Abstract
Tyrosine phosphorylation plays a central role in eukaryotic signal transduction. In yeast, MAP kinase pathways are regulated by tyrosine phosphorylation, and it has been speculated that other biochemical processes may also be regulated by tyrosine phosphorylation. Previous genetic and biochemical studies demonstrate that protein tyrosine phosphatases (PTPases) negatively regulate yeast MAP kinases. Here we report that deletion of PTP2 and PTP3 results in a sporulation defect, suggesting that tyrosine phosphorylation is involved in regulation of meiosis and sporulation. Deletion of PTP2 and PTP3 blocks cells at an early stage of sporulation before premeiotic DNA synthesis and induction of meiotic-specific genes. We observed that tyrosine phosphorylation of several proteins, including 52-, 43-, and 42-kDa proteins, was changed in ptp2Δptp3Δ homozygous deletion cells under sporulation conditions. The 42-kDa tyrosine-phosphorylated protein was identified as Mck1, which is a member of the GSK3 family of protein kinases and previously known to be phosphorylated on tyrosine. Mutation of MCK1 decreases sporulation efficiency, whereas mutation of RIM11, another GSK3 member, specifically abolishes sporulation; therefore, we investigated regulation of Rim11 by Tyr phosphorylation during sporulation. We demonstrated that Rim11 is phosphorylated on Tyr-199, and the Tyr phosphorylation is essential for its in vivo function, although Rim11 appears not to be directly regulated by Ptp2 and Ptp3. Biochemical characterizations indicate that tyrosine phosphorylation of Rim11 is essential for the activity of Rim11 to phosphorylate substrates. Our data demonstrate important roles of protein tyrosine phosphorylation in meiosis and sporulation
INTRODUCTION
Protein tyrosine phosphorylation is a universal mechanism of cellular regulation in eukaryotes (Hunter, 1995). Tyr phosphorylation has been implicated in the regulation of cell proliferation, differentiation, development, and turmorgenesis in a variety of model eukaryotic systems (Eisenmann and Kim, 1994). Although no conventional tyrosine kinases exist in Saccharomyces cerevisiae, Tyr phosphorylation has been shown to be essential in controlling the activity of MAP (mitogen-activated protein) kinases in yeast (Herskowitz, 1995). MAP kinase activation requires phosphorylation on Tyr and Thr residues catalyzed by the dual-specificity protein kinase MEK (MAP kinase kinase) (Cobb and Goldsmith, 1995). Activations of yeast MAP kinases are required for mating and zygote formation, stress response, cell wall integrity, filamentous and pseudohyphael growth, and spore wall formation (Gustin et al., 1998).
In addition to MAP kinases, biochemical purification of Tyr-phosphorylated proteins in yeast cellular lysates has identified Mck1, a member of the GSK3 (glycogen synthase kinase 3) family of kinases (Lim et al., 1993). GSK3 has been found to regulate glycogen metabolism and gene expression by phosphorylating a range of substrates, including metabolic enzymes and transcription factors (Woodgett et al., 1993; Woodgett, 1994). A growing amount of evidence suggests this family of enzyme is also involved in the regulation of development and cellular differentiation (Woodgett, 1994; Dale, 1998). Among all GSK3 members there is a highly conserved Tyr residue, and it has been shown that mammalian GSK3 is phosphorylated on this residue (Hughes et al., 1993). However, it is unclear what is the physiological significance of GSK3 Tyr phosphorylation and whether the Tyr phosphorylation is regulated. The S. cerevisiae genome contains four GSK3 genes, MCK1, RIM11, MRK1, and open reading frame YOL128C (Neigeborn and Mitchell, 1991; Shero and Hieter, 1991; Lim et al., 1993; Bowdish et al., 1994; Puziss et al., 1994; Hardy et al., 1995). Although the functions of the latter two genes are unknown, both MCK1 and RIM11 have been implicated in meiosis and sporulation of diploid cells (Neigeborn and Mitchell, 1991; Bowdish et al., 1994). In addition, MCK1 is involved in maintaining chromosomal stability (Shero and Hieter, 1991), and disruption of MCK1 results in pleiotropic phenotypes. In contrast, Rim11 has been implicated to function in meiosis and sporulation only. Under sporulation conditions, Rim11 phosphorylates a ubiquitously expressed transcription factor, Ume6, and a meiosis-inducible transcription factor, Ime1, thereby promoting the formation of a Ume6/Ime1 complex (Bowdish et al., 1994; Malathi et al., 1997). The Ume6/Ime1 complex plays a critical role in inducting sporulation-specific genes (Rubin-Bejerano et al., 1996). However, it is unclear how Rim11 activity is regulated, whether it is also phosphorylated on Tyr, and what is the physiological significance of Tyr phosphorylation.
Because the balance of protein tyrosine phosphorylation is maintained by the actions of kinases and phosphatase, protein tyrosine phosphatases play an equally critical role in controlling cellular signaling. Three protein tyrosine phosphatase (PTPase) genes, PTP1, PTP2, and PTP3, have been identified in S. cerevisiae (Guan et al., 1991, 1992; Zhan et al., 1997). PTP1 encodes a 38-kDa protein containing just the PTPase catalytic domain (Guan et al., 1991). The function of Ptp1 is not clear, although a potential substrate for Ptp1 has been reported (Wilson et al., 1995). In contrast, PTP2 and PTP3 encode much larger proteins with conserved C-terminal PTPase domains and a less-well-conserved N-terminal noncatalytic domain. Genetic and biochemical studies have demonstrated a critical requirement of Ptp3 and, to a lesser extent, Ptp2 in the dephosphorylation and inactivation of Fus3 MAP kinase in the mating pheromone response of haploid yeast cells (Zhan et al., 1997). In parallel, Ptp2 plays a more important role than Ptp3 in the regulation of Hog1 MAP kinase in high-osmotic stress response (Jacoby et al., 1997; Wurgler-Murphy et al., 1997). Therefore, protein tyrosine phosphatases have distinct as well as overlapping functions in S. cerevisiae. However, the functions of these PTPases besides their role in MAP kinase regulation have not been characterized.
In this report, we have found that homozygous deletion of both PTP2 and PTP3 greatly decreased sporulation efficiency. Diploid ptp2Δptp3Δ cells were blocked before premeiotic DNA replication, and expression of meiosis- and sporulation-specific genes were significantly reduced, suggesting that the ptp2Δ/−/ptp3Δ/− cells are defective in initiation of the sporulation program. Immunoblotting with anti-phosphotyrosine (anti-Pi-Tyr)-specific antibodies revealed that Tyr phosphorylation of several proteins p52, p43, and p42, named according to apparent molecular mass, were elevated in the ptp2Δ/−/ptp3Δ/− double disruption cells but not in wild-type or single disruption cells during sporulation. The major tyrosine-phosphorylated protein, p42, was identified as Mck1 protein kinase, a known yeast GSK3 kinase phosphorylated on Tyr. Disruption of a related yeast GSK3 gene, Rim11, reduced Tyr phosphorylation of p43. Further studies demonstrated that in vivo Rim11 was Tyr phosphorylated at the Tyr-199 residue, and this phosphorylation does not appear to be regulated by Ptp2 and Ptp3. Biochemical analysis of Rim11 immunoprecipitated from yeast cells or recombinant Rim11 purified from Escherichia coli has shown that the Tyr phosphorylation of Rim11 is mediated by autophosphorylation and is required for the kinase activity of Rim11 on physiological substrates. Furthermore, we have demonstrated that Tyr phosphorylation is essential for both Rim11 kinase activity and in vivo functions.
MATERIALS AND METHODS
Yeast Strains and Media
S. cerevisiae strains used in this study are listed in Table 1. Standard recipes were used for SC dropout, YPED, YPEAc, PSP2, and 1% potassium acetate (KAc) (Rose et al., 1990; Guthrie and Fink, 1991). All experiments were conducted at 30°C. Sporulations were routinely carried out as described (Guthrie and Fink, 1991). Genetic manipulation of yeast and preparation of medium were performed as described (Rose et al., 1990; Guthrie and Fink, 1991). PCR-based gene replacements were performed to delete the complete open reading frames of MCK1, RIM11, and SMK1 in isogenic strains (Wach et al., 1994). Deletions were confirmed by genomic PCR analysis.
Table 1.
