Abstract
We have developed a method that extracts structured information about specimens and their related findings in free-text surgical pathology reports. Our method uses regular expressions that drive a state-automaton on top of XSLT and Java. Text fragments identified are coded against the UMLS®. This paper describes the technical approach and reports on a preliminary evaluation study, designed to guide further development. We found that of 275 reviewed reports, 91% were coded at least so that all specimens and their critical pathologic findings were represented in codes.
Introduction
The Shared Pathology Informatics Network (SPIN) establishes an Internet-based virtual database that will allow investigators to locate appropriate human tissue specimens for their research.1 Surgical pathology reports are the primary source for tissue information; and while laboratory and microbiology reports are now quite commonly available in the form of structured data, surgical pathology reports are generally only available as free-text reports. In this paper we discuss our approach of transforming surgical pathology reports into structured data and we discuss experiences made based on a preliminary evaluation of these methods.
Text reports can be searched either by text indexing (word search) or by concept indexing (concept search). Word search is a generic easy to implement approach to locate information and such text indexing technology is readily available. Concept search has the advantages that one can find synonyms and related concepts without necessarily using the same words, but is more difficult to implement because free-text expressions must first be mapped to a canonical terminology.
Common approaches to free text are deep natural language understanding and information extraction. Medical investigators have applied language understanding techniques most commonly to discharge summaries, clinical notes, and radiology reports, and – outside of individualized patient care – to biomedical journal articles.2,3,4 Given the freedom in expression typical of these types of medical texts, parsing such texts requires very deep and detailed language models and results in highly generic data structures that contain much variability and are therefore hard to query.
Conversely, information extraction is an approach that only captures key information, which can be done with simpler language models. Noun phrase detection techniques have been used to find concepts suitable for indexing (tissue, diagnoses, etc. are all noun phrases.) In the domain of surgical pathology certain information extraction methods have become known as “autocoders” since they assign codes (e.g., SNOMED codes) to free text reports.5 More generic information extraction tools (such as the ones demonstrated at the Message Understanding Conference (MUC) are often geared to finding nouns with simple relationships, but typically cannot recover nested relationships.6
Special Consideration for Surgical Pathology
We hypothesized that surgical pathology reports would be easier to parse than radiology reports, clinical notes or discharge summaries, because they often conform to a certain structure that can be defined in considerable detail. This structure has been informally codified by the Association of Directors of Anatomic and Surgical Pathology (ADASP) publication of a Standardization of the Surgical Pathology Report7. This standard defines the common sections Gross Description, Microscopic Description, Final Diagnosis, etc. In turn, the standard defines the Final Diagnosis section as a list of all separately identified tissues specified by (a) organ, (b) site, and (c) procedure, with diagnoses and other observations listed under each such tissue. The standard even goes into such detail as prescribing that observations be set off from their tissue subject using a dash or a colon.
Methods
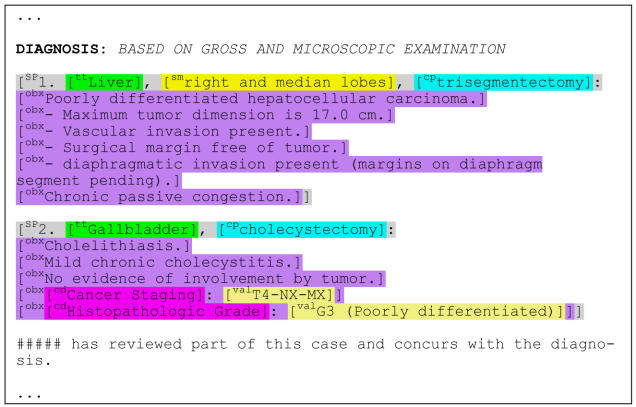
To assess the performance of a simple approach to parsing and coding diagnosis sections, based primarily on common text-report formatting style elements, we took a convenience sample of 622 reports from one pathology laboratory. We de-identified the reports using a scrubbing method that we developed as part of the SPIN project under IRB approval (#0103-36). This scrubber was based on the Thomas-algorithm,8 but with enhanced scrubbing specificity and an automatic removal of all text except for report-sections whose title contained the word “diagnosis”. The resulting scrubbed report-fragments contained no patient identifying data. Among these report-fragments 347 (56%) did not contain any explicit specimen information but only vague “descriptive diagnosis” (e.g. with the text “normal” or “see comments”.) These were excluded them from further analysis. The remaining 275 report-fragments were the subject of this study. Figure 1 shows a typical such final diagnosis section.
