Abstract
Aip3p/Bud6p is a regulator of cell and cytoskeletal polarity in Saccharomyces cerevisiae that was previously identified as an actin-interacting protein. Actin-interacting protein 3 (Aip3p) localizes at the cell cortex where cytoskeleton assembly must be achieved to execute polarized cell growth, and deletion of AIP3 causes gross defects in cell and cytoskeletal polarity. We have discovered that Aip3p localization is mediated by the secretory pathway. Mutations in early- or late-acting components of the secretory apparatus lead to Aip3p mislocalization. Biochemical data show that a pool of Aip3p is associated with post-Golgi secretory vesicles. An investigation of the sequences within Aip3p necessary for Aip3p localization has identified a sequence within the N terminus of Aip3p that is sufficient for directing Aip3p localization. Replacement of the N terminus of Aip3p with a homologous region from a Schizosaccharomyces pombe protein allows for normal Aip3p localization, indicating that the secretory pathway–mediated Aip3p localization pathway is conserved. Delivery of Aip3p also requires the type V myosin motor Myo2p and its regulatory light-chain calmodulin. These data suggest that one function of calmodulin is to activate Myo2p's activity in the secretory pathway; this function is likely the polarized movement of late secretory vesicles and associated Aip3p on actin cables.
INTRODUCTION
Cell polarity is a culmination of a set of process by which cells create specialized domains at their cortex. The ability of cells to initiate, maintain, or modify their polarity is critical to the execution of a number of important cellular functions, some examples of which include cell motility, neurite outgrowth, phagocytosis, epithelial cell differentiation, and asymmetric cell division. The regulation of cell polarity is achieved through the action of a number of regulatory proteins that lead to the assembly of a polarized and specialized cortical actin cytoskeleton. These polarized actin networks are then responsible for mediating the sorting and delivery of factors required to execute and maintain cell polarity; frequently this is achieved by actin-mediated polarization of the secretory pathway.
The simple, single-cell eukaryote Saccharomyces cerevisiae has proven to be an excellent model for the study of the regulation of cell and cytoskeletal polarity (for review, see Pringle et al., 1995; Drubin and Nelson, 1996). S. cerevisiae displays three related cell polarity pathways: one is an asymmetric cell division called budding, the second is the formation of a mating projection, and the third is the formation of pseudohyphae. In all three cases, extension of the cell surface is preceded by the polarized organization of two actin filament–containing structures: actin cortical patches and actin cables. The actin cortical patches are invaginations of plasma membrane encircled by actin filaments and actin-associated proteins (Mulholland et al., 1994), and the actin cables are bundles of actin filaments that frequently terminate at or attach to cortical patches. The organization of these actin structures during the budding cell cycle and filamentous growth has been examined by three-dimensional microscopy (Amberg, 1998; Cali et al., 1998). During the budding cycle the cortical patches at the beginning of G1 organize into a ring at the presumptive bud site, and the actin cables orient along what will be the long axis of the soon to be budding cell. The bud then emerges through the ring of cortical patches, and soon thereafter cortical patches can be observed in the cortex of the small bud. The cortical patches remain concentrated in the bud until late in the cell cycle, at which time they reorganize into two rings in the neck immediately before, during, and for a short period after septation. These changes in cytoskeletal polarity are preceded by the cell-cycle–controlled localization of several regulatory proteins, including the small GTPases Cdc42p, Rho1p, Rho2p, Rho3p, Rho4p, the formin Bni1p, and Aip3p/Bud6p. For a more complete description of regulators of yeast cell polarity see Pringle et al. (1995). The coordinate localization of these proteins and actin to sites of cell growth then leads to polarization of the secretory pathway, allowing for the delivery of large amounts of biosynthetic material for new wall synthesis and plasma membrane insertion. It is commonly believed and well supported that secretory vesicles are moved by motor proteins on the actin cables to the sites of cell growth (Pruyne et al., 1998), but the role of cortical patches in this process remains unclear.
Several observations have suggested that the relationship between establishing and maintaining a polarized cytoskeleton and a polarized secretory pathway is complex. For example, mutations in the gene encoding profilin, a regulator of actin filament assembly, have been found to display synthetic lethality with mutations in late-acting secretory pathway genes (Haarer et al., 1996). In addition, it has been observed that cells bearing mutations in secretory pathway genes have severe defects in actin cytoskeleton polarity (Lillie and Brown, 1994; Haarer et al., 1996; Mulholland et al., 1997), suggesting that an operational secretory pathway is required to maintain or perhaps even to help establish a polarized actin cytoskeleton. Furthermore, Rho1p, a conserved regulator of cell and cytoskeletal polarity, has been found to be associated with post-Golgi secretory vesicles (McCaffrey et al., 1991).
AIP3 has been identified in two-hybrid screens as encoding a protein that interacts with actin (Amberg et al., 1997) and with the formin Bni1p (Evangelista et al., 1997) and as a gene required for proper diploid bud site selection (Zahner et al., 1996). Deletion of AIP3 causes severe defects in cell polarity, including depolarization of actin cortical patches, disorganization or loss of actin cables, depolarization of the secretory pathway, and accumulation of secretory membrane intermediates, depolarization of cell growth, as well as defects in septation and nuclear positioning (Amberg et al., 1997). Actin-interacting protein 3 (Aip3p) localization is regulated during the cell cycle. First it localizes in a cap at the bud site and remains at the tip of the bud until the bud reaches approximately one-third the size of the mother cell body. Later in the cell cycle, Aip3p forms a single ring on the daughter side of the neck. Soon thereafter, a ring of Aip3p forms on the mother side of the neck, and then both rings persist through septation and for a short period after cell separation (Amberg et al., 1997). This localization pattern is strikingly similar to that for Bni1p, and both proteins also colocalize to the tip of mating projections (Evangelista et al., 1997). Temporally, Aip3p localization to the bud site and the neck precedes polarization of actin cortical patches and actin cables to these same regions of the cortex, and it has been reported that Aip3p localization cannot be completely disrupted by latrunculin A, a drug that disassembles yeast actin filaments in vivo (Ayscough et al., 1997). These results indicate that at least some of a cell's pool of Aip3p can be localized by an actin-independent mechanism.
An important problem in cell biology is how the cell cycle apparatus communicates temporal–spatial information to the actin cytoskeleton. We have investigated the mechanism of localization of Aip3p to learn more about how this process occurs. We have discovered that Aip3p delivery to the cell surface is mediated by the secretory pathway. Mutations in genes that disrupt the secretory pathway (either at early or late stages) profoundly affect Aip3p localization. Furthermore, cell fractionation, velocity gradient analysis, and density gradient analyses have shown that a pool of Aip3p is associated with post-Golgi secretory vesicles. The sequences within Aip3p sufficient for secretory pathway–mediated delivery of Aip3p are located within the N terminus of Aip3p and are conserved in a Schizosaccharomyces pombe protein of unknown function. Evidence is presented showing that distinct and conserved interactions are made within the N terminus of Aip3p, arguing for conservation of the mechanism of Aip3p localization. These data may provide a molecular explanation for the complex interrelationship between polarization of the actin cytoskeleton and polarization of the secretory pathway.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods
Yeast strains are listed in Table 1. FY23 and FY86 were provided by Fred Winston (Harvard University). Standard methods were used for growth, sporulation, and tetrad dissection of yeast. Yeast transformations were performed by electroporation or by LiOAc.
Table 1.