S. cerevisiae strains used in this work
| Strains | Genotype | Reference of source |
|---|---|---|
| Y264 | MATa/α trp1/− his3/− leu2/− ura3/− | Lab stock |
| Y162 | MATa/α trp1/− his3/− leu2/− ura3/− ptp2Δ::LEU2/ptp2Δ::LEU2 | This study |
| Y163 | MATa/α trp1/− his3/− leu2/− ura3/− ptp3Δ::URA3/ptp3Δ::URA3 | This study |
| Y164 | MATa/α trp1/− his3/− leu2/− ura3/− ptp2Δ::LEU2/ptp2Δ::LEU2 ptp3Δ::URA3/ptp3Δ::URA3 | This study |
| Y165 | MATa/α trp1/− his3/− leu2/− ura3/− ptp2Δ::LEU2/ptp2Δ::LEU2 ptp3Δ::URA3/ptp3Δ::URA3 mck1Δ::KanR /mck1Δ:: KanR | This study |
| Y166 | MATa/α trp1/− his3/− leu2/− ura3/− ptp2Δ::LEU2/ptp2Δ::LEU2 ptp3Δ::URA3/ptp3Δ::URA rim11Δ:: KanR /rim11Δ:: KanR | This study |
| KB600 | MATa/α HA-RIM11/HA-RIM11 met4/MET4 his4/HIS4 IME2/ime2-4-lacZ::LEU2 ura3/− leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG | Bowdish et al., 1994 |
| KB268 | MATa/α rim11::LEU2/rim11:LEU2 met4/MET4 his4/HIS4 IME2/ime2-4-lacZ::LEU2 ura3/− leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG | Bowdish et al., 1994 |
Plasmid Constructions
Plasmids are listed in Table 2. Standard molecular cloning techniques were used for plasmid constructions and DNA manipulation. Details of plasmid constructions are available upon request. All DNA fragments generated by PCR were verified by sequencing analysis.
Table 2.
Plasmids used in this study
| Plasmid | Markera | Reference of source |
|---|---|---|
| Phosphatase plasmid | ||
| pXZ209 | HIS3 CEN PTP3 | This study |
| pXZ210 | HIS3 CEN ptp3C814G | This study |
| pXZ223 | HIS3 CEN PTP2 | This study |
| pXZ134 | URA3 2μ PADH1-GST | Zhan and Guan, 1999 |
| pXZ110 | URA3 2μ PADH1-GST-PTP1 | Zhan and Guan, 1999 |
| pXZ113 | URA3 2μ PADH1-GST-PTP2 | Zhan and Guan, 1999 |
| pXZ123 | URA3 2μ PADH1-GST-PTP3 | Zhan and Guan, 1999 |
| pXZ136 | URA3 2μ PADH1-GST-ptp3C814G | Zhan and Guan, 1999 |
| Kinase plasmid | ||
| Flag-MCK1 | TRP1 2μ PMCK1-FLAG-MCK1 | This study |
| pKB166 | URA3 2μ PRIM11-HA-RIM11 | Bowdish et al., 1994 |
| pKB199 | URA3 2μ PRIM11-HA-rim11K68A | Bowdish et al., 1994 |
| pKB201 | URA3 2μ PRIM11-HA-rim11Y199F | Bowdish et al., 1994 |
| pXZ356 | URA3 2μ PRIM11-HA-rim11Y199E | This study |
| pXZ352 | HIS3 CEN PRIM11-HA-RIM11 | This study |
| pXZ353 | HIS3 CEN PRIM11-HA-rim11K68A | This study |
| pXZ354 | HIS3 CEN PRIM11-HA-rim11Y199F | This study |
| pXZ355 | HIS3 CEN PRIM11-HA-rim11Y199E | This study |
| pXZ118 | GST-HA-RIM11 in E. coli expression vector | This study |
| pXZ119 | GST-HA-rim11K68A in E. coli expression vector | This study |
| pXZ120 | GST-HA-rim11Y199F in E. coli expression vector | This study |
PADH1, PMCK1, and PRIM11, are ADH1, MCK1, and RIM11 promoters, respectively.
Sporulation and DNA Synthesis
For the quantitation of sporulation efficiency, cells were first grown in prespore medium to middle log phase, harvested, and resuspended in 1% Ac sporulation medium. At least 200 cells were scored by microscopic examination. Meiotic nuclei were stained by DAPI and visualized with fluorescent microscopy as described (Friesen et al., 1994). Cells that appeared binucleate, trinucleate, or tetranucleate were considered to have completed meiosis I. Cells that appeared trinucleate or tetranucleate were considered to have completed meiosis II. Premeiotic DNA synthesis was monitored as described (Guthrie and Fink, 1991). In brief, cells were first grown in PSP2 and [2-14C]uracil (0.8 μCi/ml, 60 mCi/mmol) to middle log phase, harvested, washed once with 1% KAc, and resuspended in sporulation medium (1% KAc without labeled uracil). Samples (0.5-ml culture) were taken in triplicate during sporulation, DNA was precipitated by trichloroacetic acid, and incorporation of radioactivity was determined by scintillation counting.
RNA Analysis
Cells growing in sporulation medium were collected, and total RNA was prepared by the glass bead-phenol extraction procedure (Zhu and Thiele, 1996). Total RNA was fractionated in a 1.5% agarose-formaldehyde gel and transferred to a Nytran membrane (Schleicher & Schuell, Keene, NH). Membranes were hybridized with 32P-labeled DNA probes generated by the random-priming method. The probes used were as follows: IME1, a 2.2-kb HindIII–SalI fragment from pAM504 (Smith et al., 1990); IME2, a 3.13-kb PCR fragment amplified by 5′-CTGTGACAGATAAACCC-3′ and 5′-TGGACCCGGGGAATAAACGCAAAG-3′; SPS2, a 0.8-kb PstI–HindIII fragment from p18 (Friesen et al., 1994); and DIT1, a 1.8-kb HindIII–HindIII fragment from pPB-13 (Friesen et al., 1994).
Immunoblotting
Yeast cell cultures were directly lysed in SDS sample buffer. The cell lysates were resolved on 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The membrane was blocked in 5% BSA in TBST (15 mM Tris-Cl, pH 7.6, 150 mM NaCl, and 0.2% Tween 20) and probed with anti-Pi-Tyr antibody (1:10,000 dilution; Upstate Biotechnology, Lake Placid, NY) in 5% BSA in TBST. The Western blot was developed with HRP-conjugated anti-mouse second antibody and ECL methods (Amersham, Arlington Heights, IL). For phosphotyrosine competition, the anti-Pi-Tyr antibody was preincubated with 1 mM phosphotyrosine for 30 min before Western blot. The Western blot solution also contained 1 mM phosphotyrosine during incubation.
Immunoprecipitation and Rim11 Kinase Assay
Yeast cells harboring pRS426, pKB166, pKB199, or pKB201 were grown (OD600 nm, ∼0.8) in SC-Ura. Yeast total cellular lysate was prepared and hemagglutinin (HA)-tagged protein was immunoprecipitated with monoclonal anti-HA mAb (12CA5; Babco, Richmond, CA) as described (Zhan et al., 1997). For Rim11 kinase assay, immunoprecipitants were further washed with kinase buffer (25 mM HEPES, pH 7.2, 15 mM MgCl2, 5 mM EGTA, 1 mM DTT, 0.1 mM orthovanadate, and 15 mM pNPP). Reactions were started by addition of 20 μl of a mixture containing 100 μM phospho-GS peptide (Upstate Biotechnology), 100 μM ATP, and 5 μCi of [γ-32P]ATP (7000 Ci/mmol; ICN Biochemicals, Costa Mesa, CA) in kinase buffer. After a 10-min incubation at 30°C with shaking, reactions were terminated by addition of 5 μl of 50 mM EDTA, and mixtures were spotted on p81 filter paper (Whatman, Maidstone, United Kingdom). Filter paper was washed with 180 mM phosphoric acid, dried, and subjected to scintillation counting.
Purification of Glutathione S-Transferase (GST)-Rim11 Fusion Proteins and In Vitro Autophosphorylation
GST-Rim11, GST-Rim11(K68A), GST-Rim11(Y199F), and GST-Ume6 (amino acid residues 1–200) were expressed in E. coli and purified by glutathione-agarose affinity chromatography (Guan and Dixon, 1991) followed by mono-Q fast protein liquid chromatography (FPLC). In vitro kinase assays were performed as described above. When GST-Ume6 was used as a substrate, phosphorylation of GST-Ume6 was analyzed by SDS-PAGE and autoradiography. For phosphatase treatment, GST-Rim11 (200 ng) was incubated with 1 μg of GST-Ptp1B in kinase buffer without ATP at 30°C for 30 min and then subjected to autophosphorylation for another 30 min by adding 16.7 μCi of [γ-32P]ATP (7000 Ci/mmol; ICN), cold ATP (5 μM final concentration), and vanadate (1 mM final) to inhibit the phosphatase. After electrophoresis on 10% SDS-PAGE and electrotransfering to a polyvinylidene difluoride membrane, phosphorylation was detected by autoradiography and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Bands corresponding to GST fusion proteins were excised from membrane and subjected to phosphoamino acid analysis as previously described (Kamps, 1991).
