Abstract
Although huge amounts of unstructured text are available as a rich source of biomedical knowledge, to process this unstructured knowledge requires tools that identify concepts from free-form text. MetaMap is one tool that system developers in biomedicine have commonly used for such a task, but few have studied how well it accomplishes this task in general. In this paper, we report on a study that compares MetaMap’s performance against that of six people. Such studies are challenging because the task is inherently subjective and establishing consensus is difficult. Nonetheless, for those concepts that subjects generally agreed on, MetaMap was able to identify most concepts, if they were represented in the UMLS. However, MetaMap identified many other concepts that people did not. We also report on our analysis of the types of failures that MetaMap exhibited as well as trends in the way people chose to identify concepts.
Biomedical Concept Identification
Much of biomedical knowledge is represented in textual form; yet such unstructured representations of information are difficult for computers to process in a consistent and meaningful way. To address this problem, many systems that rely on text as an information source use a tool to identify concepts as single or multi-word phrases from within the text. Because the concept identification step is critical in converting free-form text into a computable representation, we need to understand how people identify concepts from text and how well tools are able to match that concept identification process. In this study, we provide a start to achieving such an understanding. We report on both how people identify biomedical concepts from text and how well MetaMap, a commonly used tool for identifying biomedical concepts, performs compared to people.
MetaMap
Researchers at the National Library of Medicine have created a tool called MetaMap that identifies biomedical concepts from free-form textual input and maps them into concepts from the Unified Medical Language System (UMLS) Metathesaurus.1, 2
MetaMap first breaks the text into phrases and then, for each phrase, it returns the mapping options ranked according to the strength of mapping. Researchers have used MetaMap for a variety of tasks including information retrieval,3–5 text mining,6, 7 and extraction of specific kinds of concepts, such as anatomical terms,8 and molecular binding sites.9 Although MetaMap is a critical component of those systems, no one has published an evaluation of MetaMap’s ability to identify biomedical concepts in general.
Study Design
To conduct such an evaluation, we chose to compare MetaMap’s results to the results of multiple people identifying biomedical concepts from the same text.
Subjects
Through an email request, we recruited subjects who had some clinical experience. Subjects were volunteers and received no compensation for participating in the survey. In our pilot studies, subjects spent between 30 and 60 minutes on the task, but because we used a web-based survey tool, we were not able to record the time that final subjects spent on identifying concepts.
Test Text
We chose to evaluate the concept identification task on the titles of articles from MEDLINE. Our motivation for this choice was influenced by several factors. First, a wide variety of information is available from documents in the MEDLINE collection, and their titles are often informative reflections of the content of those documents. Second, our work on text mining relies on concepts identified from titles, and we needed to assess how well MetaMap would work for that task 7. Finally, no one else has evaluated concept identification on this general and widely used kind of text. To get a breadth of coverage without overwhelming our subjects, we used 20 titles about a disease (i.e., migraine), 20 titles about a treatment (i.e., beta-blockers), and 20 titles about a diagnostic test (i.e., EKG) for a total of 60 titles. For each of our search terms, specified in the parenthetical phrases above, our test set consisted of the first 20 titles from a MEDLINE search on that term, but letters, guidelines, and editorials were excluded.
Procedures
We ran MetaMap on each title and stored the results in a MySQL database for later comparison against subjects’ responses.
To collect the subject-identified biomedical concepts, we instructed each subject to use an anonymous, web-based questionnaire to specify his/her selection of concepts from the titles. On the web-based survey, subjects were first asked to specify both their medical background, and area of specialization (optional to retain anonymity). Then, the survey presented each title as a separate question followed by an empty text box beneath each title. Subjects could submit the web-based survey only after they entered at least one concept for each title. The subjects were given the following instructions for identifying concepts:
For each of the following titles (referred to as Questions 3–62 on this form), please list all biomedical concepts (single words or multi-word phrases) in the box below each title. WRITE EACH CONCEPT ON A NEW LINE. Note that you may choose to enter a concept name that does not exactly match a phrase in the title. For example, if “Breast and Ovarian Cancer” were in the title, you could list two concepts “Breast Cancer” and “Ovarian Cancer”. Feel free to cut and paste words or phrases from the title into the text box if that is easiest.
MetaMap Parameters
Many configuration options affect the execution of MetaMap as well as the display of its outputs. In particular, MetaMap provides three different types of data models that differ from each other by the level of filtering they do on the UMLS Knowledge Sources. We used their strict model, which includes all types of filtering and which they claim to be the most appropriate model for semantic-processing applications1. We used only its top-ranked terms from the output.
