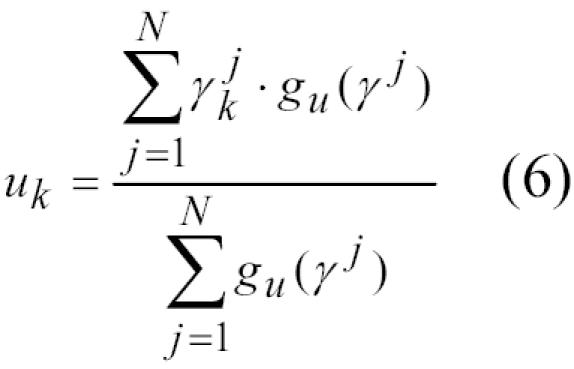 |
We refined the parameters with multiple iterations of the expectation and maximization steps, (equations 3–6) to establish estimates for the proportion of true links (p) and identifier agreement rates (mk and uk). Initial values for mk, uk, and p were 0.9, 0.1 and 0.5, respectively; values for all parameters in both registries converged to 5 decimal places after approximately 15 iterations.
Match likelihood scores for each set of 6,000 were then calculated using (1). Assuming conditional independence between identifiers, the estimated true-positive and true-negative rates for each comparison vector (γj) were calculated using:
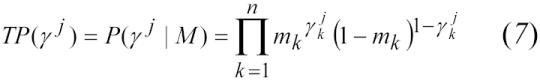 |
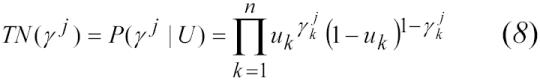 |
For a data set with n identifiers per record, there are 2(n) unique agree/disagree comparison vectors (γj). As before,1 we used last name, NYSIIS15 transformed first name (FNY), middle initial, gender, month, day, and year of birth to establish linkage status; therefore, there were 2(7) or 128 unique comparison vectors. SSN was not used for this analysis because we used it to block the record pairs; therefore, it agreed for all record-pairs. (Blocking refers to the process of grouping similar record pairs together. It is analogous to sorting socks by color before pairing them.)
We calculated the estimated true-positive and true-negative rates for each unique vector using equations (8) and (9). We ordered the vectors by ascending scores and calculated the estimated sensitivity and specificity as a function of record-pair score. We then compared the estimated sensitivity and specificity at various match likelihood score thresholds with both the manually reviewed results and the deterministic method.
RESULTS
Sensitivities/Specificities
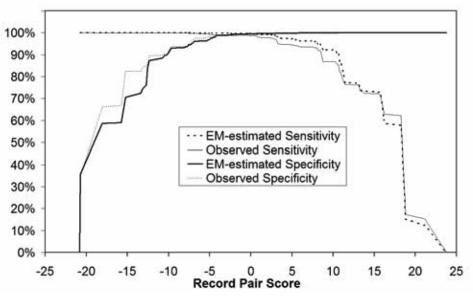
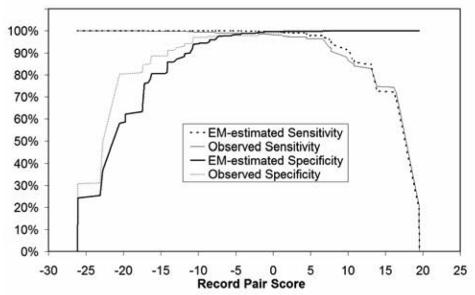
The EM-estimated and observed sensitivities and specificities for registries A and B are shown in Figure 3 and Figure 4, respectively. Table 1 shows the estimated and observed values for an estimated specificity of 99.95% and an estimated sensitivity of 99.95%. The results from the earlier deterministic algorithm are included for comparison as well. At these thresholds, the EM estimates closely reflect the observed values.
Figure 3. Registry A sensitivities and specificities as a function of match likelihood score.
The EM-estimates are compared with manually reviewed (observed) values.
Figure 4. Registry B sensitivities and specificities as a function of match likelihood score.
The EM-estimates are compared with manually reviewed (observed) values.
Table 1. Estimated and observed sensitivities and specificities for two threshold estimates and the deterministic algorithm.
(SPECest/obs = estimated and observed specificity and SENSest/obs = estimated/observed sensitivity)
| Registry A | Registry B | ||
|---|---|---|---|
| SPECest = 99.95 | SPECobs | 99.43 | 99.42 |
| SENSest | 97.39 | 98.02 | |
| SENSobs | 94.64 | 96.50 | |
| SENSest = 99.95 | SENSobs | 99.19 | 98.99 |
| SPECest | 97.04 | 98.79 | |
| SPECobs | 98.29 | 97.97 | |
| Deterministic | SENS | 87.5 | 90.9 |
| SPEC | 100 | 100 |
The area under the curve (AUC) values for the receiver-operator characteristics (ROC) curves are shown in Table 2. The estimated values very closely approximate the manually reviewed results.
Table 2.