RESULTS
Deletion of PTP2 and PTP3 Results in a Sporulation Defect
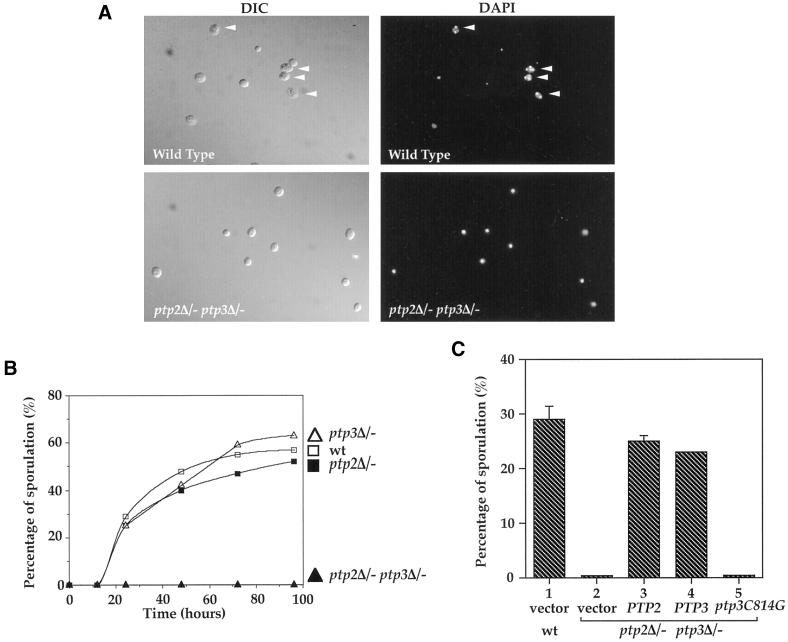
We have previously demonstrated that Ptp2 and Ptp3 play an important role in the regulation of Fus3 MAP kinase in the S. cerevisiae mating pheromone response pathway (Zhan et al., 1997). Ptp2 and Ptp3 are also involved in the negative regulation of the Hog1 MAP kinase (Jacoby et al., 1997; Wurgler-Murphy et al., 1997). To investigate whether Ptp2 and Ptp3 carry out additional physiological functions in yeast, we examined the phenotypes of ptp2Δptp3Δ deletion strains under a variety of conditions. We found that a diploid strain with homozygous deletion of both PTP2 and PTP3 genes had a defect in sporulation (Figure 1, A and B). Microscopic examination of isogenic wild-type cells revealed that a high percentage of cells had formed asci with a triad or tetrad of spores after 24 h in sporulation medium. These asci contain well-organized bi-, tri-, or tetranuclei when stained with DAPI (Figure 1A, top right, arrowheads). In contrast, ptp2Δ/− ptp3Δ/− double deletion cells were arrested as unbudded cells, and no spores were observed in sporulation medium (Figure 1A, bottom left). DAPI staining showed that cells contain a single nucleus (Figure 1A, bottom right).
Figure 1.
Diploid ptp2Δ/−ptp3Δ− double disruption cells are defective in sporulation. (A) PTP2 and PTP3 genes are required for sporulation. Wild-type (Y264, top) or ptp2Δ/− ptp3Δ/− double deletion diploid (Y164, bottom) cells were sporulated in liquid sporulation media for 2 d. Cells were fixed, stained with DAPI, and examined by Normaski phase-contrast microscopy for spores (left panels) or fluorescence microscopy for DAPI-stained nuclei (right panels). A high proportion of wild-type cells have formed asci with a triad or tetrad of spores visible (top left panel, arrowheads) and these asci containing bi-, tri-, and tetranuclei (top right panel, arrowheads). No mature spores were found in ptp2Δ/− ptp3Δ/− double deletion cells, and most of the cells only contain a single nucleus (lower right panel). (B) Defective sporulation in ptp2Δ/− ptp3Δ/− double deletion cells but not single deletion of either phosphatase gene. Wild-type (Y264, open square), ptp2Δ/− (Y162, closed square), ptp3Δ/− (Y163, open triangle), or ptp2Δ/− ptp3Δ/− (Y164, closed triangle) cells from liquid sporulation at the indicated time points were examined by phase-contrast microscopy. The percentage of sporulation was determined by counting the numbers of asci containing four mature spores. (C) Ptp3 Tyr-phosphatase activity is required for its function in sporulation. Wild-type (Y264, column 1) or ptp2Δ/− ptp3Δ/− cells (Y164, column 2) harboring vector, single-copy PTP2 (pXZ223, column 3), PTP3 (pXZ209, column 4), or ptp3C814G (pXZ210, column 5), which encodes a catalytic-deficient phosphatase, were shifted to sporulation medium, and the sporulation efficiency was determined by counting the number of sporulated cells.
When the sporulation of wild-type, ptp2Δ/− single, ptp3Δ/− single, or ptp2Δ/− ptp3Δ/− double deletion cells was monitored over the time course of 4 d, wild-type, ptp2Δ/− single, and ptp3Δ/− single deletion cells displayed similar sporulation efficiency and kinetics, whereas no spores were found in ptp2Δ/− ptp3Δ/− double deletion cells (Figure 1B). The defect was only observed with ptp2Δ/− ptp3Δ/− cells, whereas deletion of a homologous yeast phosphatase gene, PTP1, singly or in combination with either PTP2 or PTP3 had no effect on sporulation efficiency (our unpublished data). In addition, there is no significant difference of growth rate and viability between the ptp2Δ/− ptp3Δ/− double deletion cells and wild-type cells (our unpublished results). Introduction of either single-copy PTP2 or PTP3 gene into ptp2Δ/− ptp3Δ/− cells can effectively rescue the sporulation defect (Figure 1C), confirming that the sporulation defect phenotype is caused by ptp2Δ ptp3Δ deletion. We also investigated whether Ptp3 phosphatase activity is required for its function in sporulation by assessing the ability of a mutant ptp3C814G gene, which encodes a catalytically inactive PTPase (Cys-814 to Gly substitution), to complement ptp2Δ ptp3Δ deletion. Sporulation was not restored in ptp2Δ/− ptp3Δ/− cells harboring ptp3C814G, indicating lack of complementation (Figure 1C, column 5). Based on these results, we conclude that PTP2 and PTP3 have redundant functions in sporulation, and Ptp3 phosphatase activity is required for sporulation.
ptp2Δptp3Δ Deletion Blocks Premeiotic DNA Synthesis and Reduces Expression of Sporulation-specific Genes
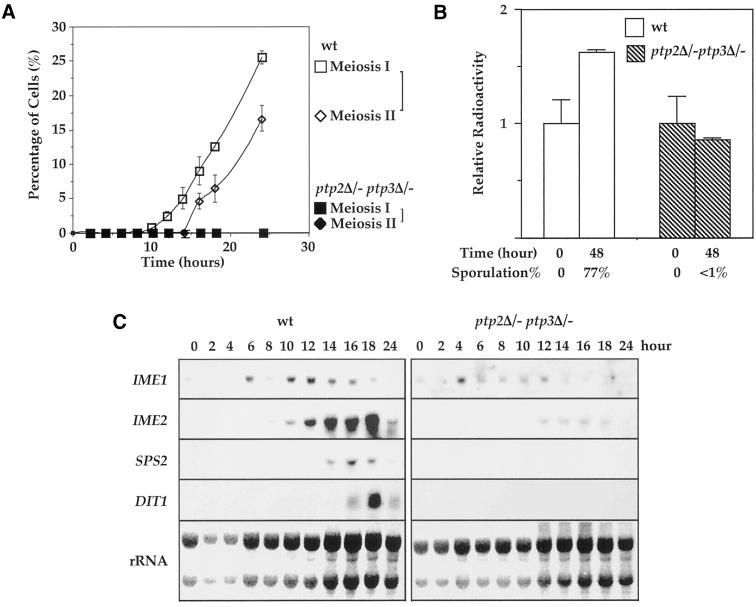
S. cerevisiae sporulation is proceeded through several well-established landmark events, including induction of meiosis-specific genes, one round of premeiosis DNA synthesis, and meiotic recombination. Cells then go through meiosis I (reductional) and meiosis II (equational) division. Finally four spores are packed into an ascus. We investigated which step of meiosis is affected in ptp2Δ/− ptp3Δ/− cells, resulting in the terminal phenotype of sporulation defect. We first examined meiosis I and II in wild-type and ptp2Δ/− ptp3Δ/− cells under a microscope after nuclear staining with DAPI. The appearance of binucleate cells and tetranucleate cells are considered as landmarks for completion of meiosis I and meiosis II, respectively. Binucleate cells were found in wild-type cells 10 h after shifting to sporulation medium, indicating completion of meiosis I (Figure 2A, open squares). After 14 h meiosis II started with accumulation of tri- and tetranucleate cells (Figure 2A, open diamonds). In contrast, few binucleate or tetranucleate cells were found in ptp2Δ/− ptp3Δ/− cells (closed squares and diamonds). These observations indicate that ptp2Δ/− ptp3Δ/− double deletion blocked sporulation before meiosis I.