Saccharomyces cerevisiae strains
| Name | Genotype | Source |
|---|---|---|
| FY23 | a ura3-52 leu2Δ1 trp1Δ63 | Fred Winston |
| FY86 | α ura3-52 leu2Δ1 his3Δ200 | Fred Winston |
| FY23×86 | a/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/TRP1 HIS3/his3Δ200 | Unpublished |
| DBY4934 | α cdc42-1 ura3 his4 leu2 gal2 RDN1∷LEU2 | David Botstein |
| DBY5889 | a sec6-4 ura3-52 | David Botstein |
| DBY5891 | a sec15-1 ura3-52 | David Botstein |
| DBY5856 | a sec17-1 ura3-52 his4-619 | David Botstein |
| DBY5876 | α sec2-41 ura3-52 leu2-3, 112 | David Botstein |
| DBY5894 | a sec4-8 ura3-52 | David Botstein |
| DBY5877 | a sec3-2 ura3-52 leu2-3, 112 | David Botstein |
| DBY2061 | α ura3-52 leu2-3, 112 | David Botstein |
| DAY155 | α sec14-3 ura3-52 leu2Δ1 | David Amberg |
| LWY2753 | a ura3-52 myo2-66 leu2 | Lois Weissman |
| LWY2599 | a myo2-2 his3Δ200 lys2-801 leu2 trp1Δ901 suc2Δ9 ade8∷HIS3 | Lois Weissman |
| DBY5711 | a ura3 ade2 lys2 his3 trp1 leu2 cmd1-Δ1∷TRP1 ade3∷HIS3∷cmd1-231 | David Botstein |
| DBY5719 | a ura3 ade2 lys2 his3 trp1 leu2 cmd1-Δ1∷TRP1 ade3∷HIS3∷cmd1-239 | David Botstein |
| DBY5706 | a ura3 ade2 lys2 his3 trp1 leu2 cmd1-Δ1∷TRP1 ade3∷HIS3∷cmd1-226 | David Botstein |
| DBY5708 | a ura3 ade2 lys2 his3 trp1 leu2 cmd1-Δ1∷TRP1 ade3∷HIS3∷cmd1-228 | David Botstein |
| DBY5713 | a ura3 ade2 lys2 his3 trp1 leu2 cmd1-Δ1∷TRP1 ade3∷HIS3∷cmd1-233 | David Botstein |
| DBY5701 | a ura3 ade2 lys2 his3 trp1 leu2 ade3∷HIS3∷cmd1-221 | David Botstein |
| DBY5705 | a ura3 ade2 lys2 his3 trp1 leu2 ade3∷HIS3∷cmd1-225 | David Botstein |
| Y822 | a ura3-1 leu2-3, 112 ade2-1 can1-100 trp1 his3∷FUS1-HIS3 Mfa2Δ∷FUS1-lacZ bni1Δ∷∶LEU2 | Charlie Boone |
| Y609 | a ura3-52 lys2-801 ade2-101 trp1Δ spa2-Δ2∷TRP1 his3Δ200 | Mike Snyder |
| Y147 | a ura3 his3 lleu2 cdc24-4 | Doug Johnson |
| TZY33 | α vrp1-1 sup11 ade2-1 mod5-1 ura3-1 lys2-1 leu2-3, 112 his4-519 | Anita Hopper |
| T65.1D | α vrp1Δ∷LEU2 ura3-52 ade1 leu2-3, 112 ile MEL | Anita Hopper |
| JCY1364 | a/α ura3-52/ura3-52 leu2-112/leu2-112 his3Δ200/HIS3 lys2/lys2 trp1-1/trp1-1 ade2/ade2 las17∷LEU2/LAS17 cla4∷TRP1/CLA4 | George Sprague |
| HJY1 | las 17Δ∷LEU2 ura3-52 leu2-3, 112, lys2, trp1-1 ade2 his3 | This study |
| HJY3 | a sec6-4 ura3-52 pep4Δ∷Kanr | This study |
The PEP4 deletion allele was constructed by double-fusion PCR (Amberg et al., 1995). The sequences of the primers was as follows: HJo-PEP4-1 (5′-ATCAACTGGTTAAGGGAACAGA-3′), HJo-PEP4-2 (5′-TTAATTAACCCGGGGATCCGACTTTTGCAGCAACTTGGTT-3′), HJo-PEP4-3 (5′-GTTTAAACGAGCTCGAATTCTTGGCCAAAGC-AATTTGAG-3′), and HJo-PEP4-4 (5′-GGCCTCCTTAAGCTTGGG-3′). The kanamycin resistance marker was amplified using primers F1 and R1 and plasmid pFA6a-kanMX6 as the template (Longtine et al., 1998). The resulting pep4Δ::Kanr cassette was used to replace PEP4 in strain DBY5889. Loss of Pep4p activity was confirmed using an N-acetyl-dl-phenylalanine β-napthyl ester overlay test (Jones, 1977).
Strain HJY1 was constructed by sporulation and tetrad dissection of strain JCY1364.
Plasmid Constructions and DNA Manipulations
Plasmid pDA254 was constructed by digesting the green fluorescent protein (GFP)-Aip3p–encoding plasmid pRB2190 (Amberg et al., 1997) with SacI (New England BioLabs, Beverly, MA), resecting the overhang with mung bean nuclease and religating the plasmid. Plasmid pDA257 was constructed by PCR amplifying part of AIP3 with primers DAo-GFP/AIP3-1 (5′-GAAGATCTATGAAGATGGCCGTGGAT-3′) and DAo-AIP3-13 (5′-GCAGATCTTTACCGCTAATTGCAGAACCAAA-3′), digesting the PCR product with BglII, and ligating it into plasmid pRB2138 (Doyle and Botstein, 1996), which had been digested with BamHI and dephosphorylated with calf intestinal phosphatase. Plasmid pDA259 was constructed by digesting plasmid pDA257 with BamHI, resecting the overhang with mung bean nuclease, and religating the plasmid. Plasmid pHJ9 was constructed by PCR amplification of AIP3 from genomic DNA using primer Hjo-GFP/AIP3-1.1 (5′-GCGGCAGATCTTTGAGGGAACCACCATCTG-3′) and primer HJo-AIP3-2.1 (5′-GGCGCAAGCTTTTAGCCGCTAATTGCAGAACCAAA-3′). The PCR product was digested with BglII and HindIII and ligated into plasmid pRB2138, which had been digested with BamHI and HindIII. Plasmid pHJ6 was constructed by PCR amplification of AIP3 from genomic DNA using primer DAo-GFP/AIP3-1 and primer DAo-AIP3-15 (5′-GCAGATCTTTATGCGTTTGGAGCATTGTTAG-3′). The PCR product was digested with BglII and cloned into BamHI-digested plasmid pRB2138. Plasmid pHJ34 was constructed by PCR amplification of AIP3 from genomic DNA using primer DAo-GFP/AIP3–1 and primer HJo-AIP3-6 (5′-GCGGCAAGCTTTTAACGGGTCCCTTCACTACCC-3′). The PCR product was digested with BglII and HindIII and ligated into BamHI- and HindIII-digested plasmid pRB2138. The GST-Aip3 fusion encoding plasmid pHJ29 was constructed by PCR amplification of AIP3 from genomic DNA using primer Dao-AIP3-20 (5′-GCGGCTAGCGATGAAGATGGCCGTGG-3′) and primer HJo-AIP3-4 (5′-GCGCTCGAGTTAACGGGTCCCTTCACTACCC-3′). The PCR product was digested with NheI–XhoI and ligated into XbaI–SalI-digested plasmid pEG(KT) (Mitchell et al., 1993).
Plasmid pHJ36 was constructed by PCR amplification of AIP3 from genomic DNA using primer HJo-AIP3-7 (5′-GCAGATCTGAGCTCATGCAGCAAGAGCATCGGATA-3′) and primer HJo-AIP3-2.1. The PCR product was digested with BglII and HindIII and cloned into BamHI- and HindIII-digested plasmid pTD125. Plasmid pHJ37 was constructed by PCR amplification of the AIP3 homologue from S. pombe genomic DNA using primer HJo-POM-4 (5′-GCGGCGAGCTCATGTTTAATAACGGCGATAA-3′) and primer HJo-POM-5 (5′-GCGGCGAGCTCGGATTTAACCAGAGATTGCTT-3′). The PCR product was digested with SacI and cloned into SacI-digested plasmid pHJ36.
Cell Fractionation
For the crude fractionations a 250-ml culture was grown to 1–2 × 107 cells/ml, and the cells were pelleted, washed with 250 ml H2O, and resuspended in 25 ml of 0.1 M Tris, pH 8, plus a protease inhibitor mixture (5 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mM PMSF, 2 mM benzamidine, 1 mM EDTA, 2 μg/ml chymostatin). The cells were then passed twice through a French Press (American Instrument, Silver Spring, MD) set at 1200 pounds per square inch on the gauge. The cell extract (the fraction designated T for total) was centrifuged at 12,000 rpm for 20 min at 4°C in a Beckman JA20 rotor/Beckman J2–21 centrifuge (Beckman Intruments, Palo Alto, CA) to create fractions S1 (supernatant 1) and P1 (pellet 1). Five milliliters of fraction S1 were then centrifuged in a Beckman Ti75 rotor/Beckman L8–70 ultracentrifuge at 35,000 rpm for 1 h at 4°C to create fraction S2 (supernatant 2) and P2 (pellet 2). Samples from each fraction were loaded on SDS-PAGE gels, transferred to nitrocellulose (Bio-Rad, Hercules, CA), and blotted with rabbit anti-GFP antibodies (gift of Pam Silver, Harvard University) diluted to 1:1000, rabbit anti-Snc1p antibodies (gift of Pat Brennwald, Cornell University Medical College) diluted to 1:1000, or rabbit anti-Pma1p antibodies (gift of Amy Chang, Albert Einstein College of Medicine) diluted to 1:500. The Western blots were then developed by incubation in protein A-peroxidase conjugate (Pierce, Rockford, IL) diluted 1:10,000 and Pierce SuperSignal chemiluminescent substrate.