Area under the curve (AUC) values for both estimated and observed results.
| Registry A | Registry B | |
|---|---|---|
| Manual Review | .9998 | .9999 |
| EM-Estimate | .9980 | .9989 |
True links and identifier agreement
Manual review found 5,298 true links out of 6,000 record pairs for registry A and 5,655 true links from 6,000 for registry B. The EM algorithm estimated 5,254 and 5,597 true links for registries A and B, respectively. Table 3 shows the estimated and observed agreement rates for individual identifiers, from which all estimated data were derived.
Table 3. EM-estimated identifier agreement rates compared with observed agreement rates.
EM accurately estimates parameters for most cases. The lower uest accuracy in registry B is related to the relatively small number of non-links in that data.
| Registry A | Registry B | |||||||
|---|---|---|---|---|---|---|---|---|
| Identifier | mest | mobs | uest | uobs | mest | mobs | uest | uobs |
| LN | .937 | .935 | .246 | .216 | .977 | .976 | .466 | .388 |
| FNY | .890 | .886 | .047 | .014 | .925 | .924 | .131 | .008 |
| MI | .225 | .223 | .009 | .010 | .283 | .283 | .046 | .008 |
| G | .802 | .799 | .390 | .391 | .825 | .825 | .320 | .240 |
| MB | .970 | .964 | .090 | .084 | .989 | .980 | .092 | .098 |
| DB | .917 | .910 | .053 | .047 | .955 | .945 | .049 | .052 |
| YB | .920 | .912 | .042 | .041 | .966 | .957 | .065 | .057 |
Vectors
Table 4 shows two sample agreement vectors (γj) from registry A and their corresponding parameters. The estimated values are derived using the agreement rates from Table 3. The first vector represents record-pair agreement on all except one identifier; the second vector agrees on only one identifier. The match likelihood score for the first vector is 18.8, indicating a highly probable link relative to other pairs in the data set. The second vector has a low score and thus will likely not be considered a true link.
Table 4. Two agreement vector examples and their corresponding linkage parameters.
The individual components in the vector represent agreement-disagreement between last name, NYSIIS transformed first name, middle initial, gender, month, day, and year of birth, respectively.
| Agreement Vectors (γ j): | ||
|---|---|---|
| {1 1 0 1 1 1 1} | {0 1 0 0 0 0 0} | |
| True-link rate, estimated | 0.424 | 3.1x10−6 |
| SENSest | 15.36% | 100% |
| SENSobs | 17.26% | 99.92% |
| True non-link rate, estimated | 9.3x10−7 | 0.117 |
| SPECest | 100% | 70.64% |
| SPECobs | 100% | 82.45% |
| Link Score | 18.8 | (−15.2) |
DISCUSSION
The probabilistic method represents an improvement over the deterministic method for a number of reasons. First, sensitivities were substantially improved by 6% to 7% with minimal decrease in the specificity (Table 1). Second, although the deterministic method achieved a numerical value for specificity of 100%, this was from a sample size of 6,000. In a much larger sample we may detect false positives. Third, deterministic sensitivities may decrease in data with different identifier characteristics such as different ethnic names or greater typographical error rates. Fourth, the probabilistic method automatically adapts to the specific data set while the deterministic model does not. Fifth, while false positives and false negatives are not completely eliminated, one can select an estimated level of linkage sensitivity or specificity with reasonable accuracy.
The estimated sensitivity and specificity differ most from observed values at the extremes of scores (Figure 3 and Figure 4). This is due in part to the assumption of conditional independence (CI) in both the EM algorithm and probabilistic scoring method. That is, the models assume that identifiers such as first name and gender are independent, when in fact there is dependence. For example, there are few males where the first name is Mary.
Further, agreement/disagreement values (m and u) were estimated using exact-match rules, while manual review used all informational cues available to the reviewer. For example, a true-link record pair failing to exactly match on several identifiers may fall below the estimated true link threshold, while a manual review may reveal name misspellings and numerical nearness in birth dates that provide sufficient evidence to conclude the two records are indeed a true link.
String comparator functions16, 17 can be used to improve the accuracy of comparison vectors and thus improve linkage estimate accuracy. String comparators allow for minor spelling variations and typographical errors in data. Linkage accuracy may also improve if we include other information such as zip code, race, or other accurately recorded, stable identifiers.
This study is limited by the fact that we blocked record-pairs on SSN alone. Records that agree at the outset on SSN will have a high proportion of true-links. Record pairs formed with additional blocking schemes may produce different results.
Additionally, it is notable that the Felligi-Sunter probabilistic record linkage method can be used as a systematic method to predict the performance of individual identifier agreement/disagreement vectors. Because these vectors represent exact match, or deterministic decisions, this method theoretically can be used to automatically discover the most accurate linkage combinations for a deterministic linkage algorithm without any human review.
Future work will analyze the effect of including additional identifier blocking combinations. Additionally, many of the steps involved in probabilistic linkage can be performed in parallel fashion, greatly reducing processing time for large data sets.18 We will explore developing a parallel version of the software. We will also expand functionality to include string comparators with corresponding probability density functions.