Figure 1.
Example a surgical pathology report’s final diagnosis section. The colors and brackets are added by the parser, but the original words and spacing are preserved.
The Parser
We used a regular-expression-based parser that searched for specimen “headers,” i.e., that part of the diagnosis section, that identifies the tissue- type (organ, e.g., Liver) site-modifiers (e.g., right lobe), and collection-method (procedure, e.g., segmentectomy). When the parser finds such a specimen header, it assumes that the immediately following text describes observations about that specimen. The observations may be diagnoses, assessments of surgical margins, invasion, grading and staging, etc. Figure 2 shows the structured information that the parser generates.
Figure 2.
XML output for the example in Figure 1.
We implemented this parser entirely in XSLT9, using Saxon10, with some calls to standard Java class libraries for regular expression matching. For example, a single regular expression matches an entire specimen header using capturing groups to extract the components for tissue-type, site modifiers, and collection-method. When a specimen header is found, other regular expressions find diagnostic sentences. The regular expressions are applied depending on context, which is controlled by a pushdown state automaton. The “grammar” is defined in XML that is a hybrid of XSLT with blended-in extensions defining the parser’s states and transitions. A “compiler-compiler” transforms the grammar definition into pure executable XSLT, where states are mapped to XSLT modes and transitions to XSLT templates. The events are defined by regular expressions.
The Coder
As a phrase coder we use the National Library of Medicine’s MMTx, the Java rewrite of the MetaMap UMLS coder.11 Because the MMTx system, although written in Java, is designed as a stand-alone program and relatively hard to move, we have wrapped a simple XML Web-Server around MMTx, so that we only need a single coder server installation. The coder is used strictly as a phrase coder, i.e., the parser sends small text fragments to the coder as HTTP GET request using the XPath function document with the phrase to be coded as part of the URL argument. The response to these requests is a small XML document of mappings and concepts sorted by descending score.
Because the parser is aware of specific parts of the text, it can guide the coding process by accepting only certain UMLS semantic types that fit the expected meaning of the phrase. Based on experience of coding and reviewing a training set, we allowed the UMLS semantic types listed in Table 1 for the tissue-type, site modifiers, collection-method, and diagnosis components. Particularly for tissue-type, we accept a wide range of semantic types because pathologists often use terms metaphorically, e.g., mentioning “shoulder” when they mean “skin of shoulder” and often the difference is so slight that the UMLS does not include the appropriate tissue concepts. For example, the word “tumor” is classified in the UMLS as a “neoplastic process” which is a “biologic function”. However, when used as a specimen type it is meant as an “anatomic structure” for which the UMLS lists no concept. Note that we have excluded the semantic type “finding” from coding observations because in our experience it terms such as “maximum”, and “present” which are UMLS “findings”, have shadowed more useful terms in the phrase, thereby generating many useless codes.
Table 1.
Matrix showing the semantic types expectations for the fields tissue-type (tt), site-modifier (sm), collection-method (cm), and diagnosis (dx), 1 means expected, blank or 0 means rejected.
| Semantic Type | tt | sm | cm | dx |
|---|---|---|---|---|
| Acquired Abnormality (acab) | 1 | 1 | ||
| Anatomical Abnormality (anab) | 1 | 1 | ||
| Body System (bdsy) | 1 | |||
| Body Location or Region (blor) | 1 | 1 | ||
| Biomedical or Dental Material (bodm) | 1 | |||
| Body Part (bpoc) | 1 | 1 | ||
| Body Space or Junction (bsoj) | 1 | |||
| Cell or Molecular Dysfunction (comd) | 1 | |||
| Diagnostic Procedure (diap) | 1 | |||
| Disease or Syndrome (dsyn) | 1 | 1 | ||
| Embryonic Structure (emst) | 1 | |||
| Finding (fndg) | 1 | 0 | ||
| Neoplastic Process (neop) | 1 | 1 | ||
| Organ (orga) | 1 | |||
| Pathologic Function (patf) | 1 | |||
| Substance (sbst) | 1 | |||
| Sign or Symptom (sosy) | 1 | 1 | ||
| Spatial Concept (spco) | 1 | |||
| Tissue (tisu) | 1 | |||
| Therapeutic or Preventive Procedure (topp) | 1 |
The observations under each specimen contain more complexity besides simple diagnostic assertions, including (a) negation, (b) uncertainty, (c) surgical margins, (d) invasiveness, (e) dimensions and (f) histological grading (of cancerous tissue), which are all relevant for us. Currently, our parser finds negation, uncertainty and observations of margins by the following simple approach (prototypical for detecting the other kinds of information): First the parser cuts observation statements into smaller phrases separated by comma, semicolon, and by the words “and”, “with”, or “without”. Then, when the word “margin” or “margins” is mentioned in the phrase, it assumes that the phrase makes a statement about a margin. Likewise, a negating word “no”, “not”, “none”, “negative”, “exclude( d)”, “free” and “without” signals that the entire phrase is negated. (Initially we did not include “denies” and “absence”12.) The words “cannot”, “question(able)”, or “doubt(ful)” signal that the phrase is uncertainty. For example: “a firm diagnosis of malignancy […] cannot be made” was correctly marked as uncertain.