In the sorbitol velocity gradient analysis, strain HJY3 (sec6-4 pep4Δ) was grown overnight at 25°C in YPD to midlog phase and shifted to 37°C for 2 h. Cells (300 U at OD599) were harvested and washed with 10 mM Tris, pH 7.5, 10 mM sodium azide. Cells were converted to spheroplasts (Walworth and Novick, 1987) and lysed in 3 ml ice-cold lysis buffer (10 mM triethanolamine acetate, pH 7.2, 1 mM EDTA, 0.8 M sorbitol, protease inhibitors) in a 7-ml Dounce homogenizer (Wheaton, Millville, NJ). Cell extracts were clarified by centrifugation at 450 × g for 3 min, and 0.6 ml of the supernatant was loaded on the top of a sorbitol gradient made with 1.22-ml steps of 40, 37.5, 35, 32.5, 30, 27.5, 25, 22.5, and 20% sorbitol (wt/vol) in 10 mM triethanolamine acetate, pH 7.2, 1 mM EDTA. The gradient was centrifuged in an SW40 rotor (Beckman Instruments) at 24,000 rpm for 1 h at 4°C. Sixteen fractions (720 μl/fraction) were collected from the top of the gradient, and the pellet was dissolved in the last fraction. Samples from each fraction were loaded on SDS-PAGE gels and transferred to nitrocellulose. Aip3p, Snc1p, and Pma1p were detected as described above for the crude fractionations.
For the sucrose density floating gradient, the cell extract was prepared as for the velocity gradient. After clarification of the cell extract, it was adjusted to 60% sucrose (wt/vol), and 1 ml of sample was underlayed in a sucrose gradient made with 1-ml steps of 50, 47.5, 45, 40, 35, 30, 25, and 20% sucrose (wt/vol) in 20 mM triethanolamine acetate, pH 7.2, 1 mM EDTA, protease inhibitors. The gradients were centrifuged in an SW40 rotor at 27,000 rpm for 20 h at 4°C. Thirteen fractions (690 μl/fraction) were collected from the top of the gradient, and the pellet was dissolved in the last fraction. Samples from each fractions were loaded on SDS-PAGE gels and transferred to nitrocellulose. Aip3p, Snc1p, and Pma1p were detected as described above for the crude fractionations. Quantitation of the fractionation data was performed by densitometry using ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Microscopy
Microscopic analysis was performed on a Zeiss Axioskop (Carl Zeiss, Oberkochen, Germany) with a Plan-APOCHROMAT 100× objective. Cells were visualized either by differential interference contrast (DIC) or epifluorescence with a standard fluorescein isothiocyanate filter set. Images were captured with a SPOT2 camera (Diagnostic Instruments, Sterling Heights, MI) and downloaded directly into Adobe Photoshop (Adobe Systems, San Jose, CA).
Two scoring strategies were used to quantify GFP-Aip3p localization defects. In one scheme, the percentage of cells with some GFP-Aip3p in the correct location (bud site, bud cortex, or neck cortex) was determined. In the other scheme, the percentage of cells with some Aip3p in the wrong location was determined. In both schemes, 200–350 cells were scored for each strain.
RESULTS
The Secretory Pathway Is Required for Aip3p Localization
Our interest in identifying proteins at the cell surface involved in Aip3p localization led us to focus our analysis on proteins that display cell cycle–regulated localization patterns similar to that of Aip3p. Likely candidates were the septins; however, we had previously shown that septins were not involved in Aip3p localization (Amberg et al., 1997), and so we began to focus on other proteins whose localization also has been shown to be septin independent (see Figures 1–3). Strains carrying mutations in these genes were obtained and transformed with the plasmid encoding a GFP-Aip3p fusion protein pDAb204. We reasoned that this was a good reporter for Aip3p localization, as we had previously shown that the fusion protein can complement a deletion allele and that its localization is identical to that of the endogenous protein (Amberg et al., 1997).
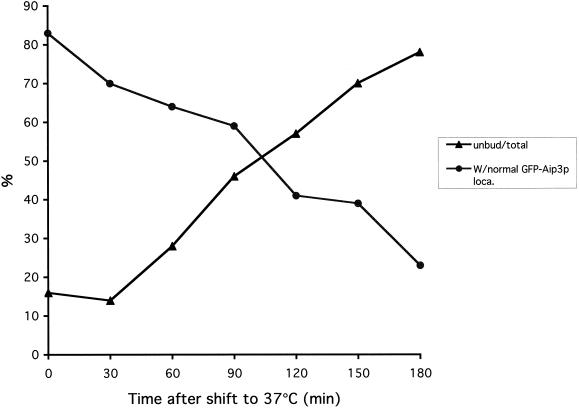
Figure 1.
Cdc42p is required at start for Aip3p localization. The cdc42-1 strain DBY4934 expressing GFP-Aip3p from plasmid pDAb204 was shifted to 37°C for 3 h. The percentage of cells in the population able to correctly polarize at least some of their pool of GFP-Aip3p was scored, and the percentage of unbudded cells in the population was scored and both were plotted versus time at 37°C.
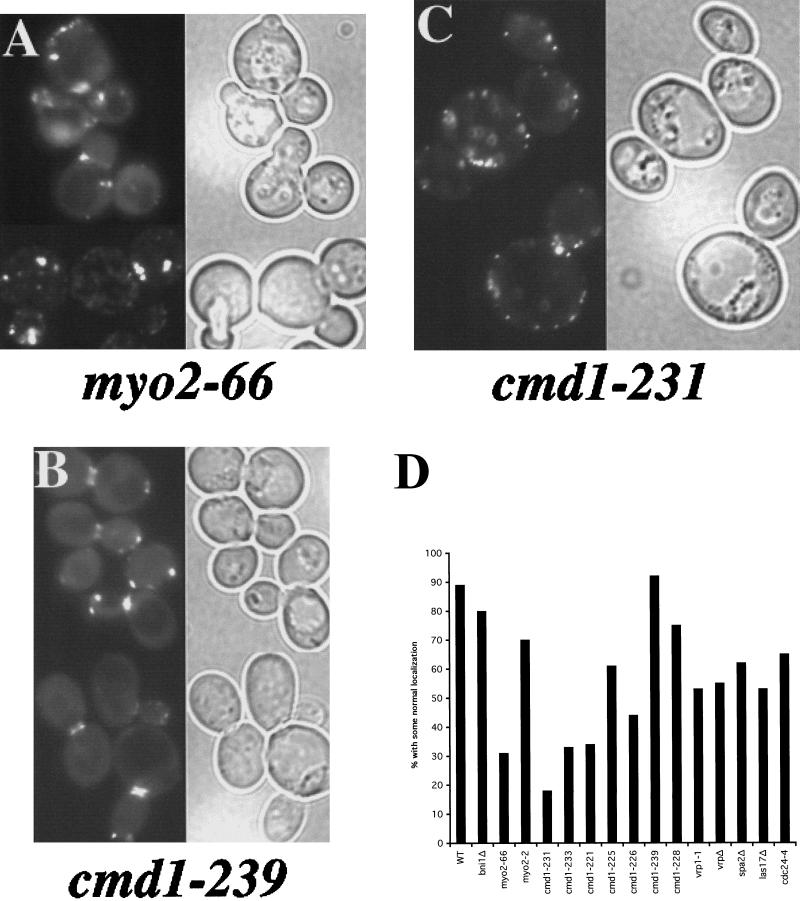
Figure 3.
The role of cell polarity proteins in Aip3p localization. The myo2-66 strain LWY2753 (A), the cmd1-239 strain DBY5719 (B), and the cmd1-231 strain DBY5711 carrying the GFP-Aip3p–expressing Cen vector pDAb204, were shifted to 37°C for 90 min and examined by fluorescence microscopy. GFP fluorescence is shown on the left of each panel, and a DIC view of the same cells is shown on the right of each panel. GFP-Aip3p localization was quantified in several cell polarity mutants after a shift to 37°C for 90 min (D). Individual cells were scored for the correct localization of some of their pool of GFP-Aip3p, and the values reported are derived from counts of 200–350 cells.
The small GTPase Cdc42p is thought to play a central role in establishing cell polarity, and likewise, we found it was critical for GFP-Aip3p localization. cdc42-1 cells expressing GFP-Aip3p were shifted to 37°C and examined over time for GFP-Aip3p localization defects. Interestingly, Aip3p localization was normal in most cells after the temperature shift until they had completed their last cell cycle, at which time the cells arrested in an unbudded state with GFP-Aip3p aberrantly localized in nonpolarized spots and large aggregates. As shown in Figure 1, we quantified this effect by scoring the percentage of cells able to correctly localize at least some of their pool of GFP-Aip3p, compared with the percentage of unbudded cells in the population. As can clearly be seen, there is an inverse relationship between normal Aip3p localization and the presence of unbudded cells. These observations suggest that Cdc42p behaves like a trigger; once pulled at the start of the cell cycle it is not required again until the start of the next cell cycle.