CONCLUSION
In our hospital registry data, the EM algorithm accurately estimated linkage parameters without human intervention. Such a methodology may be used where record linkage is required, but human intervention is not possible or practical. The software tools used to perform linkage may be obtained by contacting the author (SJG).
Acknowledgments
This research was performed at the Regenstrief Institute in Indianapolis, Indiana and was funded, in part, by National Cancer Institute grant U01CA91343, a Cooperative Agreement for The Shared Pathology Informatics Network; the National Library of Medicine grant T15 LM-7117-05; and the Indiana Genomics Initiative (INGEN) of Indiana University, which is supported in part by Lilly Endowment Inc. Thanks to Sean Thomas MD, Queen's Medical Center, John A. Burns School of Medicine, University of Hawai'i and Dr. Paul Dexter, Regenstrief Institute, Indiana University School of Medicine for reviewing this manuscript.
REFERENCES
- 1.Grannis S, Overhage J, McDonald C. Analysis of Identifier Performance using a Deterministic Linkage Algorithm. In: American Medical Informatics Association; 2002 2002; San Antonio, TX; 2002. [PMC free article] [PubMed]
- 2.McDonald C, Overhage J, Dexter P, Blevins L, Meeks-Johnson K. Canopy Computing: Using the Web in Clinical Practice. Journal of the American Medical Association. 1998;280(15):1325–1329. doi: 10.1001/jama.280.15.1325. [DOI] [PubMed] [Google Scholar]
- 3.Gill L. Methods for Automatic Record Matching and Linking and their use in National Statistics. Norwich: Her Majesty's Stationary Office; 2001.
- 4.Newman TB, Brown AN. Use of commercial record linkage software and vital statistics to identify patient deaths. J Am Med Inform Assoc. 1997;4(3):233–7. doi: 10.1136/jamia.1997.0040233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Victor TW, Mera RM. Record linkage of healthcare insurance claims. Medinfo. 2001;10(Pt 2):1409–13. [PubMed] [Google Scholar]
- 6.Pates RD, Scully KW, Einbinder JS, Merkel RL, Stukenborg GJ, Spraggins TA, et al. Adding value to clinical data by linkage to a public death registry. Medinfo. 2001;10(Pt 2):1384–8. [PubMed] [Google Scholar]
- 7.Winkler W. The State of Record Linkage and Current Research Problems. Washington, D.C.: Statistical Research Division, U.S. Bureau of the Census; 1999. Report No.: RR1999/04.
- 8.Felligi I, Sunter A. A Theory for Record Linkage. Journal of the American Statistical Association. 1969;64(328):1183–1210. [Google Scholar]
- 9.Howe G, Lindsay J. A Generalized Iterative Record Linkage Computer System for Use in Medical Follow-Up Studies. In: Record Linkage Techniques - 1985 May 9–10,1985; Arlington, VA: Washington Statistical Society and Federal Committee on Statistical Methodology. p. 97–111.
- 10.Arellano M, Weber G. Issues in Identification and Linkage of Patient Records Across an Integrated Delivery System. Journal of Health Care Information Management. 1993;12(3):43–52. [PubMed] [Google Scholar]
- 11.Winkler W. Using the EM Algorithm for Weight Computation in the Felligi-Sunter Model of Record Linkage. Washington, DC: Statistical Research Division, Methodology and Standards Directorate, U.S. Bureau of the Census; 2000. Report No.: RR2000/05.
- 12.Jaro M. Advances in Record-Linkage Methodology as Applied to Matching the 1985 Census of Tampa, Florida. In: Record Linkage Techniques: Proceedings of an International Workshop and Exposition; 1997; Arlington, VA: National Academy Press; 1997. p. 351–357.
- 13.Yancey W. Improving EM Algorithm Estimates for Record Linkage Parameters. Proceedings of the Section on Survey Research Methods, American Statisitcal Association 2002(to appear).
- 14.Dempster A, Laird N, Rubin D. Maximum Likelihood from Incomplete Data via the EM Algorithm. Royal Statistical Society. 1976;39(Series B):1–38. [Google Scholar]
- 15.Lynch B, Arends W. Selection of a surname encoding procedure for the Statistical Reporting Service record linkage system. Washington, DC: United States Department of Agriculture; 1977.
- 16.Porter E, Winkler W. Approximate String Comparison and its Effect on an Advanced Record Linkage System. In: Record Linkage Techniques: Proceedings of an International Workshop and Exposition; 1997; Arlington, VA: National Academy Press; 1997. p. 190–199.
- 17.Sidelli R, Friedman C. Validating Patient Names in an Integrated Clinical Information System. In: Symposium on Computer Applications in Medical Care; 1991; Washington, DC; 1991. p. 588–592. [PMC free article] [PubMed]
- 18.Christen P, Zhu J, Hegland M, Roberts S, Nielsen O, Churches T, et al. High Performance Computing techniques for Record Linkage. In. Canberra, Australia: ANU Data Mining Group, Australian National University; 2002.