Rating of Results
The results were reviewed in a web-based review application, where an excerpt of the XML encoding was displayed side-by-side with the relevant scrubbed freetext sections. The coded phrases were highlighted by color codes and enclosed in brackets (see Figure 1.)
For each report-fragment, the reviewer – who, for this preliminary evaluation was the same person as the developer and author of this paper (an M.D. without special training in pathology) – marked issues and then rated the overall coding on the following scale:
Excellent: all interesting findings are coded. Even benign or normal findings must be coded
Good: all critical findings are coded. Minor or benign descriptive concepts need not be coded.
Sufficient: most critical findings are coded. Some critical findings may not be coded if they are represented by other findings that are coded. This includes if one specimen part among others has been missed
Defective: major findings are not coded or negations not detected.
Results
Table 2 summarizes the results of the rating. It shows that about 90% of the coding was at least sufficient, i.e., had all of their critical findings represented by codes. Table 3 lists 5 kinds of errors identified through the review along with their frequency.
Table 2.
Distribution of Ratings
| rating | n | n/ 275 |
|---|---|---|
| excellent | 81 | 29% |
| good | 117 | 43% |
| sufficient | 51 | 19% |
| defective | 26 | 9% |
Table 3.
Distribution of Error Types
| type of error | n | n/ 26 |
|---|---|---|
| UMLS coding | 12 | 46% |
| semantic type neglect | 5 | 19% |
| unexpected format | 5 | 19% |
| negation scope | 2 | 8% |
| unknown (caused by a bug) | 2 | 8% |
In 4 cases, the UMLS in that version did not contain an appropriate concept for the phrase (e.g., “fibrocystic change”, “squamous proliferation”, “lipofibroma”, and also “lipoma”.) Note that missing histo-pathologic concepts were the cause of less than optimal coding in a number of other cases as well but did not come to light in our review to date (since we did not rate the coding quality with more scrutiny, see discussion below.)
In 2 cases, the UMLS lexicon list did not contain a synonym or the coder did not find a synonym that could have been found with word stemming (“ethmoids” for “ethmoid bone” and “content” for “contents”.)
In 5 cases the coder could not handle exceptional situations (e.g., “unknown site, procedure not specified”) or unusual expressions and typos. E.g., “disorder proliferative endometrium” (for the histologic substrate of dysfunctional uterine bleeding) was coded simply as “disease”. A common type of problems is the coding of description of normal findings (e.g., “Benign endocervical elements”) or inconclusive findings (e.g.,”Necro-inflammatory debris and food fragments”.)
In one case “Right upper lobe of lung” (as tissue type) was coded as “Right upper zone pneumonia” because this coding had received a slightly better score than a more appropriate coding. This problem with spurious or sub-optimal codes can be seen more frequently in report-fragments otherwise satisfactorily coded.
In 1/5 of the cases the key concept was coded but the coding was rejected because of an unexpected semantic type. We failed to add “Body Substance” (bdsu) to the set of semantic types acceptable for tissue- type and therefore failed to code “Urine” and “CSF”. In one case we missed “acute cellular rejection” because we do not accept “Organ or tissue function” (ortf) as observations. The reason we rejected this semantic type was that it would have caused the very common reference “See note” to be coded as the concept of “vision”.
1/5 of the errors were due to unexpected text format that the parser was not prepared to handle. This includes interjected notes inside the diagnosis sections, and specimen headers broken into two lines of text.
Two of the defective results were due to missed negation (just 2% of 143 properly detected negations.) In these two cases the negated concept was separated from the negation keyword by a comma (e.g., “no evidence of granuloma, dysplasia or malignancy” coded as negated “granuloma” but affirmed “malignant neoplasm”.)