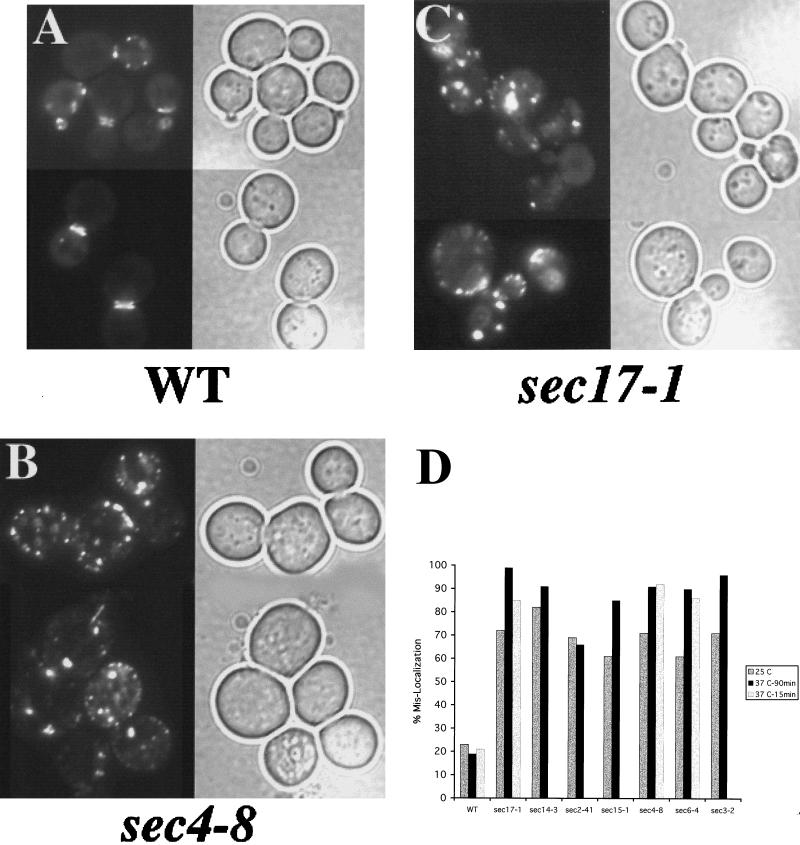
Next we examined components of the exocyst, a complex of proteins involved in the polarized delivery and fusion of late secretory vesicles to the plasma membrane (Bowser et al., 1992; TerBush and Novick, 1995). We discovered that mutations in several different components of the exocyst (for example, Sec6p and Sec15p) led to severe defects in GFP-Aip3p localization (see Figure 2D). We sought to determine whether these defects reflected a general requirement of the secretory pathway for Aip3p localization by examining strains defective in other, nonexocyst components of the secretory pathway. In Figure 2B are shown sec4-8 cells expressing GFP-Aip3p at 37°C. As can be seen, the GFP-Aip3p is largely accumulating in a nonpolarized distribution at the cell cortex and in aggregates that appear to be located in the interior of the cell. This is in contrast to the wild-type congenic control strain shown in Figure 2A. Sec4p is a small GTPase that is associated with late secretory vesicles and is required at late stages of the secretory pathway (Novick et al., 1981; Goud et al., 1988; Kabcenell et al., 1990). The involvement of the secretory pathway in Aip3p localization was not restricted to late-acting components, as we also found similarly severe (although qualitatively different) defects in mutants defective at early (endoplasmic reticulum to Golgi) stages of the secretory pathway. Figure 2C shows a representative sample of cells carrying a sec17-1 allele, and as can be seen, the GFP-Aip3p accumulated in a few large aggregates that appear to reside in the interior of the cell. We then quantified the defects in Aip3p localization by scoring the percentage of cells that display aberrant GFP-Aip3p localization patterns (Figure 2D). This is a slightly less stringent criterion than that used to quantify Aip3p localization defects in the cdc42-1 strain; it reflects the importance of a pathway for proper Aip3p localization as opposed to the necessity of a pathway for the ability to localize any Aip3p. All of the secretory pathway mutants cause misaccumulation of Aip3p in comparison with the wild-type congenic control. Interestingly, all of the sec mutants examined were quite defective for GFP-Aip3p localization even at the permissive temperature of 25°C. This may indicate that the secretory pathway is at least partially compromised at permissive temperature in these mutants and/or that GFP-Aip3p localization may be a very sensitive reporter of the efficiency of the secretory pathway.
Figure 2.
The secretory pathway is required for proper Aip3p localization. The wild-type strain DBY2061 (A), sec4-8 strain DBY5894 (B), and sec17-1 strain DBY5856 (C) carrying the GFP-Aip3p–expressing Cen vector pDAb204, were shifted to 37°C for 90 min and examined by fluorescence microscopy. GFP fluorescence is shown on the left of each panel, and a DIC image of the same cells is shown on the right of each panel. GFP-Aip3p localization was quantified in several secretory pathway mutants after a shift to 37°C for 90 min (D). Individual cells were scored for the mislocalization of some of their pool of GFP-Aip3p, and the values reported are derived from counts of 200–350 cells.
We next examined the potential role in Aip3p localization of several proteins that colocalize with Aip3p (see Figure 3D). We thought that these proteins were possible candidates for being Aip3p receptors at the cortex. We transformed mutant strains with the GFP-Aip3p–encoding plasmid and scored defects in Aip3p localization (Figure 3D). In this case we used the more stringent scoring strategy that was used to quantify the GFP-Aip3p localization defect in the cdc42-1 strain. We reasoned that mutations in a putative Aip3p receptor should be unable to place any Aip3p at the correct location (0% by our scoring strategy). None of the mutants examined met this criterion. However, two additional proteins that are extremely important for proper Aip3p localization were uncovered by this approach: the type V myosin Myo2p and its regulatory light chain calmodulin (Cmd1p). For both genes we observed allele specificity with respect to the severity of Aip3p localization defects. myo2-66 encodes a protein with a mutation in the actin-interacting motor domain (Lillie and Brown, 1994), and the strain carrying this allele had severe defects in Aip3p localization (see Figure 3, A and D), whereas the tail domain mutant myo2-2, which causes defects in vacuolar inheritance (Catlett and Weisman, 1998), displayed rather mild defects in Aip3p localization (Figure 3D). For calmodulin, the cmd1-231 strain showed <20% of its cells with some normal Aip3p localization (see Figure 3, C and D), whereas the cmd1-239 (see Figure 3, B and D) strain was indistinguishable from the wild-type control strain.
Intragenic complementation between calmodulin mutants has identified four separable functions of yeast calmodulin (Ohya and Botstein, 1994a,b). In our analysis we looked at mutants from each complementation group (cmd1-231, cmd1-226, cmd1-239, and cmd1-228) and found that the function defined by cmd1-231 seemed most important for Aip3p localization whereas the function defined by cmd1-239 was not important at all. This was not surprising, because the cmd1-239 strain is phenotypically defective for a nuclear function of calmodulin (spindle pole body duplication), whereas cmd1-231 strains are defective for bud emergence. In agreement with the intragenic complementation analysis, we also found that strains bearing a mutation in the same intragenic complementation group as cmd1-231 (cmd1-233) also displayed severe defects in Aip3p localization (Figure 3D). Because cmd1-231 encodes for a protein with two phenylalanine to alanine mutations (F89A and F12A), we then asked whether one mutation contributed more than the other to the Aip3p mislocalization phenotype. The cmd1-221 (F12A) strain was most defective, with 34% showing some normal localization compared with 61% for the cmd1-225 (F89A) strain. Furthermore, cmd1-231 and cmd1-233 both share the F12A mutation, providing additional support for the importance of this residue of calmodulin for Aip3p localization.
Finally, it is notable that the Aip3p-interacting protein Bni1p is absolutely dispensable for proper Aip3p localization (Figure 3D), and we also have found that Aip3p and the secretory pathway are similarly unimportant for Bni1p localization (our unpublished observations). Therefore, although Aip3p and Bni1p interact with each other and appear to be involved in the same process, they are delivered to the cell surface by independent mechanisms.