Figure 2.
ptp2Δ/− ptp3Δ/- deletion cells were blocked before pre-meiotic DNA synthesis and induction of sporulation specific genes was decreased in double deletion cells. (A) ptp2Δ/− ptp3Δ/− double deletion cells are arrested before meiosis I in sporulation medium. Wild-type (Y264) and ptp2Δ/− ptp3Δ/− (Y164) cells were withdrawn from sporulation medium at the indicated time points. Cells were stained with DAPI and examined by fluorescence microscopy to determine the percentage of cells that had completed meiosis I or meiosis II. Tetranucleate cells (also include trinucleate cells) were counted as cells that completed meiosis II, whereas cells with more than one nucleus were counted as cells that completed meiosis I. Open square, meiosis I in wild type; open diamond, meiosis II in wild type; closed square, meiosis I in ptp2Δ/− ptp3Δ/−; closed diamond, meiosis II in ptp2Δ/− ptp3Δ/−, respectively. (B). Premeiotic DNA synthesis is absent in ptp2Δ/− ptp3Δ/− double deletion cells. Wild-type diploid yeast cells (Y264, open bars) or ptp2Δ/− ptp3Δ/− (Y164, closed bars) were grown in presporulation medium with [14C]uracil to label the endogenous pool of nucleotides. Cells were shifted into sporulation medium for 48 h in the absence of [14C]uracil. DNA was isolated, and total radioactivity incorporated in DNA was determined by scintillation counting. In parallel, the sporulation efficiency of the same cell samples was determined by microscopic examination. Average and SD from duplicated assays are shown. (C) Expression of sporulation-specific genes is decreased in ptp2Δ/− ptp3Δ/− double deletion cells. RNA was prepared from either wild-type (Y264, left panels) or ptp2Δ/− ptp3Δ/− double deletion cells (Y164, right panels) taken at various times after transferred to sporulation medium. Northern hybridization was performed on total RNA with probes derived from IME1, IME2, SPS2, and DIT1 genes as indicated. The amount of total RNA loaded in each lane was visualized with methylene blue staining for rRNA (bottom panels).
A critical event before meiosis I is one round of premeiotic DNA synthesis. We compared premeiotic DNA synthesis for wild-type and ptp2Δptp3Δ cells (Figure 2B). Premeiotic DNA synthesis differs from mitotic DNA synthesis in that it uses nucleotides derived from hydrolysis of existing nucleic acids. Thus, premeiotic DNA synthesis can be biochemically distinguished from mitotic DNA synthesis. Wild-type or ptp2Δ/− ptp3Δ/− cells were grown in presporulation medium with [14C]uracil to label the endogenous pool of nucleotides. Cells were then shifted into sporulation medium for 48 h in the absence of 14C-uracil. DNA was isolated, and total radioactivity incorporated in DNA was determined by scintillation counting. In wild-type cells, the amount of radioactivity incorporated into DNA was increased (Figure 2B, open bars) to 1.7-fold after 48 h in sporulation medium, indicating premeiotic DNA synthesis. The sporulation efficiency was 77% for wild-type cells. In contrast, no new DNA synthesis was observed in ptp2Δ/− ptp3Δ/− double deletion cells (Figure 2B, hatched bars) after 48 h in sporulation medium. These results suggest that the defect of ptp2Δptp3Δ in sporulation occurs before premeiotic DNA synthesis.
Several classes of temporally distinct sporulation-specific genes, referred to as early, middle, and late genes (Mitchell, 1994), are sequentially expressed as the sporulation program proceeds. To further characterize the sporulation defect of ptp2Δ ptp3Δ deletion stains, the expression pattern of sporulation-specific genes was determined (Figure 2C). Induction of IME1 and IME2, two early sporulation-specific genes, was significantly diminished in ptp2Δ/− ptp3Δ/− deletion cells (Figure 2C). The expressions of middle and late sporulation-specific genes, including SPS2 and DIT1, were eliminated in ptp2Δ/− ptp3Δ/− deletion cells (Figure 2C). Because the earliest sporulation-specific gene, IME1, is affected by ptp2Δ/− ptp3Δ/− deletion, our results indicate that PTP2 and PTP3 function early in sporulation and possibly play roles in the initiation of the meiotic program.
Elevated Protein Tyr Phosphorylation in ptp2Δ/− ptp3Δ/− Double Deletion Cells during Sporulation
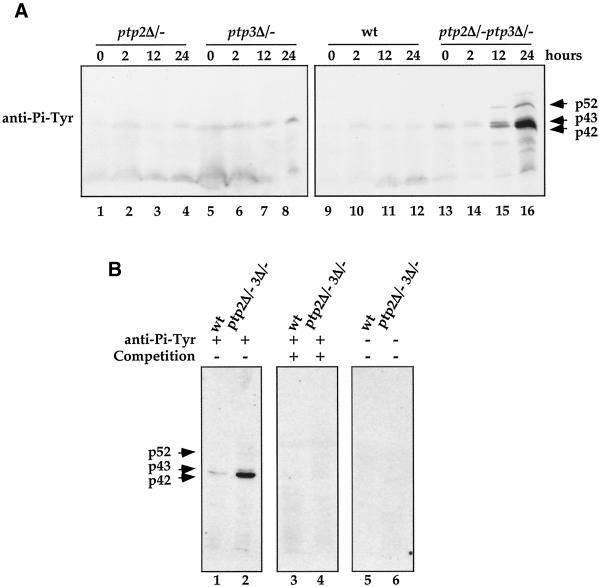
We compared protein Tyr phosphorylation patterns in wild-type (Figure 3A, lanes 9–12), ptp2Δ/− single (lanes 1–4), ptp3Δ/− single (lanes 5–8), and ptp2Δ/− ptp3Δ/− double deletion (lanes 13–16) cells during sporulation (Figure 3A). Tyr-phosphorylations of 52-, 43-, and 42-kDa proteins were induced in ptp2Δ/− ptp3Δ/− cells after 12 h in sporulation medium. The signal detected by the anti-Pi-Tyr antibody was specific, because it was completely abolished if the primary antibody was first incubated with 1 mM phosphotyrosine (Figure 3B, lanes 3–4) or when the primary antibody was omitted in the incubation (lanes 5–6). Our data demonstrate that Tyr phosphorylation of p52, p43, and p42 is elevated in ptp2Δ/− ptp3Δ/− deletion cells during sporulation, suggesting that PTPases regulate Tyr phosphorylation during sporulation.
Figure 3.
Elevated protein tyrosine phosphorylation in ptp2Δ/− ptp3Δ/− double deletion cells during sporulation. (A) Tyrosine phosphorylation of 52-, 43-, and 42-kDa proteins is up-regulated in ptp2Δ/− ptp3Δ/− double deletion cells during sporulation. Total cellular lysates were prepared from wild type (Y264, lanes 9–12), ptp2Δ/− single deletion (Y162, lanes 1–4), ptp3Δ/− single deletion (Y163, lanes 5–8), and ptp2Δ/− ptp3Δ/− double deletion (Y164, lanes 13–16) grown in sporulation medium for various times. Equal amount of lysates were resolved on SDS-PAGE and subjected to immunoblotting with anti-Pi-Tyr antibody. The prominent tyrosine-phosphorylated proteins were denoted p42, p43, and p52 according to their apparent molecular masses. (B) Competition of anti-Pi-Tyr Western blots. Cell lysates prepared from wild-type (Y264) or ptp2Δ/− ptp3Δ/− double deletion cells (Y164) incubated in sporulation medium for 24 h were subjected to anti-Pi-Tyr Western blotting (lanes 1 and 2). The immunoreactive signals were specifically competed if the anti-Pi-Tyr antibody was preincubated with 1 mM phosphotyrosine (lanes 3 and 4). As a control, no signal was detected on Western blotting without primary anti-Pi-Tyr antibody (lanes 5 and 6).
Mck1 and Rim11 Kinases Are Tyr Phosphorylated
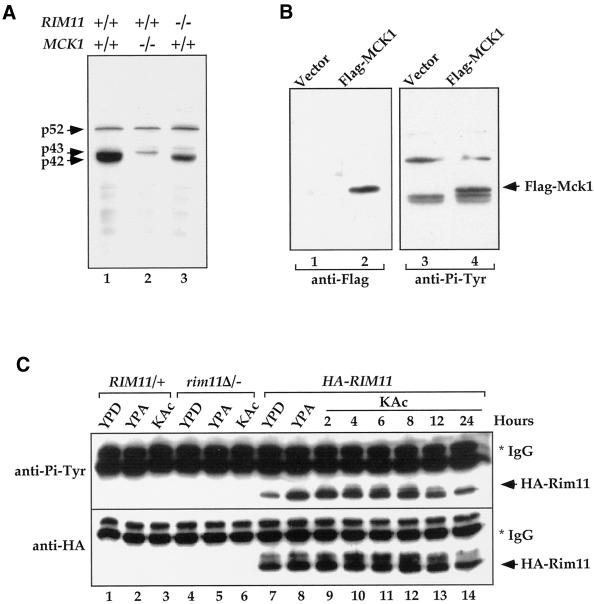
We attempted to biochemically purify p52, p43, and P42 proteins from ptp2Δ/− ptp3Δ/− cells cultured in sporulation medium. The chromatography properties on phosphocellulose and Mono-Q FPLC columns (our unpublished data), and the apparent molecular mass of 42 and 43 kDa prompted us to test whether Mck1, which is known to be Tyr phosphorylated (Lim et al., 1993), is one of the proteins. The yeast MCK1 gene encodes a member of the GSK3 family of protein kinases. Genetic study of MCK1 indicates that it is involved in meiosis and sporulation (Neigeborn and Mitchell, 1991) and chromosomal stability (Shero and Hieter, 1991). The MCK1 gene was deleted in ptp2Δ/− ptp3Δ/− cells, and lysate prepared from the triple deletion cells was subjected to anti-Pi-Tyr immunoblotting. Deletion of MCK1 resulted in a selective loss of the major band of p42 Tyr-phosphorylated protein (Figure 4A, compare lanes 1 and 2), indicating that p42 is either Mck1 or its tyrosine phosphorylation is dependent on Mck1. To distinguish between these two possibilities, a Flag epitope-tagged Mck1 was introduced into the ptp2Δ/− ptp3Δ/− deletion cells, and anti-Pi-Tyr immunoblotting was performed (Figure 4B). We observed a new Tyr-phosphorylated band that appeared above the p42/43 doublet in cells expressing Flag-Mck1 (Figure 4B, lane 4) but not in vector control (lane 3), consistent with the notion of FLAG-Mck1 being phosphorylated on Tyr. Immunoblotting with an anti-Flag antibody confirmed that the new Tyr-phosphorylated band was Flag-Mck1 (lanes 1 and 2). Therefore, our results indicate that the p42 is Tyr-phosphorylated Mck1.