Data Cleaning
After collecting data from the subjects, we noticed that they often identified the same medical concepts, but they represented them in slightly different ways. These syntactic variations made it difficult to determine a consensus automatically for any of the specified concepts. Thus, to decrease the variability in the subjects’ answers, we checked and cleaned all the subjects’ responses through the following actions:
Correction of spelling errors
Elimination of extra punctuation and spaces
Elimination of determiners (e.g. removing a from a pilot study or the from the pilot study)
Elimination of extraneous definitions that were not part of the original text (e.g. removing as target location from Spain as target location or removing as target decade from 1990–2000 as target decade)
Separation of the concepts connected by the conjunction and (e.g. removing and from Migraine and Heart Arrest and specifying it as the two different concepts Migraine and Heart Arrest)
One person did all the data cleaning according to the specified guidelines, and a second person double-checked all the cleaned results for consistency and accuracy.
Comparison Process
After cleaning the data, for each title, we designated the reference standard or gold standard as those concepts that at least half of the subjects listed. To evaluate MetaMap’s performance against that of our subjects, we needed to specify what constituted a match between terms. Given the variety of concepts identified, we decided to note two types of matches: an exact match, and a partial match.
Exact Match
We considered a concept an exact match if MetaMap identified it, and it exactly matched the reference standard or any of its synonyms from the UMLS Metathesaurus. For example, migraine and headache, migraine are synonyms in the UMLS. Thus, it would constitute an exact match even if the reference standard was migraine and MetaMap listed headache, migraine as the extracted concept. For other concepts that were not defined in the UMLS Metathesaurus, we used only plural and singular forms of the concepts as their synonyms. We ignored case differences.
Partial Match
We considered a concept a partial match if MetaMap identified it and it was a subset of the reference standard. All the words of a multiword MetaMap concept must appear in the reference standard concept for it to be a partial match. For example, when the reference standard was leptomeningeal angiomatosis and MetaMap identified Angiomatosis as a concept, it was considered a partial match. In addition, to qualify as a partial match, the words from the MetaMap concept must appear in the same order as they did in the reference standard, without any additional words between them. For example, when the reference standard was trigeminal ganglia neurons, if MetaMap identified trigeminal ganglia as a concept, it was considered a partial match, but if MetaMap selected the concept trigeminal neurons instead, it was not considered a match at all.
Results
We examined the results of six subjects (three nurses and three physicians) and compared them against MetaMap’s results. We excluded results from two of our eight original subjects (one nurse and one physician) because they did not follow the directions. One subject rephrased the title rather than identifying individual concepts. The other subject took a radically different approach to the problem than all other subjects. Rather than selecting the concepts that were explicitly contained in or referred to by the title, the excluded subject appeared to read the title and generate all concepts that could be discussed in a document with such a title.
The six subjects specified 492 concepts across all the titles. Of that total, 151 qualified as the reference standard. Some concepts from the reference standard appeared in more then one title. For example, migraine was a concept selected as a reference standard for 12 different titles. If we eliminated such duplicates, 133 unique concepts were in our reference standard set. Of those 133 concepts, 73 of them were in the UMLS Metathesaurus; 60 concepts were not in the UMLS.
The primary goal of our study was to determine how well MetaMap functions as a concept-identification tool. Thus, the metrics we focused on were MetaMap’s precision and recall, rather than using a metric such as the Kappa statistic to evaluate inter-subject agreement. Future studies will examine such aspects of inter-rater reliability.
MetaMap Recall
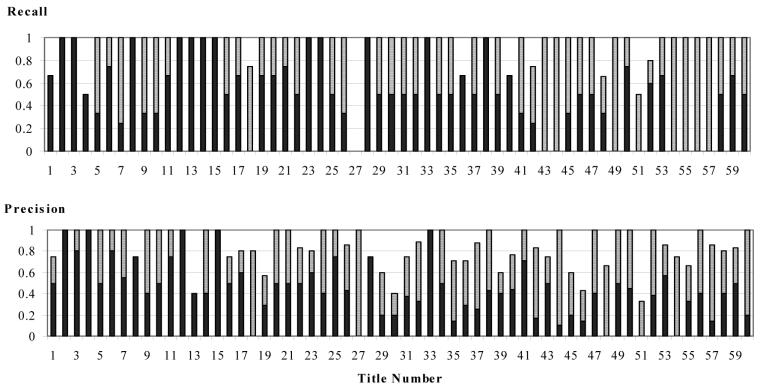
To determine how well MetaMap was able to identify all appropriate biomedical concepts, we calculated two versions of recall. For each title, the exact-match recall was calculated as the number of terms that both were identified by MetaMap and exactly matched the reference standard, divided by the total number of reference terms. The partial-match recall was calculated in the same way, except that the numerator included partial matches as well as exact matches. (Note that recall is equivalent to sensitivity.) See Figure 1 for a graphical representation of both the exact-match and partial-match results for each title. MetaMap’s average results are presented in Table we noticed that MetaMap’s performance seemed worse for the last third of the titles.