Biochemical Analysis of the Aip3p Localization Pathway
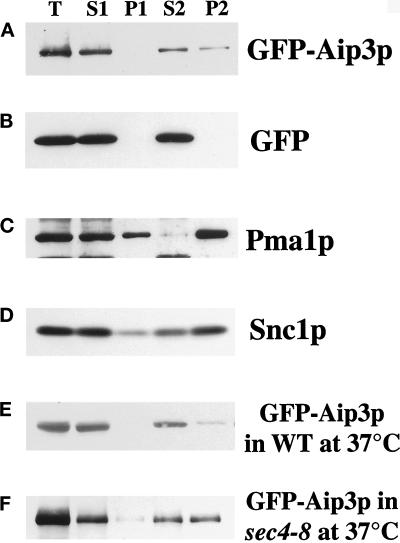
Two possible explanations for the importance of the secretory pathway for Aip3p localization are 1) either a need to frequently replenish a putative plasma membrane receptor for Aip3p or 2) that Aip3p is delivered to the cell surface by the secretory apparatus. To investigate these possibilities, we began to fractionate cells to determine whether Aip3p is associated with secretory membranes. Initially, we undertook crude fractionation of cells expressing the GFP-Aip3p fusion protein. Cell extracts were separated by differential centrifugation, first at 15,000 × g for 20 min to create supernatant 1 (S1) and pellet 1 (P1), and then S1 was further fractionated at 100,000 × g for 1 h to create supernatant 2 (S2) and pellet 2 (P2). Samples from the fractions were then separated by SDS-PAGE, transferred to nitrocellulose, and blotted with anti-GFP antibodies or antibodies for other markers (Figure 4). As can be seen in Figure 4A, an appreciable amount of GFP-Aip3p (∼25%) was consistently detected in the high-speed pellet from wild-type cells. P2 is the microsomal fraction and has been shown to be enriched in late secretory vesicles (Goud et al., 1988). This was confirmed in parallel experiments in which we detected the late vesicle-borne, integral membrane protein Snc1p (Protopopov et al., 1993) (Figure 4D) as being concentrated in P2. The presence of GFP-Aip3p in P2 was not due to the GFP sequence, because cells expressing GFP alone contained no GFP in the P2 fraction (Figure 4B). We were surprised that little if any GFP-Aip3p is detectable in the P1 plasma membrane fraction, even though large amounts of the plasma membrane protein Pma1p were detectable in that fraction (Figure 4C). Perhaps these relatively harsh methods destabilize an association between Aip3p and the plasma membrane.
Figure 4.
GFP-Aip3p is associated with microsomes. Wild-type cells (A–E) or the sec4-8 strain DBY5894 (F) transformed with the GFP-Aip3p–expressing plasmid pDAb204 (A, C, and D) or the GFP-expressing plasmid pTD125 (B) were grown at 30°C (A–D) or 37°C for 90 min (E and F) to 2 × 107 cells/ml. Cells were collected, washed, and lysed in a French Press, and a sample was collected (T). The cell extracts were then centrifuged for 20 min at 15,000 × g, and a sample of the supernatant was collected (S1), the pellet was reconstituted in the original volume, and a sample was taken (P1). The S1 fraction was then centrifuged further at 100,000 × g for 1 h, and a sample of the supernatant was collected (S2). The pellet was reconstituted in the original volume, and a sample was taken (P2). The samples were separated on an SDS-PAGE gel, transferred to nitrocellulose, and blotted with anti-GFP antibodies (A, B, E, and F), anti-Pma1p antibodies (C), or anti-Snc1p antibodies (D).
The fractionation behavior of GFP-Aip3p was similar to that of Sec4p (Goud et al., 1988), a small GTPase that cycles between a cytosolic and peripheral membrane-associated pool (Novick et al., 1993). If Aip3p is also cycling on and off secretory membranes, then we reasoned that in late secretory pathway mutants, Aip3p should accumulate in the microsomal fraction. To examine this, we fractionated both wild-type (Figure 4E) and sec4-8 (Figure 4F) cells expressing GFP-Aip3p after a 90-min shift to 37°C. The temperature shift in wild-type cells, if anything, led to a smaller percentage of the protein in fraction P2. In contrast, in the sec4-8 strain, a greater percentage of the GFP-Aip3p (∼1:1 S2:P2) was detected in fraction P2. Because this strain is known to accumulate late secretory vesicles at nonpermissive temperature (Novick et al., 1980), we believe that this shift of Aip3p into fraction P2 reflects accumulation of late secretory vesicles loaded with GFP-Aip3p.
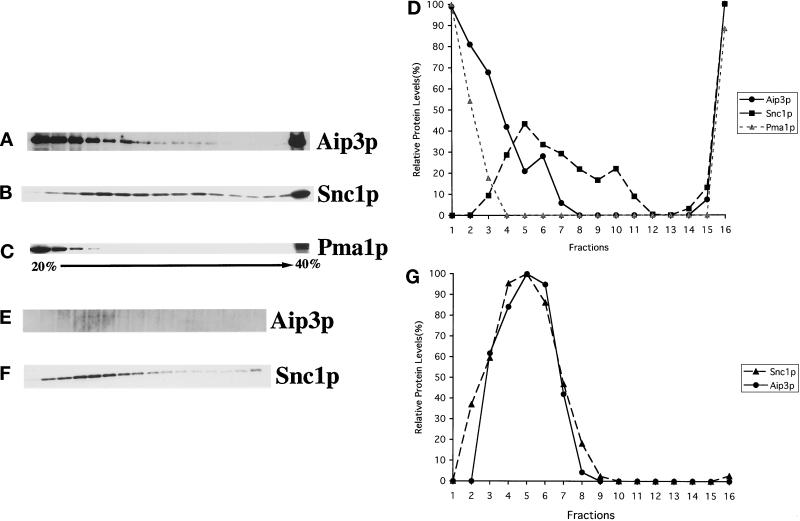
To preclude any possibility that Aip3p is pelleting at 100,000 × g because it is merely in a large multiprotein complex, we examined Aip3p's sedimentation behavior more carefully by gradient analyses. In these studies we examined endogenous Aip3p from a sec6-4 strain shifted to 37°C for 2 h. This was done to enrich the lysates for late secretory vesicles (Walworth and Novick, 1987). In previous experiments we had found that endogenous Aip3p was too unstable in cell extracts for biochemical analysis, a problem partially alleviated by fusion of GFP to Aip3p's N terminus (our unpublished observations). For these experiments we constructed a sec6-4 strain deleted for the gene encoding a major vacuolar protease Pep4p (Ammerer et al., 1986; Woolford et al., 1986). Indeed, deletion of PEP4 solved much of the Aip3p degradation problem. Cell extracts were prepared from spheroplasted cells lysed in a dounce homogenizer. The extracts were clarified by low-speed centrifugation (450 × g) and loaded either onto the top of a sorbitol velocity gradient (Figure 5, A–D) or the bottom of a sucrose density gradient (Figure 6), and fractions were isolated, examined in Western blots, and quantified by densitometry. The sorbitol velocity gradient is optimized for the separation of secretory vesicles from other cell constituents (Brennwald et al., 1994) as confirmed by the sedimentation behavior of the secretory vesicle marker Snc1p (Protopopov et al., 1993) (Figure 5B). As can be seen in Figure 5A, a pool of Aip3p does cosediment with the Snc1p peak consistent with the idea that some of the Aip3p is associated with late secretory vesicles.
Figure 5.
Aip3p fractionates with a late secretory vesicle marker in a sorbitol velocity gradient. The sec6-4 pep4Δ strain HJY3 was grown to 1 × 107 cells/ml and shifted to 37°C for 2 h, and cells were collected. Cells were then spheroplasted and lysed in a Dounce homogenizer. The cell extract was clarified by centrifugation at 450 × g for 3 min, loaded onto the top of a 20–40% sorbitol gradient, and centrifuged at 71,000 × g for 1 h (A–D), or microsomes were isolated and loaded onto the top of a 20–40% sorbitol gradient and centrifuged at 71,000 × g for 1 h (E–G). Sixteen fractions were collected from the top of the gradient, and samples from these fractions were separated on SDS-PAGE gels, transferred to nitrocellulose, and blotted with anti-Aip3p (A and E), anti-Snc1p (B and F), and anti- Pma1p (C) antibodies. Densitometry was used to quantify the relative amounts of protein in each fraction (D and G).
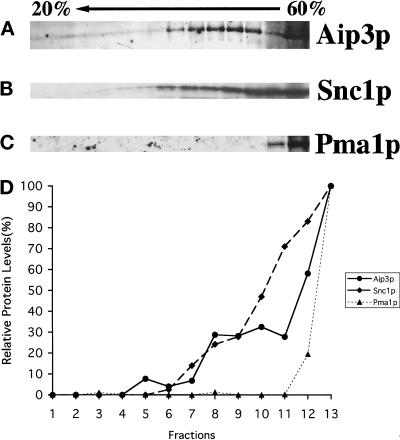
Figure 6.