Figure 4.
Tyrosine phosphorylation of Mck1 and Rim11. (A) Deletion of MCK1 and RIM11 in ptp2Δ/− ptp3Δ/− double deletion cells eliminated Tyr phosphorylation of p42 and p43, respectively. Lysates were prepared from ptp2Δ/− ptp3Δ/− (Y164, lane 1), ptp2Δ/− ptp3Δ/− mck1Δ/− (Y165, lane 2), and ptp2Δ/− ptp3Δ/− rim11Δ/− cells (Y166, lane 3) taken 24 h after being shifted to sporulation medium. Tyr phosphorylation was detected by immunoblotting with anti-Pi-Tyr antibody. (B) Flag-tagged MCK1 is phosphorylated on Tyr in vivo. Control vector (pRS424, lanes 1 and 3) or Flag epitope-tagged Mck1 (pRS424-Flag-MCK1, lanes 2 and 4) were introduced into ptp2Δ/− ptp3Δ/− double deletion cells (Y164). Total cellular lysates were subjected to immunoblotting with anti-Flag (left, lanes 1 and 2) or anti-Pi-Tyr (right, lanes 3 and 4) antibody. Yeast cells containing epitope-tagged Mck1 showed an extra Tyr-phosphorylated band as indicated (Flag-Mck1). (C) Rim11 is phosphorylated on Tyr. Wild-type (lanes 1–3), rim11Δ/rim11Δ (KB268, lanes 4–6), or wild-type cells bearing an integrated copy of HA-RIM11 (KB600, lanes 7–14) were grown in YPD, YPA (rich acetate medium), or 1% KAc (sporulation medium). HA-Rim11 was immunoprecipitated from total cellular lysates and subjected to immunoblotting with anti-Pi-Tyr (top) or anti-HA (bottom) antibody to determine Tyr phosphorylation or protein levels, respectively.
A second member of the yeast GSK3 kinase family, Rim11, which has a molecular mass of ∼43 kDa (Figure 3A, minor tyrosine-phosphorylated band slightly above p42), has been shown to be essential for yeast meiosis and sporulation (Bowdish et al., 1994; Puziss et al., 1994). Homozygous deletion of RIM11 resulted in disappearance of Tyr phosphorylation of the 43-kDa band (Figure 4A, compare lanes 1 and 3), suggesting that p43 protein Tyr phosphorylation is Rim11 dependent. However, because the p43 Tyr phosphorylation is observed only in sporulation medium, it is possible that the disappearance of p43 Tyr phosphorylation is a consequence of the inability of rim11Δ/rim11Δ cells to sporulate. We directly examined whether Rim11 is phosphorylated on Tyr by performing anti-Pi-Tyr immunoblotting on HA-tagged Rim11 immunoprecipitated from total cellular lysates (Figure 4C). Cells with a functional HA-tagged RIM11 gene integrated into chromosome were grown in YPD, YPAc (rich acetate medium), or 1% KAc sporulation medium. Tyr phosphorylation was readily detected on Rim11 when cells were grown in YPD (lane 7). Furthermore, Tyr phosphorylation was enhanced when cells were shifted from YPD to acetate medium YPAc (lane 8) and was persistent throughout sporulation (lanes 9–14). Therefore, we conclude that Rim11 is Tyr phosphorylated, and phosphorylation is induced in acetate medium.
Rim11 Tyr Phosphorylation and Activity Are Not Affected by ptp2Δptp3Δ Deletion or Ptp2 and Ptp3 Overexpression
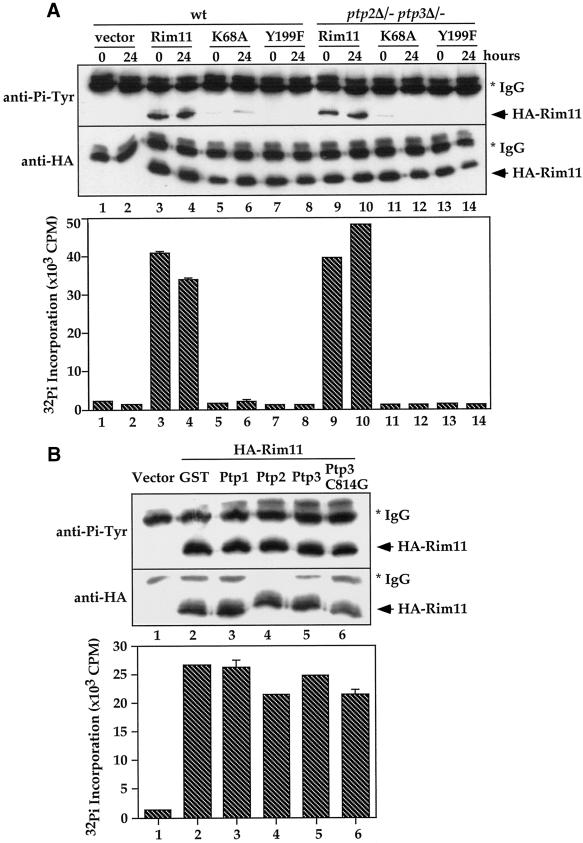
To investigate whether Rim11 is regulated by Ptp2 and Ptp3, we first compared Rim11 Tyr phosphorylation level and kinase activity in wild-type and ptp2Δ/− ptp3Δ/− cells after 0 and 24 h in sporulation medium (Figure 5A). When HA-tagged Rim11 was immunoprecipitated from cell lysates (Figure 5A, middle), similar Tyr phosphorylation levels were observed on HA-Rim11 in wild-type (top, lanes 3 and 4) and double deletion cells (lanes 9 and 10). Interestingly, Rim11 Tyr phosphorylation appears to be dependent on its own kinase activity, because when a conserved Lys-68 residue essential for kinase activity was mutated to Ala, very little Tyr phosphorylation was detected on the Rim11K68A mutant (lanes 5, 6, 11, and 12). In addition, when Tyr-199 in Rim11, which is highly conserved among GSK3 family members, was mutated to Phe, Tyr phosphorylation was abolished (lanes 7, 8, 13, and 14), suggesting that Tyr-199 is the in vivo phosphorylation site. In parallel, the kinase activity of immunoprecipitated HA-Rim11 (Figure 5A, bottom) was determined by in vitro kinase assay with a commonly used GSK3 substrate, phospho-GS peptide, which corresponds to the GSK3 recognition site in rabbit muscle glycogen synthase (Wang and Roach, 1993). There is no significant difference on Rim11 kinase activities in wild-type (columns 3 and 4) and ptp2Δ/− ptp3Δ/− cells (columns 9 and 10). The kinase activity was abolished by K68A (columns 5, 6, 11, and 12) and Y199F mutations (columns 7, 8, 13, and 14).
Figure 5.
Rim11 Tyr phosphorylation and activity are not affected by ptp2Δptp3Δ deletion or Ptp2 and Ptp3 overexpression. (A) Rim11 Tyr phosphorylation and kinase activity are not affected by ptp2Δ ptp3Δ deletion. Wild-type (Y264, lanes 1–8) or ptp2Δ/− ptp3Δ/− double deletion cells (Y164, lanes 9–14) containing vector (pRS313, lanes 1–2), HA-tagged Rim11 wild type (pXZ352, lanes 3, 4, 9, and 10), K68A (pXZ353, lanes 5, 6, 11, and 12), or Y199F (pXZ354, lanes 7, 8, 13, and 14) were harvested at 0 and 24 h after being shifted to sporulation medium. HA-tagged proteins were immunoprecipitated and subjected to anti-Pi-Tyr (top) or anti-HA immunoblotting (middle) to determine Tyr phosphorylation or protein levels, respectively. In parallel, immunoprecipitated proteins were subjected to in vitro kinase assay using phospho-GS peptide as a substrate to determine the kinase activity (bottom). The radioactivity incorporated into the peptide was measured by scintillation counting. Average results from duplicated kinase assays are shown. (B) Overexpression of Ptp2 and Ptp3 has no effect on Rim11 Tyr phosphorylation and kinase activity. Wild-type cells (Y264) harboring HA-Rim11 (pXZ352) were transformed with vector (lane 1), pXZ134 (lane 2), pXZ110 (lane 3), pXZ113 (lane 4), pXZ123 (lane 5) or pXZ136 (lane 6) to overexpress GST or the indicated PTPases. Cellular lysates were prepared from transformants growing in SC-His,Ura, and HA-Rim11 was immunoprecipitated. Tyr phosphorylation (top), protein levels (middle), and kinase activities (bottom) were determined.