Figure 1. MetaMap’s Recall and Precision.
The dark columns indicate how well MetaMap performed when only exact matches were considered. The light columns (on top of the dark ones) indicate how much MetaMap’s performance increases if we consider partial matches too.
MetaMap Precision
To determine how well MetaMap was able to identify only concepts that were in the title text, we calculated several versions of precision. For each title, the exact-match precision was calculated as the number of terms that both were identified by MetaMap and exactly matched the reference standard, divided by the total number of terms that MetaMap identified. The partial-match precision was calculated in the same way, except that the numerator included partial matches as well as exact matches. (Note that precision is equivalent to positive predictive value.) See Figure 1 for a graphical representation of the results for each title. MetaMap’s average results are presented in Figure 2.
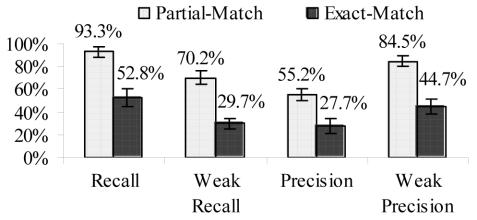
Figure 2. MetaMap’s Average Precision and Recall –
The results of MetaMap’s performance, averaged over all titles. The 95% confidence intervals are represented as the error bars on each column.
For precision, we decided that a weaker version of the calculation was necessary to account for the variation in subject responses. It did not seem fair to penalize MetaMap if a small number of subjects also identified the same concept. Thus, we also calculated what we call weak precision, where a MetaMap identified concept was considered a match if at least one subject also identified the concept, rather than requiring a match with the reference standard of at least half the subjects. With this weaker definition, MetaMap’s average exact-match precision increased.
Analysis of MetaMap’s Failures
The ways in which MetaMap failed could also provide valuable insight for MetaMap developers in determining where improvements are needed, but also for MetaMap users in deciding whether or where to use MetaMap.
For the 151 concepts in the reference standard, MetaMap matched 81 concepts exactly, 60 concepts partially, and ten not at all. Most failures were caused by missing concepts in the UMLS. However, seven of the 60 partially matched concepts and four of the ten unmatched concepts were in the UMLS. For those eleven unfound UMLS concepts, we noticed four kinds of errors: (1) four cases where MetaMap split a noun phrase incorrectly, (2) three cases where it retrieved the correct concept as a candidate phrase but failed to rank it high enough, (3) three cases where it split the noun phrase correctly but still failed to identify it as a concept, and (4) one case where MetaMap changed the original noun phrase in such a way that the identified concept was completely different from the original phrase.
Choices in Splitting a Noun Phrase
Our study also provided insight into how people identify biomedical concepts. Long, multi-word phrases can be a challenge for concept identification because people make different choices on whether to split up the phrase and how to split it up. For example, consider the title, “Statin use and leg functioning in patients with and without lower-extremity peripheral arterial disease.” Subjects chose the following ways to split the long phrases (the number of subjects who made that choice is in parentheses):
lower-extremity peripheral arterial disease (1)
lower extremity (2)
peripheral arterial disease (4)
lower-extremity arterial disease (1)
peripheral vascular disease (1)
Choices in Including General Terms
Subjects also made different choices as to whether to include a general term as a biomedical concept. For example, subjects 1, 3, 4, and 6 included the general term treatment as a concept or as part of other concepts, such as migraine treatment, for some of the four titles that contained the word treatment; the other two subjects did not include treatment for any of the four titles. MetaMap makes no distinction between general or specific terms; it identifies any term that it recognizes, and selected treatment as a term in all four titles.
A single subject sometimes was inconsistent in his or her choice about whether to include general terms. For example, subject 6 identified migraine treatment and medical treatment as medical concepts for two occurrences of treatment, but did not identify any treatment-related concept for the other two occurrences.
Further research is needed to determine both whether there are other consistent patterns in how people identify concepts and how we can use this knowledge to develop better concept-identification tools.