Aip3p fractionates with a late secretory vesicle marker in a sucrose density gradient. The sec6-4 pep4Δ strain HJY3 was grown to 1 × 107 cells/ml and shifted to 37°C for 2 h. Cells were collected and then spheroplasted and lysed in a Dounce homogenizer. The cell extract was clarified by centrifugation at 450 × g for 3 min, adjusted to 60% sucrose, underlaid in a 20–50% sucrose gradient, and centrifuged at 100,000 × g for 20 h. Thirteen fractions were collected from the top of the gradient, and samples from these fractions were separated on SDS-PAGE gels, transferred to nitrocellulose, and blotted with anti-Aip3p (A), anti-Snc1p (B), and anti- Pma1p (C) antibodies. Densitometry was used to quantify the relative amounts of protein in each fraction (D).
To remove the large peak of nonvesicle-associated Aip3p observed in the top of the velocity gradient (see Figure 5A), we clarified the extracts by centrifugation at 15,000 × g and isolated microsomes in a 100,000 × g pellet. This fraction was then loaded on the top of a sorbitol velocity gradient and analyzed as described above (Figure 5, E–G). As can be seen in Panels E–G, microsomal-associated Aip3p cofractionates with the peak of Snc1p.
The argument could be made that Aip3p is not on vesicles but in a complex of a size and shape that would sediment, similarly to secretory vesicles, through a velocity gradient. To convince ourselves that this was not the case, we underlaid the cell extract (adjusted to 60% sucrose) in a 20–50% sucrose gradient, centrifuged the gradient at 100,000 × g for 20 h to equilibrium, collected fractions, and analyzed the fractions in Western blots (Figure 6). In this gradient, cell constituents are separated by density, and membrane-associated proteins should float upward (Diaz et al., 1997; Roberg et al., 1999). As can be seen, that Aip3p does float upward in this gradient (Figure 6A) to the same extent as Snc1p (Figure 6B) strongly suggests that Aip3p is in fact lipid/vesicle associated.
Identification of the Aip3p Addressing Domain
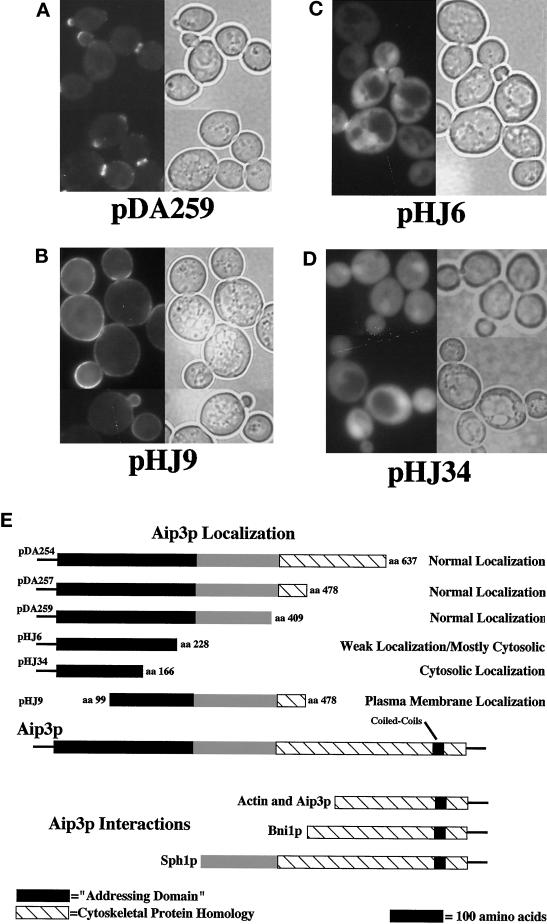
Next we sought to identify a minimal sequence within Aip3p sufficient for allowing secretory pathway–mediated localization of the protein. First we constructed a set of truncation mutants, removing portions of the C terminus of the protein while retaining fusion of GFP to the N terminus of the remaining Aip3p sequence (see Figure 7E). Expression of the N-terminal 409 amino acids of Aip3p was sufficient for a localization pattern that was indistinguishable from the full-length protein (see Figure 7A; cf. Figure 2A). Further truncation from the C terminus (clone pHJ6), into a region conserved with a S. pombe gene (see Figure 8A), had a noticeable effect on Aip3p localization; much of the Aip3p was localized evenly throughout the cytoplasm, and only a small amount appeared to be properly localized (see Figure 7C). Finally, we constructed a GFP fusion to the N-terminal 166 amino acids of Aip3p (pHJ34, see Figure 7E) and found that localization of this fusion protein appeared to be completely cytosolic (Figure 7D).
Figure 7.
Identification of the N-terminal–addressing domain of Aip3p. Wild-type strain FY23×86 was transformed with plasmid pDA259 (A), plasmid pHJ9 (B), plasmid pHJ6 (C), and pHJ34 (D). GFP-aip3p localization was visualized by fluorescence microscopy (A—D, left side), and the same cells were visualized by DIC microscopy (A—D, right side). The domain structure of Aip3p is displayed in E and compared with regions tested for localization as GFP fusions and regions previously reported as displaying two-hybrid interactions (Amberg et al., 1997; Evangelista et al., 1997; Roemer et al., 1998).
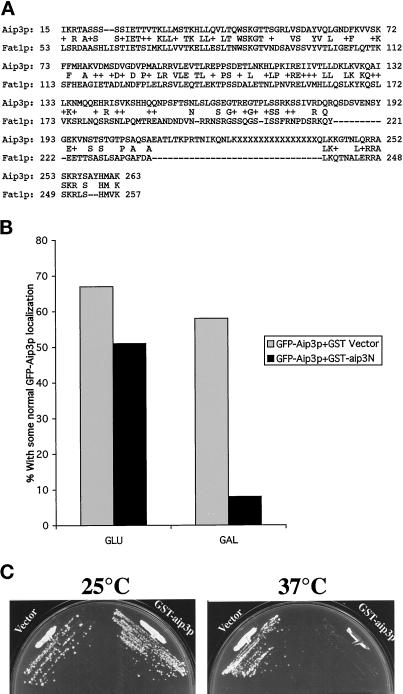
Figure 8.
The addressing domain of Aip3p is conserved. The sequence alignment tool BLAST was used to align the protein sequences of AIP3 and an S. pombe gene of unknown function (FAT1, GenBank accession number Z97208; A). The GFP-Aip3p-encoding plasmid pDA278 and the GST-aip3-encoding plasmid pHJ29 were transformed into strain FY86, and the strain was grown either continuously in glucose-containing medium (GLU) or shifted out of raffinose-containing medium into galactose-containing medium for 4 h (GAL), and the percentage of cells able to correctly localize some of their pool of GFP-Aip3p was scored (B). Strain FY86 was transformed with either the GST-expressing plasmid pEG(KT) (Vector) or the GST-aip3p–expressing plasmid pHJ29 (GST-aip3p), and the transformants were grown on galactose-containing plates at 25 or 37°C (C).
We then constructed a truncation mutant lacking the N terminus of Aip3p (amino acids 99–478, clone pHJ9, see Figure 7E). As can be seen in Figure 7B, the protein expressed from this construct localized over the entire surface of the plasma membrane, sometimes with a slightly higher concentration in the neck or bud. This striking result suggests that the N terminus of Aip3p is critical for Aip3p localization and that loss of this sequence either leads to an inability to efficiently target the protein to specific sites at the plasma membrane or defects in retaining the protein at specific sites on the plasma membrane. In either case, secretory pathway–mediated delivery of this protein to the cell surface does not appear to be impaired. Consistent with this assertion, the protein encoded by pHJ9 was associated with microsomes in a crude fractionation experiment to the same extent as the full-length fusion protein, and expression of the truncated protein in secretory pathway mutants led to aberrant, internal accumulation similar to that observed for the full-length protein (our unpublished observations).
The significance of our truncation analysis is threefold. First, we were able to remove (without affecting localization) the region of Aip3p defined by two-hybrid analysis as being sufficient for the Aip3p–Aip3p, Aip3p–actin, and Aip3p–Bni1p interactions (Amberg et al., 1997; Evangelista et al., 1997), further supporting that these interactions are unimportant for Aip3p localization. Second, in all cases constructs lacking the C terminus of AIP3 were unable to complement the cytoskeletal and cell polarity defects of an aip3Δ strain, emphasizing the importance of the C terminus for full Aip3p function (our unpublished observations). Third, database searches have identified a S. pombe homologue of Aip3p of previously unknown function (GenBank accession number Z97208). We have named this gene FAT1 for fission yeast AIP three. Fat1p is most similar to Aip3p in ∼263 amino acids at their N termini (32% identity with p = 4 × 10−23; see Figure 8A). As discussed above, truncation of this domain of Aip3p from either the N- or C-terminal ends affected Aip3p localization. For these reasons, we believe this sequence motif may identify a conserved set of protein–protein interactions involved in subcellular protein localization.