Discussion
Our parser is obviously handicapped by the fact that it relies only on structural clues (described by regular expressions.) This leads to great difficulties with reports that are less consistently formatted. For instance, we need to have a clear distinction between specimen headers and observation statements underneath, which often is given through the placement of colons or dashes and clues such as indention. We would like to move towards more semantic constraint checking during parsing. For example, when the a text assumed to be a specimen header does not have the semantic content of at least a tissue-type (organ, analyzed substance), we have to attempt coding an observation under the prior specimen instead.
In order to drive the parser state-machine by semantics, however, we need to improve on our use of the UMLS. Particularly we need to subset the UMLS into different sets of source code systems and semantic types when coding, so as to reduce the cases of spurious codes assigned to phrases when more appropriate candidates are available given our semantic expectations. We should use the MetamorphoSys program, which is distributed with the UMLS Knowledge Source and subset our UMLS database to only include the relevant code systems. This would likely reduce the number of spurious codes. However, with MetamorphoSys, the subset is static and cannot be dynamically changed to accommodate changing semantic expectations during the coder process.
Of course, a successor to our prototype needs to undergo more thorough evaluation with a better assessment of the quality of the coding and the detail of what has been found. These reviews should best be carried out by pathologists not involved in the development of our coder. However, we believe that before this is useful, we should at least improve on the terminology model so that we can implement more normalization (e.g., mapping “left hand” and “hand, left” to the same concept “hand” with site-modifier “left”.)
We also plan to translate the UMLS CUIs generated during coding into terms from a single source vocabulary that is appropriate for a single kind of concept. For instance, if we translate the tissue diagnosis to ICD-O, we can then cross-reference and validate our automatically generated codes with cancer registry data. Often we also find SNOMED codes in the pathology reports which we should recognize and compare our UMLS coding against those SNOMED codes.
Conclusion
Our coder method is simple and the grammar is small and maintainable, based mostly on regular expressions and produces an output that has the right level of structure, detail of information but also uniformity. Based on our performance review we are cautiously optimistic that our approach makes sense to pursue further. In order to improve our parser’s performance, we need to better adapt the UMLS to the specific needs of surgical pathology reports.
Acknowledgments
This research was performed at the Regenstrief Institute and funded, by National Cancer Institute grant U01CA91343, a Cooperative Agreement for The Shared Pathology Informatics Network. We thank our colleagues of the SPIN consortium. Special thanks to Drs. M. Becich and J. Gilbertson (University of Pittsburgh) for the discussions that have sparked the inspiration to this approach. Thanks also to Alan Aronson and the MMTx team for their cooperation.
References
- 1.Shared Pathology Information Network (SPIN): http://www.cancerdiagnosis.nci.nih.gov/spin/
- 2.Sager N, Lyman MS, Bucknall C, Nhan NT, Tick LJ. Natural Language Processing and the Representation of Clinical Data. JAMIA. 1994;1(2):142–160. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122(9):681–8. doi: 10.7326/0003-4819-122-9-199505010-00007. [DOI] [PubMed] [Google Scholar]
- 4.Haug PJ, Koehler S, Lau LM, Wang P, Rocha R, Huff SM. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care. 1995:284–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Carter KJ, Rinehart S, Kessler E, Caccamo LP, Ritchey NP, Erickson BA, Castro F, Poggione MD. Quality assurance in anatomic pathology: automated SNOMED coding. JAMIA. 1996 Jul–Aug;3(4):270–2. doi: 10.1136/jamia.1996.96413134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Institute for Standards and Technology (NIST). Message Understanding Conference Proceedings. http://www.itl.nist.gov/iaui/894.02/related_projects/muc/
- 7.Association of Directors of Anatomic and Surgical Pathology. Standardization of the Surgical Pathology Report. http://www.panix.com/~adasp/standSPrep.htm
- 8.Thomas SM, Mamlin B, Schadow G, McDonald C. A successful technique for removing names in pathology reports using an augmented search and replace method. Proc AMIA Symp. 2002:777–81. [PMC free article] [PubMed] [Google Scholar]
- 9.World Wide Web Consortium. XSL Transformations XSLT) 2.0. [W3C Working Draft]. http://www.w3.org/TR/xslt20/XSLT
- 10.Kay M. SAXON: The XSLT Processor. http://saxon.sf.net
- 11.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. See also http://mmtx.nlm.nih.gov/ [PMC free article] [PubMed]
- 12.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: a quantitative study using the UMLS. JAMIA. 2001 Nov–Dec;8(6):598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]