Related Work
Many other systems extract biomedical concepts from text, but most systems attempt to extract only certain types of concepts, depending on the desired task or relating to a particular area of medicine. For example, MedLEE has been used to determine diagnostic codes for radiological reports,10 GENIES (a modified version of MedLEE) was used to identify molecular pathways,11 the Linguistic String Project was used to identify quality assurance parameters in discharge summaries for asthma management cases,12 and SPRUS was used to identify coded findings from radiology reports.13
In contrast, MetaMap tries to identify all biomedical concepts from free-form textual input. This more general goal is much more difficult to evaluate effectively because there is such variability in what is identified as a biomedical concept when the concept-identification task is considered independent of the application goal or medical specialization. In designing our study, we based our ideas on the methods and criteria described by Friedman and Hripcsak.14 With the exception of a calculation of inter-rater reliability, our study conformed to their 20 criteria for a well-designed study of natural-language tools. Because our task of general concept identification was much more open-ended than the natural-language tasks that they reported on, such as identifying diagnoses, we could not provide the subjects with an exhaustive list of all possible concepts. Thus, our evaluation resulted in considerably more variability in subjects’ responses. However, the study also afforded us the opportunity to study people’s strategies in concept identification more thoroughly than in previous studies.
Conclusions
Although getting complete and exact consensus on the concept identification task proved impossible, our evaluation clearly indicates that MetaMap does an excellent job at extracting common biomedical concepts from free-form text. Most of the concepts from the reference standard that MetaMap did not identify were terms that were not present in the UMLS. Thus, MetaMap’s recall performance is determined largely by the coverage of biomedical terms in the UMLS, and can only be increased substantially by a corresponding increase in the UMLS vocabulary.
MetaMap’s weakest point is its lack of precision. However, people showed a great deal of variation in the concepts that they identified, and when the weaker version of precision was used, MetaMap’s performance increased.
One limitation of this study is that it examined MetaMap’s performance on only title phrases; we have no data to examine its performance on other types of text. However, because MEDLINE titles contain such a variety of concepts and phrasings, our study provides convincing evidence that MetaMap meets its goals of identifying most biomedical concepts from free-form text without identifying too many extraneous concepts.
This study also furthered our knowledge of how people select biomedical concepts from text. We learned that people do agree on a substantial portion of biomedical concepts, but the task is a highly subjective one. Complete and exact agreement will always be difficult to find, but further studies in this area could help us design even more accurate tools for biomedical concept identification.
Acknowledgments
We thank the physicians and nurses who participated in this study. Thanks also to Lelia Arnheim for the data entry and consistency checking. This work was supported by a grant from the National Science Foundation.
References
- 1.Aronson, A. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program in Proc AMIA Symp 2001. 17–21. [PMC free article] [PubMed]
- 2.Aronson, A.R., MetaMap: Mapping Text to the UMLS Metathesaurus 1996.
- 3.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus. Proc AMIA Symp. 1997;36(1):485–9. [PMC free article] [PubMed] [Google Scholar]
- 4.Pratt, W. and H. Wasserman. QueryCat: Automatic Categorization of MEDLINE Queries in Proc AMIA Symp 2000. Los Angeles, CA. p. 655–659. [PMC free article] [PubMed]
- 5.Wright LW, et al. Hierarchical Concept Indexing of Full-Text Documents in the Unified Medical Language System Information Sources Map. Journal of the American Society for Information Science. 1998;50(6):514–523. [Google Scholar]
- 6.Weeber, M., et al. Text-based discovery in biomedicine: the architecture of the DAD-system in Proc AMIA Symp 2000. 903–7. [PMC free article] [PubMed]
- 7.Pratt, W. and M. Yetisgen-Yildiz. A Knowledge-Based, Text-Mining Approach to Finding Connections in the Biomedical Literature in SIGIR-03: International ACM Conference on Research and Development in Information Retrieval 2003 (submitted). Toronto, Canada.
- 8.Sneiderman, C., T. Rindflesch, and C. Bean. Identification of anatomical terminology in medical text in Proc AMIA Symp 1998. 428–32. [PMC free article] [PubMed]
- 9.Rindflesch, T., L. Hunter, and A. Aronson. Mining molecular binding terminology from biomedical text. in Proc AMIA Symp 1999. 127–31. [PMC free article] [PubMed]
- 10.Hripcsak G, et al. A reliability study for evaluating information extraction from radiology reports. J Am Med Inf Assoc. 1999;6:143–150. doi: 10.1136/jamia.1999.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman, C., et al. GENIES: a natural-langauge processing system for the extraction of molecular pathways from journal articles in Bioinformatics suppl 2001. 74–82. [DOI] [PubMed]
- 12.Sager N, et al. Natural Language Processing and the Representation of Clinical Data. J Am Med Inf Assoc. 1994;1(2):142–60. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haug P, Ranum D, Frederick P. Computerized Extraction of Coded Findings from Free-text Radiologic Report. Radiology. 1990;174:543–8. doi: 10.1148/radiology.174.2.2404321. [DOI] [PubMed] [Google Scholar]
- 14.Friedman C, Hripcsak G. Evaluating natural language processors in the clinical domain. Methods Inf Med. 1998;37(4–5):334–44. [PubMed] [Google Scholar]