We next tested whether Rim11 Tyr phosphorylation and kinase activity are affected by overexpression of Ptp2 and Ptp3 (Figure 5B). GST (lane 2), GTS-Ptp1 (lane 3), GST-Ptp2 (lane 4), GST-Ptp3 (lane 5), or catalytically deficient GST-Ptp3C814G (lane 6) was overexpressed (from a strong ADH1 promoter on multicopy plasmids) in cells expressing only an endogenous amount of HA-Rim11. Although all GTS-PTPases were expressed and functional in yeast cells (Zhan and Guan, 1999), there is no significant difference on the Tyr phosphorylation (Figure 5B, top, lanes 2–6) and kinase activity (bottom, columns 2–6) of Rim11 in PTPase-overexpressing cells.
Therefore, our results demonstrate that Rim11 is Tyr phosphorylated in vivo on Tyr-199. The Tyr phosphorylation is dependent on Rim11's own kinase activity and most likely is due to autophosphorylation (see Figure 7). Our observations with PTPase deletion and overexpression suggest that Rim11 Tyr phosphorylation and kinase activity do not appear to be regulated by Ptp2 and Ptp3 during sporulation. Rim11 is unlikely to be the tyrosine-phosphorylated p43 observed in ptp2Δ/− ptp3Δ/− deletion cells.
Figure 7.
Tyrosine phosphorylation of Rim11 at the Tyr-199 residue is required for Rim11 kinase activity and its in vivo function. (A) Tyrosine phosphorylation and kinase activity are essential for RIM11 function in sporulation. Wild-type (KB600, column 1) cells harboring control vector or rim11Δ/rim11Δ deletion cells (KB268) containing vector (pRS426, column 2), HA-tagged wild-type RIM11 (pKB166, column 3), rim11K68A (pKB199, column 4), rim11Y199F mutant (pKB201, column 5), or rim11Y199E (pXZ356, column 6) were cultured in sporulation medium. The percentage of sporulated cells after 24 h was determined by microscopic counting. (B) Tyrosine phosphorylation and kinase activity are essential for Rim11 function in ime2-lacZ induction. An a/α rim11Δ/rim11Δ ime2-lacZ/IME2 diploid was transformed with vector (column 1), HA-tagged Rim11 wild-type (pKB166, column 2), K68A (pKB199, column 3), Y199F (pKB201, column 4), or Y199E mutant (pXZ356, column 5) and cultured in glucose and acetate or 8 h after cells were shifted to sporulation media. The expressions of ime2-lacZ in glucose (SC-Ura, closed bar), acetate (SAc-Ura, open bar), and sporulation medium (Spo, hatched bar) were determined by β-galactosidase assay. Activities are in Miller units and are averages of two duplicated assays. Y199F and Y199E mutations eliminate Rim11 in vivo kinase activity. HA-tagged Rim11 (pKB166), Rim11K68A (pKB199), Rim11Y199F (pKB201),or Rim11Y199E (pXZ356) was expressed in rim11Δ/rim11Δ deletion cells (KB268). The HA-tagged proteins were immunoprecipitated from yeast cell lysates with anti-HA antibody. The Tyr phosphorylation (top), protein levels (middle), and kinase activity (bottom) were determined by anti-Pi-Tyr, anti-HA immunoblotting, or in vitro kinase assay.
Tyrosine Phosphorylation Is Important for Rim11 to Phosphorylate Substrates but Not for Autophosphorylation
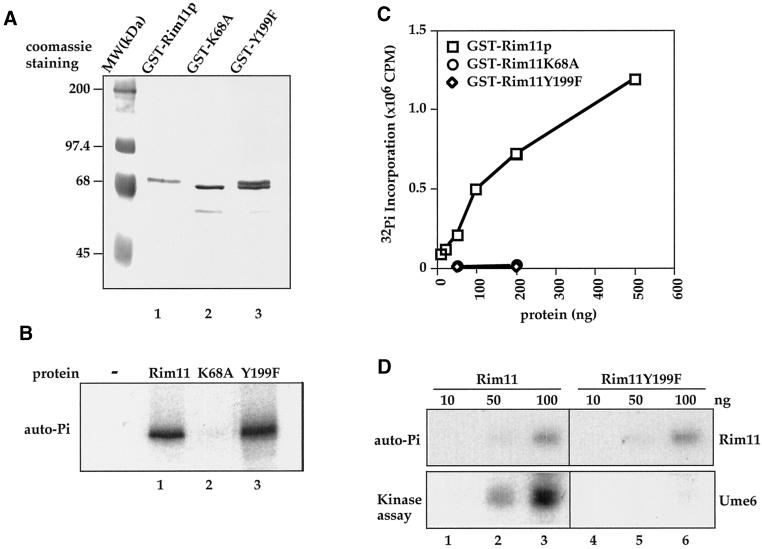
To understand the biochemical significance of Rim11 Tyr phosphorylation, we characterized the catalytic properties of recombinant Rim11 proteins. Wild-type Rim11, K68A, and Y199F were expressed as GST fusion proteins in E. coli and purified by glutathione-agarose affinity chromatography followed by Mono Q ion exchange FPLC (Figure 6A). In SDS-PAGE, the wild-type GST-Rim11 (lane 1) migrated slower than the K68A mutant (lane 2), likely because of autophosphorylation of the wild-type enzyme. GST-Rim11Y199F migrated as a doublet (Figure 6A, lane 3), which has a slower-migrating species that matches the wild-type Rim11 (lane 1) and a faster-migrating species that matches the Rim11K68A (lane 2), suggesting that Rim11Y199F exists as a mixture of hyperphosphorylated (top band) and hypophosphorylated (bottom band) forms. When the purified proteins were incubated with [γ-32P]ATP (Figure 6B), we observed that GST-Rim11 (lane 1) and Y199F (lane 3) proteins showed similar autophosphorylation activity, whereas no autophosphorylation was detected with K68A (lane 2).
Figure 6.
Tyrosine residue 199 in Rim11 is required for the recombinant kinase to phosphorylate substrates but not for autophosphorylation. (A) Coomassie blue staining of purified GST-Rim11, K68A and Y199F protein. GST fusion proteins were expressed in E. coli and purified by glutathione-agarose affinity chromatography and Mono-Q FPLC. (B) Autophosphorylation of GST-Rim11. Equal amounts of GST-Rim11 (lane 1), K68A (lane 2), and Y199F (lane 3) were incubated with [γ-32P]ATP to allow autophosphorylation. Proteins are analyzed on SDS-PAGE and followed by autoradiography. Rim11 and Y199F showed similar autophosphorylation activity, whereas no autophosphorylation was detected on K68A. (C) Kinase activity of recombinant GST-Rim11. The indicated amounts of purified GST fusion Rim11 wild type, K68A, and Y199F were used in in vitro kinase assays with phospho-GS peptide as an artificial substrate. The radioactivity incorporated into peptide was determined by scintillation counting. Square, activity of Rim11; circle, K68A; diamond, Y199F. (D) Tyr-199 is required for Rim11 to phosphorylate Ume6, a physiological substrate of Rim11. Recombinant GST-Ume6 was used as a substrate for GST-Rim11 (lanes 1–3) or GST-Rim11Y199F (lanes 4–6) in the in vitro kinase assay. GST-Ume6 was incubated with the indicated amount of GST-Rim11 or GST-Rim11Y199F in the presence of [γ-32P]ATP. The proteins were resolved by SDS-PAGE and subjected to autoradiography to visualize Rim11 autophosphorylation (top) or phosphorylation of Ume6 (bottom).
To test whether recombinant Y199F mutant protein still retains the kinase activity toward substrates, we incubated Rim11 wild-type, K68A, and Y199F with phospho-GS peptide in the in vitro kinase assays (Figure 6C). The purified wild-type GST-Rim11 was capable of phosphorylating the phospho-GS peptide (open squares). Mutation of the conserved lysine residue in the ATP binding pocket of Rim11 (GST-Rim11K68A) abolished the ability to phosphorylate the peptide substrate (open circle) as well as autophosphorylation activity (Figure 6B, lane 2). In contrast to normal autophosphorylation activity (Figure 6B, lane 3), the activity of GST-Rim11Y199F toward substrate was abolished (Figure 6C, open diamond). Similar results were found with HA-tagged Rim11 immunoprecipitated from yeast (see Figure 5A, bottom, columns 7, 8, 13, and 14, and Figure 7C, bottom, column 4).