To address this possibility, we cloned an N-terminal region of Aip3p that displays strong similarity to Fat1p (amino acids 1–166), as a fusion to GST into a 2-μm plasmid that can be maintained at extremely high copy levels. The fusion protein, under the control of the GAL promoter, was induced in wild-type cells also expressing GFP-Aip3p, and effects on Aip3p localization were monitored by fluorescence microscopy. The GST-aip3p fusion interfered with normal localization of the full-length GFP-Aip3p in a manner similar to that observed in the secretory pathway mutants. We quantified this effect in the same manner that we quantified the defects in cell polarity mutants and found that when the conserved N terminus of Aip3p was overexpressed, only 8% of the cells were able to localize some their pool of Aip3p normally, compared with 58% of the cells overexpressing GST alone (see Figure 8B). In addition to causing mislocalization of Aip3p, we also found that overexpression of the Aip3p-addressing domain in a wild-type strain made cells temperature sensitive (Figure 8C). Because temperature sensitivity is part of the phenotype of aip3Δ deletion alleles, we believe that mislocalization of Aip3p is phenocopying the complete loss of the protein (Amberg et al., 1997).
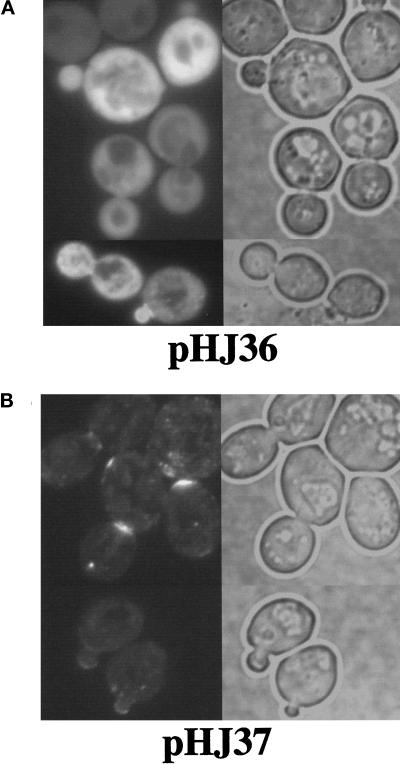
Finally, we asked whether the N terminus of Fat1p could functionally replace the N terminus of Aip3p for localization. For these experiments, we focused on the first 134 amino acids of Aip3p because it shows the greatest sequence identity to Fat1p (44% identity). First we constructed a GFP–aip3p fusion in which the N-terminal 134 amino acids were removed (encoded on plasmid pHJ36). In wild-type cells this fusion protein largely accumulated in the cytosol, with a small amount of the protein accumulating in the buds and bud necks of a few cells (Figure 9A). We then examined the localization of a fusion protein in which the N-terminal 134 amino acids of Aip3p were replaced with the N-terminal 175 amino acids of Fat1p (encoded on plasmid pHJ37). In contrast to the truncation mutant, the chimeric protein localized much more like the wild-type, full-length Aip3p (see Figure 9B). The chimeric protein showed strong localization to the bud and bud neck, a reduction in cytosolic signal over the truncation mutant but some aberrant accumulation in small spots. This experiment indicates that not only is the Aip3p localization pathway conserved, but the protein–protein interactions required for secretory pathway-mediated localization of Aip3p are conserved as well.
Figure 9.
The N terminus of Fat1p can function in the secretory pathway-mediated localization pathway. Wild-type strain FY23×86 was transformed with plasmid pHJ36 encoding a fusion of GFP to amino acids 135–478 of Aip3p (A) and plasmid pHJ37 encoding a fusion of GFP to the N-terminal 175 amino acids of Fat1p, followed by amino acids 135–478 of Aip3p (B). The transformants were examined by fluoresence microscopy (left) and DIC microscopy (right).
DISCUSSION
It is commonly believed that the assembly of a polarized actin cytoskeleton is achieved by concentrating regulators of actin assembly in subdomains at the cell cortex. A complex and poorly understood interplay between these regulators leads to actin assembly and the recruitment of factors that stabilize and specialize the network. One such regulator is Aip3p (Amberg et al., 1997). Aip3p is critical to the establishment and/or maintenance of a polarized actin cytoskeleton. Spatially and temporally, Aip3p localization precedes actin polarization during the budding cell cycle of S. cerevisiae, and loss of AIP3 leads to gross defects in many (if not all) aspects of cell polarity (Amberg et al., 1997). Because little is known about mechanisms underlying the polarized localization of regulators of the actin cytoskeleton, we have focused on understanding the mechanism behind the regulation of Aip3p localization. Here we present genetic and biochemical data that strongly support the model that Aip3p is delivered to sights of polarized actin assembly by the secretory pathway.
Aip3p localization was scrutinized in strains mutant for proteins whose localization behavior mimicked that of Aip3p. We examined several such mutants and those that affected Aip3p localization were all tied directly to the function of the secretory pathway. These included the bona fide “sec” mutants, sec2, sec3, sec4, and sec6 (see Figure 2), as well as mutants in the type V myosin Myo2p and its regulatory light-chain calmodulin (see Figure 3). Some of the secretory pathway components we examined possess a polarized localization pattern like Aip3p, such as components of the “exocyst” (TerBush and Novick, 1995), the small GTPase Sec4p (Brennwald and Novick, 1993), and Sec3p (Finger et al., 1998); however, we found that early acting Sec proteins, such as Sec14p and Sec17p, are also essential for Aip3p localization. This argues that the entire secretory pathway must be operational for normal Aip3p delivery. Mutations in VRP1, SPA2, LAS17, or CDC24 only mildly affected GFP-Aip3p localization, suggesting that they are not directly involved in the Aip3p localization pathway.
MYO2 was first identified as a mutant defective in cell morphology (Johnston et al., 1991), but the protein it encodes has since been determined to be involved in late secretory vesicle delivery (Lillie and Brown, 1994; Govindan et al., 1995), probably by transporting late secretory vesicles on actin cables that terminate in the bud (Pruyne et al., 1998). Our discovery that a MYO2 mutant is defective for Aip3p localization indicates that Myo2p must act on Aip3p-bearing secretory vesicles. A mutation in the actin-binding motor domain of Myo2p (myo2-66) severely affected Aip3p localization. However, a mutation in the tail domain of Myo2p known to cause vacuolar inheritance defects (Catlett and Weisman, 1998) had a much less disruptive effect on Aip3p localization (see Figure 3). We interpret these data to indicate that Myo2p delivers Aip3p-bearing vesicles by translocating these vesicles on actin cables.
Our data also demonstrate a requirement for calmodulin in Aip3p delivery. We believe the calmodulin connection to Aip3p localization is most likely through Myo2p. It is known that calmodulin is bound to Myo2p (Brockerhoff et al., 1994) and that calmodulin in other organisms acts as a regulatory light chain for many different species of myosin motors. It is interesting that we observed strong allele specificity in calmodulin mutants with respect to defects in Aip3p localization. The construction and genetic analysis of a set of calmodulin phenylalanine-to-alanine mutants previously led to the identification of four distinct functions for yeast calmodulin (Ohya and Botstein, 1994a,b). Intragenic complementation between the mutants was used to assign them to four functional groups. We found that mutants in one of the four complementation groups were most defective for Aip3p localization and that this defect could largely be attributed to a F12A mutation in calmodulin (see Figure 3). These data indicate that the function affected in this complementation group is one that serves to activate the Myo2p motor to move late secretory vesicles (Aip3p-bearing vesicles included) on actin cables.
An argument can be made that the secretory pathway involvement in Aip3p localization is indirect and is, for example, due to a failure to deliver a transmembrane receptor for Aip3p to the cell surface. This model could explain the Aip3p localization defects in secretory pathway mutants, but it does not account for the fractionation behavior of Aip3p. By crude fractionation and differential centrifugation (Figure 4), velocity gradient analysis (Figure 5), and floating and density gradient analysis (Figure 6), a pool of Aip3p behaves like a secretory vesicle–associated protein. The sequence of Aip3p has no obvious signal peptide and no potential transmembrane domains and is in fact quite highly charged. Therefore, we believe that Aip3p is a peripherally associated membrane protein and that its association with vesicles must be through an interaction with another membrane-associated protein. Aip3p is not unique in this behavior; biochemically, Aip3p behaves similarly to the rab-like GTPase Sec4p. Sec4p cycles on and off late secretory vesicles in a mechanism that is believed to be tied to the GTPase cycle of the protein (Novick et al., 1993). In crude fractionation experiments analogous to those we performed in Figure 4, Sec4p also partitions between cytosolic and microsome-associated pools (Goud et al., 1988).
