Abstract
In order to meet the need for an expressive ontology in neuroinformatics, we have integrated the extensive terminologies of NeuroNames and Terminologia Anatomica into the Foundational Model of Anatomy (FMA). We have enhanced the FMA to accommodate information unique to neuronal structures, such as axonal input/output relationships.
Currently available controlled medical terminologies have neither the comprehensiveness nor the specificity needed by investigators in basic or clinical neuroscience, or in neuroinformatics. NeuroNames1 (NN) has been incorporated in UMLS as one of its source vocabularies. The NN includes more than 6500 neuroanatomical terms, about 4000 of which are synonyms. However, the NN is term rather than concept oriented, its hierarchy is constructed predominantly on the basis of –part of-, rather than –is a-, relationships between terms, and it excludes the spinal cord and peripheral nervous system. Terminologia Anatomica2 (TA) includes an extensive term list for macroscopic neuroanatomy, but it is neither comprehensive nor consistent in its semantic structure. Thus there exists a need to establish a new resource that comprehensively models the structure of knowledge in the neuroscience domain. To fill this need, we are developing the neuroanatomical component of the Foundational Model of Anatomy4 (FMA), consistent with its declared principles.
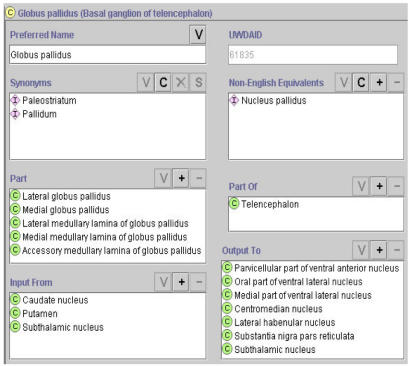
In this poster and demo we show that we have rearranged approximately 5000 NN terms to build a preliminary ontology, and have represented them in the frame-based Protégé knowledge acquisition system (Figure 1), based on explicit rather than implicit definitions. We have extended this neuroanatomical ontology to include structures of the spinal cord and the peripheral nerves. Additionally, we have incorporated approximately 4000 terms from TA into this ontology, primarily as synonyms to preferred names of existing concepts. We have integrated this neuroanatomy ontology with the class subsumption inheritance hierarchy of the FMA. We are enhancing the FMA high level frames to accommodate additional data types such as axonal input/output relationships, neuron types, neurotransmitter and receptor types, etc. The next tasks will be to formalize the explicitly stated defining attributes in Protégé frames and continue to model relationships specific for neuroanatomical concepts.
Figure 1.
Part of the frame of Globus pallidus.
Acknowledgments
Supported by Human Brain Project grant DC02310 and NIH grant LM06822.
References
- 1.Martin RF, Bowden DM, 2000. Primate Brain Maps. Elsevier: Oxford.
- 2.Federative Committee on Anatomical Terminology, 1998. Terminologia Anatomica.: Stuttgart: Thieme.
- 3.Martin RF, Mejino JLV, Bowden DM, Brinkley JF, Rosse C. Foundational Model of Neuroanatomy: Implications for the Human Brain Project. Proc AMIA Symp. 2001:438–442. [PMC free article] [PubMed] [Google Scholar]
- 4.Rosse C, Shapiro LG, Brinkley JF. The Digital Anatomist Foundational Model: Principles for Defining and Structuring its Concept Domain. J Am Med Inform Assoc. 1998:820–824. [PMC free article] [PubMed] [Google Scholar]