To further confirm the above observations and to eliminate possible artifact associated with artificial substrate, we purified a physiological substrate of Rim11, Ume6, and analyzed Rim11 kinase activity toward Ume6. Consistent with the results with peptide substrate, wild-type Rim11 can phosphorylate Ume6 (Figure 6D, bottom, lanes 1–3) in a dose-dependent manner. In contrast, Rim11Y199F did not phosphorylate Ume6 (bottom, lanes 4 and 5); even its autophosphorylation activity (Figure 6D, top, lanes 4–6) is comparable with that of wild-type kinase (top, lanes 1–3). Therefore, our in vitro biochemical data demonstrate that Tyr phosphorylation on Y199F is required for Rim11 to phosphorylate its substrates.
Tyrosine Phosphorylation of Rim11 at the Tyr-199 Residue Is Required for Rim11 Kinase Activity and In Vivo Functions
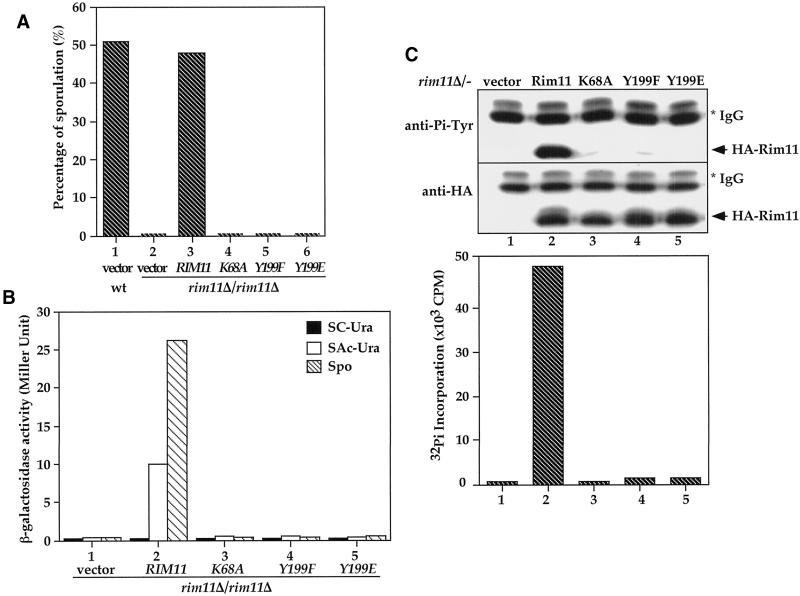
The physiological significance of Tyr phosphorylation on the Rim11 Tyr-199 residue was examined by complementation tests of the sporulation defects in rim11Δ/− deletion cells. We also created a Y199E mutant that we wished to mimic phosphorylation with a negatively charged Glu residue. Isogenic wild-type cells could effectively sporulate (Figure 7A, column 1), whereas rim11Δ/− cells carrying vector had a defect in sporulation (column 2). The sporulation defect was rescued by introducing the wild-type RIM11 gene (column 3). Neither K68A (column 4), Y199F (column 5), nor Y199E (column 6) could effectively rescue the sporulation defect, suggesting that the kinase activity and Tyr-199 phosphorylation are essential for Rim11 in vivo function. These results also indicate that Glu substitution in Rim11 could not mimic phosphorylation; therefore, the Y199E mutant behaved as a loss-of-function allele.
In addition to the complementation assay, we assessed the biological functions of RIM11 wild type and mutants by another physiological assay: induction of meiosis-specific genes. It has been shown that one of the functions for Rim11 is to control the activity of a sporulation-specific transcription factor, Ime1, which regulates the expression of early meiotic-specific genes, including IME2 (Bowdish et al., 1994). Therefore, we tested whether K68A, Y199F, and Y199E can activate IME2 promoter in vivo. An a/α rim11Δ/− IME2/ime2-lacZ diploid was transformed with vector, wild-type RIM11, rim11K68A, rim11Y199F, or rim11Y199E. The expression of the ime2-lacZ reporter gene was determined by assaying β-galactosidase activity (Figure 7B) from cells grown in SC-Ura (glucose medium, filled bars), SAc-Ura (acetate medium, open bars), or sporulation medium (hatched bars). No reporter expression was detected in control cells carrying only vector (column 1). The ime2-lacZ reporter was expressed in cells harboring wild-type RIM11 (column 2) grown in SAc-Ura acetate medium, and this expression was further induced after shifting cells to sporulation medium for 12 h. In contrast, neither rim11K68A (column 3), rim11Y199F (column 4), nor rim11Y199E (column 5) could induce ime2-lacZ reporter in acetate or sporulation medium when introduced into rim11Δ/− cells. Based on these results, we conclude that K68A, Y199F, and Y199E substitutions abolished the ability of Rim11 to activate its physiological substrate Ime1.
In parallel to the physiological assays, we examined Tyr phosphorylation and kinase activity of HA-tagged Rim11 wild type, K68A, Y199F, and Y199E when expressed in rim11Δ/− cells. Tyr phosphorylation (Figure 7C, top, lane 2) and kinase activity (bottom, column 2) were readily detected in immunoprecipitated HA-Rim11. The phosphorylation and kinase activity were essentially abolished by K68A (lane 3 and column 3), Y199F (lane 4 and column 4), and Y199E (lane 5 and column 5) substitutions, correlating to the lack of functions of these mutants. Therefore, our data strongly suggest that the Tyr phosphorylation on Tyr-199 via autophosphorylation is essential for the in vivo functions of Rim11 in meiosis and sporulation.
DISCUSSION
Roles of Protein Tyrosine Phosphatases in Yeast Meiosis and Sporulation
In S. cerevisiae, the involvement of Tyr phosphorylation in cellular signaling has been well established only in the regulation of MAP kinases. Much less is known about the tyrosine phosphorylation in regulation of other cellular responses in yeast. In this report, by analyzing the phenotypes of strains with phosphatase genes deleted, we have demonstrated that Ptp2 and Ptp3 are required for efficient meiosis and sporulation. Homozygous deletion of both ptp2Δ ptp3Δ causes no adverse growth defect but results in a significant reduction of sporulation efficiency. In ptp2Δ/− ptp3Δ/− cells, the expression of sporulation-specific genes was significantly reduced, suggesting that Ptp2 and Ptp3 are required for the initiation of meiotic differentiation. The sporulation defect phentype can be affected by strain background. Double deletion of PTP2/PTP3 in yeast strains dervied from SK background does not result in an apparent sporulation defect (our unpublished results). It is possible that in strains derived from SK background, which was selected for high sporulation effeciency, genetic variation has diminished the requirement of phosphatase in sporulation. The phosphatase activity of Ptp3 appears to be required for its function in sporulation, because a catalytically deficient Ptp3C814G mutant was unable to complement the sporulation defects of ptp2Δ/ptp3Δ in our strains. Because both Ptp2 and Ptp3 are Tyr-specific phosphatases, our results suggest an involvement of Tyr phosphorylation in regulation of meiosis and sporulation. One possible mechanism for PTPase action is that the phosphatases dephosphorylate a Tyr-phosphorylated regulator(s), thus allowing initiation of meiotic differentiation. Consistent with the notion that phosphatases are involved in the regulation of sporulation, Park et al. (1996) have recently reported that deletion of a dual-specificity phosphatase, YVH1, together with ptp2Δ resulted in a significant reduction in sporulation.
Regulation of Rim11 Tyr Phosphorylation
We have shown that there are at least three cellular proteins, p42, p43, and p52, that became Tyr phosphorylated in ptp2Δ/− ptp3Δ/− cells during sporulation. We presented data indicating that p42 is likely to be the MCK1 gene product. Mck1 was previously shown to undergo autophosphorylation on tyrosine residues (Lim et al., 1993). Because deletion of MCK1 results in a pleiotropic phenotype, whereas cells with a homologous GSK3 gene, RIM11, deleted only displays a sporulation defect, we investigated whether p43 is encoded by RIM11. Deletion of RIM11 abolished p43 Tyr phosphorylation. We observed that Rim11 is constitutively Tyr phosphorylated on the Tyr-199 residue, which is conserved among every member of the GSK3 family. Tyr phosphorylation of Rim11 was induced by shifting cells from glucose to acetate medium. However, Rim11 Tyr phosphorylation does not appear to be regulated by Ptp2 and Ptp3, because neither deletion nor overexpression of the PTPase genes had a significant effect on the Tyr phosphorylation of Rim11. Therefore, it is unlikely that Rim11 is the 43-kDa protein whose Tyr phosphorylation is up-regulated in ptp2Δ/− ptp3Δ/− cells during sporulation. Future studies are required to determine the identities of p43 and p52 and the role of tyrosine dephosphorylation of these proteins in sporulation.