What are the proteins directly involved in Aip3p delivery and targeting? We have begun to address this question by defining the sequence elements within Aip3p that are involved in the interactions required for proper protein delivery and localization (Figure 7E). Mutational analysis has identified a region at the N terminus of Aip3p that is necessary and sufficient for Aip3p localization. Within this region of Aip3p is a sequence that is conserved with the N terminus of a protein of unknown function from S. pombe (Figure 8A). Exogenous overexpression of this domain of Aip3p interferes with Aip3p localization in trans (Figure 8B), and the N terminus of the S. pombe homologue can functionally replace the N terminus of Aip3p in the Aip3p localization pathway (Figure 9B). These data lead us to conclude that the sets of protein–protein interactions and the pathways required for Aip3p delivery and localization have been conserved and are mediated through a sequence motif we designate as the Aip3p “addressing domain.”
Several previous observations have suggested that Aip3p localization does not require the actin cytoskeleton. On the basis of the work presented here, it is clear that the Aip3p-actin interaction that led to our initial discovery of Aip3p (Amberg et al., 1997) is unimportant for Aip3p localization (see Figure 7E). However, the experiments in this article strongly suggest that Aip3p is delivered to the bud site in association with secretory vesicles that are moved on actin cables. When cells are released from Go to G1 in the presence of the actin disassembly drug latrunculin A, 30% of the cells are able to localize Aip3p to the bud site (Ayscough et al., 1997). This actin-independent localization likely reflects diffusion of Aip3p to and capture by a receptor for Aip3p at the cell surface, and whether this is free Aip3p or vesicle-associated Aip3p we do not know. Similarly, the observation that Aip3p localization precedes actin polarization during the cell cycle may reflect a diffusion-based mechanism as well. Capture of a small amount of Aip3p may be sufficient to help seed actin assembly at these sites, leading to further polymerization events, including actin cable extension. Once cables have formed, then we expect that Myo2p begins the delivery of large amounts of Aip3p and associated proteins to the bud site so that a polarized actin cytoskeleton can be fully assembled and then maintained. Because actin filaments in cells are unstable and are continuously disassembling (Ayscough et al., 1997), we expect that maintaining a polarized actin cytoskeleton requires the continual delivery of positive regulators of actin assembly. This may in fact be the major function of secretory pathway–mediated delivery of Aip3p: to deliver Aip3p-associated proteins (such as actin itself) to the bud site, acting like a molecular taxi cab.
This secretory pathway–mediated, Aip3p localization pathway is not used by all cell polarity effectors. Localization of the formin Bni1p, known to interact with Aip3p (Evangelista et al., 1997), is unaffected by disruption of the secretory pathway. In addition, Aip3p and Bni1p are not dependent on each other for their localization. The phenotypic similarity between aip3Δ and bni1Δ strains, the fact that the two proteins interact with each other and show the same localization pattern, suggests they operate in the same pathway, and yet we have found that they are localized by different mechanisms. Collectively, these observations suggest a model for regulation of cell polarity in which different regulators are focused at the cell surface by independent methods and that perhaps it is the spatial overlap of these proteins that leads to the efficient assembly and maintenance of a polarized actin cytoskeleton.
ACKNOWLEDGMENTS
We thank the many people who provided strains for this study, in particular Lois Weissman, who was kind enough to send us the myo2-2 strain before publication, and Charlie Boone, who provided the GFP-Bni1p–encoding construct. Anti-GFP antibodies were provided by Pam Silver, anti-Snc1p antibodies by Pat Brennwald, anti-Pma1p antibodies by Amy Chang, and anti-Aip3p antibodies by John Pringle. We thank Pat Brennwald, Chris Kaiser, and Patty Kane for technical advice. This research was supported by National Institutes of Health grant GM-56189.
REFERENCES
- Amberg DC. Three-dimensional imaging of the yeast actin-cytoskeleton through the budding cell-cycle. Mol Biol Cell. 1998;9:3259–3262. doi: 10.1091/mbc.9.12.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Botstein D, Beasley EM. Precise gene disruption in Saccharomyces cerevisiae by double fusion PCR. Yeast. 1995;11:1275–1280. doi: 10.1002/yea.320111307. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JM, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacoular enzyme required for processing of vacoular precursors. Mol Cell Biol. 1986;6:2490–2499. doi: 10.1128/mcb.6.7.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor Latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowser R, Muller H, Govindan B, Novick P. Sec8p and Sec15p are components of a plasma membrane-associated 19.5S particle that may function downstream of Sec4p to control exocytosis. J Cell Biol. 1992;118:1041–1056. doi: 10.1083/jcb.118.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Novick P. Interactions of three domains distinguishing the Ras-related GTP-binding proteins Ypt1 and Sec4. Nature. 1993;360:560–563. doi: 10.1038/362560a0. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Stevens RC, Davis TN. The unconventional myosin, Myo2p, is a calmodulin target at sites of cell growth in Saccharomyces cerevisiae. J Cell Biol. 1994;124:315–323. doi: 10.1083/jcb.124.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cali BM, Doyle TC, Botstein D, Fink GR. Multiple functions of actin during filamentous growth of Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1873–1889. doi: 10.1091/mbc.9.7.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett NL, Weisman LS. The terminal tail region of a yeast myosin-V mediates its attachment to vacoular membranes and sites of polarized growth. Proc Natl Acad Sci USA. 1998;95:14799–14804. doi: 10.1073/pnas.95.25.14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E, Schimmoller F, Pfeffer SR. A novel Rab9 effector required for endosome-to-TGN transport. J Cell Biol. 1997;138:283–290. doi: 10.1083/jcb.138.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle T, Botstein D. Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc Natl Acad Sci USA. 1996;93:3886–3891. doi: 10.1073/pnas.93.9.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92:559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- Goud B, Salminen A, Walworth NC, Novick PJ. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer BK, Corbett A, Kweon Y, Petzold AS, Silver P, Brown SS. SEC3 mutations are synthetically lethal with profilin mutations and cause defects in diploid-specific bud-site selection. Genetics. 1996;144:495–510. doi: 10.1093/genetics/144.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston GC, Prendergast JA, Singer RA. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EW. Bipartite structure of the ADE3 locus of Saccharomyces cerevisiae. Genetics. 1977;85:209–223. doi: 10.1093/genetics/85.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell AK, Goud B, Northup JK, Novick PJ. Binding and hydrolysis of guanine nucleotide by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J Biol Chem. 1990;265:9366–9372. [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Immunofluorescence localization of the unconventional myosin, Myo2p, and the putative kinesisn-related protein, Smy1p, to the same regions of polarized growth in Saccharomyces cerevisiae. J Cell Biol. 1994;125:825–842. doi: 10.1083/jcb.125.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie Ar, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- McCaffrey M, Johnson JS, Goud B, Myers AM, Rossier J, Popoff MR, Madaule P, Boquet P. The small GTP-binding protein Rho1p is localized on the golgi apparatus and postgolgi vesicles in Saccharomyces cerevisiae. J Cell Biol. 1991;115:309–319. doi: 10.1083/jcb.115.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Marshall TK, Deschenes RJ. Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast. 1993;9:715–723. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of golgi-derived secretory vesicles. J Cell Biol. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Brennwald P, Walworth NC, Kabcenell AK, Garrett M, Moya M, Roberts D, Muller H, Govindan B, Bowser R. The cycle of SEC4 function in vesicular transport. Ciba Found Symp. 1993;176:218–228. [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for posttranslational events in the secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ohya Y, Botstein D. Diverse essential functions revealed by complementing yeast calmodulin mutants. Science. 1994a;263:963–966. doi: 10.1126/science.8310294. [DOI] [PubMed] [Google Scholar]
- Ohya Y, Botstein D. Structure-based systematic isolation of conditional-lethal mutations in the single yeast calmodulin gene. Genetics. 1994b;138:1041–1054. doi: 10.1093/genetics/138.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;LX:729–743. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Roberg KJ, Crotwell M, Espenshade P, Gimeno R, Kaiser CA. LST1 is a SEC24 homologue used for selective export of the plasma membrane ATPase from the endoplasmic reticulum. J Cell Biol. 1999;145:659–672. doi: 10.1083/jcb.145.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Vallier L, Sheu Y-J, Snyder M. The Spa2-related protein. SphIp, is important for polarized growth in yeast. J Cell Sci. 1998;111:479–494. doi: 10.1242/jcs.111.4.479. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Novick PJ. Purification and characterization of constitutive secretory vesicles from yeast. J Cell Biol. 1987;105:163–174. doi: 10.1083/jcb.105.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford CA, Daniels LB, Park FJ, Jones EW, Van Arsdell JN, Innis MA. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacoular hydrolases. Mol Biol Cell. 1986;6:2500–2510. doi: 10.1128/mcb.6.7.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud-site selection in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]