In mammalian cells, GSK3 has been shown to be constitutively phosphorylated on Tyr (Hughes et al., 1993). However, it is unclear whether the Tyr phosphorylation is mediated by upstream kinases or by autophosphorylation. In our investigation on the regulation of Rim11 Tyr phosphorylation, we found that the in vivo Tyr phosphorylation of Rim11 is dependent on its own kinase activity, suggesting that autophosphorylation is responsible for the Tyr phosphorylation. It is formally possible that Rim11 Tyr phosphorylation was catalyzed by another kinase whose activity depends on Rim11 in vivo. Our data support a model that the tyrosine phosphorylation of Rim11 is autocatalytic. When kinase-deficient Rim11K68A is expressed in wild-type cells, its Tyr phosphorylation is mostly abolished, arguing against the dependence on another kinase. The autophosphorylation mechanism is further supported by the observation that purified recombinant GST-Rim11 undergoes tyrosine phosphorylation in vitro. Given the high degree of sequence homology between Rim11 and mammalian GSK3 (∼50∼60% homology), it is conceivable that the autophosphorylation mechanism may also be responsible for Tyr phosphorylation of mammalian GSK3 in vivo.
Function of Rim11 Tyr Phosphorylation
No kinase activity toward substrates was detected on Rim11Y199F mutant immunoprecipitated from yeast cells. Loss of kinase activity of Rim11Y199F could be due to either tyrosine phosphorylation-activating Rim11 kinase activity or the mutation causing an inappropriate global folding of Rim11. When Rim11Y199F mutant was purified as recombinant protein from E. coli, the autophosphorylation activity was intact, although no kinase activity toward substrates was detected. Given the efficient GST-Rim11Y199F autophosphorylation activity, we believe that Rim11Y199F should have a correct global structure compared with wild type. Thus our data support the model that phosphorylation of Tyr-199 is required for specific activity of Rim11 toward substrate, possibly playing a role in substrate recognition and phosphorylation rather than kinase activity per se. One of Rim11 in vivo function is to phosphorylate two key meiotic regulators, Ime1 (Bowdish et al., 1994) and Ume6 (Malathi et al., 1997). Phosphorylation of Ime1 and Ume6 by Rim11 promotes the complex formation of Ime1 and Ume6, which induces the expression of sporulation specific genes (Rubin-Bejerano et al., 1996; Malathi et al., 1997). We propose that the function of tyrosine phosphorylation is to activate Rim11 kinase, thereby enhancing the ability of Rim11 to phosphorylate physiological substrates, including Ume6 and Ime1. Consistent with the in vivo requirement of Tyr-199 phosphorylation for Rim11 activity, Y199F mutation, which eliminated the phosphorylation site, completely abolished in vivo functions of Rim11.
In summary, this report demonstrates essential roles of tyrosine phosphorylation in yeast meiosis and sporulation by both protein Tyr phosphatases and kinase. Our results show for the first time that Tyr phosphorylation of Rim11 is required for its kinase activity toward substrates and plays an essential role in the regulation of meiosis and sporulation. The functional significance of tyrosine phosphorylation may also apply to members of mammalian GSK3 family that tyrosine phosphorylation enhances the ability of GSK3 to phosphorylate physiological substrates.
ACKNOWLEDGMENTS
We thank Dr. J. Segall for generous gifts of plasmids and Drs. T. Lanigan, S.J. Stewart, and K. Orth for comments on the manuscript. This work was supported by grant GM89570 from National Institutes of Health (K.-L.G.) and the MacArthur Fellowship Program (K.-L.G.).
REFERENCES
- Bowdish KS, Yuan HE, Mitchell AP. Analysis of RIM11, a yeast protein kinase that phosphorylates the meiotic activator IME1. Mol Cell Biol. 1994;14:7909–7919. doi: 10.1128/mcb.14.12.7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb MH, Goldsmith EJ. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- Dale TC. Signal transduction by the Wnt family of ligands. Biochem J. 1998;329:209–223. doi: 10.1042/bj3290209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Kim SK. Signal transduction and cell fate specification during Caenorhabditis elegans vulval development. Curr Opin Genet Dev. 1994;4:508–516. doi: 10.1016/0959-437x(94)90065-b. [DOI] [PubMed] [Google Scholar]
- Friesen H, Lunz R, Doyle S, Segall J. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 1994;8:2162–2175. doi: 10.1101/gad.8.18.2162. [DOI] [PubMed] [Google Scholar]
- Guan K, Deschenes RJ, Dixon JE. Isolation and characterization of a second protein tyrosine phosphatase gene, PTP2, from Saccharomyces cerevisiae. J Biol Chem. 1992;267:10024–10030. [PubMed] [Google Scholar]
- Guan KL, Deschenes RJ, Qiu H, Dixon JE. Cloning and expression of a yeast protein tyrosine phosphatase. J Biol Chem. 1991;266:12964–12970. [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink RG. Guide to Yeast Genetics and Molecular Biology. Vol. 194. 1991. [PubMed] [Google Scholar]
- Hardy TA, Wu D, Roach PJ. Novel Saccharomyces cerevisiae gene, MRK1, encoding a putative protein kinase with similarity to mammalian glycogen synthase kinase-3 and Drosophila Zeste-White3/Shaggy. Biochem Biophys Res Commun. 1995;208:728–734. doi: 10.1006/bbrc.1995.1398. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hughes K, Nikolakaki E, Plyte SE, Totty NF, Woodgett JR. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 1993;12:803–808. doi: 10.1002/j.1460-2075.1993.tb05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Jacoby T, Flanagan H, Faykin A, Seto AG, Mattison C, Ota I. Two protein-tyrosine phosphatases inactivate the osmotic stress response pathway in yeast by targeting the mitogen-activated protein kinase, Hog1. J Biol Chem. 1997;272:17749–17755. doi: 10.1074/jbc.272.28.17749. [DOI] [PubMed] [Google Scholar]
- Kamps MP. Determination of phosphoamino acid composition by acid hydrolysis of protein blotted to Immobilon. Methods Enzymol. 1991;201:21–27. doi: 10.1016/0076-6879(91)01005-m. [DOI] [PubMed] [Google Scholar]
- Lim MY, Dailey D, Martin GS, Thorner J. Yeast MCK1 protein kinase autophosphorylates at tyrosine and serine but phosphorylates exogenous substrates at serine and threonine. J Biol Chem. 1993;268:21155–21164. [PubMed] [Google Scholar]
- Malathi K, Xiao Y, Mitchell AP. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/mr.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neigeborn L, Mitchell AP. The yeast MCK1 gene encodes a protein kinase homolog that activates early meiotic gene expression. Genes Dev. 1991;5:533–548. doi: 10.1101/gad.5.4.533. [DOI] [PubMed] [Google Scholar]
- Park HD, Beeser AE, Clancy MJ, Cooper TG. The S. cerevisiae nitrogen starvation-induced Yvh1p and Ptp2p phosphatases play a role in control of sporulation. Yeast. 1996;12:1135–1151. doi: 10.1002/(sici)1097-0061(19960915)12:11<1135::aid-yea11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Puziss JW, Hardy TA, Johnson RB, Roach PJ, Hieter P. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol Cell Biol. 1994;14:831–839. doi: 10.1128/mcb.14.1.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shero JH, Hieter P. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1) Genes Dev. 1991;5:549–560. doi: 10.1101/gad.5.4.549. [DOI] [PubMed] [Google Scholar]
- Smith HE, Su SS, Neigeborn L, Driscoll SE, Mitchell AP. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wang Y, Roach PJ. Inactivation of rabbit muscle glycogen synthase by glycogen synthase kinase-3. Dominant role of the phosphorylation of Ser-640 (site-3a) J Biol Chem. 1993;268:23876–23880. [PubMed] [Google Scholar]
- Wilson LK, Benton BM, Zhou S, Thorner J, Martin GS. The yeast immunophilin Fpr3 is a physiological substrate of the tyrosine-specific phosphoprotein phosphatase Ptp1. J Biol Chem. 1995;270:25185–25193. doi: 10.1074/jbc.270.42.25185. [DOI] [PubMed] [Google Scholar]
- Woodgett JR. Regulation and functions of the glycogen synthase kinase-3 subfamily. Semin Cancer Biol. 1994;5:269–275. [PubMed] [Google Scholar]
- Woodgett JR, Plyte SE, Pulverer BJ, Mitchell JA, Hughes K. Roles of glycogen synthase kinase-3 in signal transduction. Biochem Soc Trans. 1993;21:905–907. doi: 10.1042/bst0210905. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy SM, Maeda T, Witten EA, Saito H. Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol. 1997;17:1289–1297. doi: 10.1128/mcb.17.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan XL, Deschenes RJ, Guan KL. Differential regulation of FUS3 MAP kinase by tyrosine-specific phosphatases PTP2/PTP3 and dual-specificity phosphatase MSG5 in Saccharomyces cerevisiae. Genes Dev. 1997;11:1690–1702. doi: 10.1101/gad.11.13.1690. [DOI] [PubMed] [Google Scholar]
- Zhan XL, Guan KL. A specific protein-protein interaction accounts for the in vivo substrate selectivity of Ptp3 toward the Fus3 MAP kinase. Genes Dev. 1999;13:2811–2827. doi: 10.1101/gad.13.21.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Thiele DJ. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell. 1996;87:459–470. doi: 10.1016/s0092-8674(00)81366-5. [DOI] [PubMed] [Google Scholar]