Abstract
To better serve an antibiotic guidance program, we hypothesized that the relatively few antibiotic susceptibility measurements conducted in the microbiology laboratory could be extended to predict antibiotic susceptibilities for all antibiotics on the hospital formulary using expert infectious disease logic. With the assistance of infectious disease specialists, we developed these logic rules and then applied them to 26,196 unique patient culture specimens and the accompanying 334,131 antibiotic susceptibility measurements generating 804,809 additional predicted bug-drug susceptibility data points. From the resulting data set, the antibiotic susceptibility profile for one pathogen, Streptococcus pneumoniae, is highlighted herein. We then incorporated the extended susceptibility profiles into a computerized antibiotic guidance program that matches current patients of interest with the positive cultures from past similar patients and calculates predicted effective antibiotic therapy. We conclude that this method successfully derives antibiotic predictions and merits further testing to evaluate its potential use in the hospital environment.
INTRODUCTION
Physicians must frequently initiate antibiotic therapy in infected patients prior to knowledge of the infecting pathogen and its susceptibility profile, a course of action known as providing empiric therapy. Physicians have handbooks, medical literature, experts in infectious disease, and their own experience as guides on the best antibiotic choices for a given scenario. The provision of inadequate empiric therapy has been found to place the patient at two-fold increased risk of death in adult ICU patients,(1) and a five-fold increased risk of death in pediatric patients.(2) In such circumstances, physicians may elect to initiate broad spectrum therapy to provide antibiotic coverage against as many potential pathogens and potential resistance as possible. However, the pressure to provide broad therapy is tempered by antibiotic costs, toxicities, and the increased rate of antibiotic resistance engendered by their use.
To assist physicians with the best targeting of empiric antibiotic therapy, investigators have developed computerized antibiotic decision support tools that query large databases of microbial cultures, mathematically summarize the antibiotic resistance data, and provide antibiotic therapy suggestions to physicians for given clinical scenarios. Leibovici, et al, reported the results on an offline analysis of one computer program, documenting a potential improvement in targeting of empiric antibiotic therapy.(3) The work by Leibovici follows earlier work by Evans, et al, which documented an improvement in empiric targeting of antimicrobials in an offline study(4) and followed with an online evaluation of one-year’s use of the system in an intensive care unit.(5) The portion of this antimicrobial management program that calculates empiric therapy from the past bacterial cultures benefits from a microbiology laboratory practice of testing the majority of bacterial isolates against a large panel of antibiotics to determine the pattern of antibiotic susceptibilities.(6) This large panel of antibiotic susceptibility tests provides a rich database of results and mathematically maximizes the potential to find a common effective antibiotic across a broad range of diverse bacterial culture results. Unfortunately, not all microbiology labs test all isolates against a broad panel of antibiotics. Costs are a limiting resource, and therefore, many labs only test and publish antibiotic susceptibilities that are clinically relevant for the infected patient.
To address this issue of limited antibiotic susceptibility testing, we describe herein a method derived to safely amplify the susceptibility data for pathogens, using simple expert infectious disease rules. The rules are captured in a pathogen-antibiotic logic matrix, called the bug-drug logic table. This method uses the measured antibiotic susceptibility data to provide predictions of antibiotic susceptibility for all other antibiotics on formulary. This technique amplifies the knowledge base available to find common effective one- and two-drug combinations for a given clinical scenario.
The hypothesis of this current work is that a bug-drug logic matrix founded on expert rules can be used to successfully amplify the limited antibiotic susceptibility testing common to many hospitals. The overall clinical hypothesis driving this work is that a computer program founded on the local microbiology cultures database can improve empiric targeting of antimicrobials while reducing costs, toxicities, and environmental impact of antibiotic use.
METHODS
The last eleven years of microbiology data from the Sunquest laboratory information system has been stored at WVU Hospital. For this project, five years of culture results and susceptibilities data were downloaded into a password-protected Microsoft Access database. Infectious disease physicians partnered with the authors to develop a system of expert rules that would take a limited amount of information about a bacterial isolate’s antibiotic susceptibilities and extend or amplify this data to predict the pathogen’s susceptibility to all other antibiotics on the hospital’s formulary. For instance, Streptococcus pneumoniae is tested against 6 antibiotics in our microbiology lab, yet 44 different antibiotics are available on formulary. One rule of thumb that physicians use can be stated in a simple if-then statement. If Streptococcus pneumoniae is susceptible to penicillin, then it is susceptible to other beta-lactam antibiotics such as ampicillin, amoxicillin, and cefuroxime. This type of logic can be developed for all pathogens of interest, generating predictions for all antibiotics on formulary. Another example, if Escherichia coli is measured susceptible to piperacillin, then it is also susceptible to piperacillin/tazobactam.
These logic statements were developed through extensive knowledge engineering sessions held with the infectious disease experts. In generating these statements, we used a conservative, or pessimistic, guiding principle: never assign an antibiotic a substitute, or surrogate, that may be more effective, only allow surrogates that are less effective. All if-then decisions generated using were incorporated into a pathogen-antibiotic logic matrix call the bug-drug logic table. Table 1 shows an example of a subset of entries for Streptococcus pneumoniae. In the matrix, at the intersection of Streptococcus pneumoniae and penicillin, is an “M” for “measured,” signifying that the susceptibility of the pneumococcus isolate against penicillin is commonly measured in the lab. However, at amoxicillin, “~penicillin” is placed signifying that amoxicillin is not tested, and that the susceptibility result for penicillin (susceptible or resistant) can be used as a surrogate for amoxicillin. Some antibiotics are never effective against a certain pathogen (“R” for resistant), while some are always effective (“S” for susceptible).
Table 1.
A subset of entries in the bug-drug logic table. M = “measured” - the susceptibility of the isolate for this antibiotic is measured in the lab. R = “resistant” - this antibiotic is never effective for this pathogen. S = “susceptible” - currently, this antibiotic is always effective for this pathogen. The entry “~penicillin” signifies that the result for penicillin serves as a surrogate for this antibiotic.
| Drug | Streptococcus pneumoniae |
|---|---|
| penicillin | M |
| ampicillin | ~penicillin |
| amoxicillin | ~penicillin |
| cefuroxime | ~penicillin |
| ceftriaxone | M |
| cefotaxime | M |
| ceftazidime | R |
| vancomycin | S |
The bug-drug logic table is then used to extend the measured antibiotic susceptibilities for all cultured pathogens against all antibiotics on formulary for the entire dataset from the last five years. This enlarged antibiotic susceptibilities results set can then be used to drive the antibiotic guidance program.
Antibiotic recommendations are derived from the database using a simple tally of past measured and predicted antibiotic susceptibilities profiles. To generate these recommendations, a database query form was developed with language and principles designed to be understandable to the physician end user. The interface allows one to find the pathogens and antibiotic susceptibility profiles from patients in the past five years who match current patients of interest. Potential matching parameters include: infected specimen of interest (blood, urine, wound, etc), location in the hospital (MICU, PICU, etc), age category, gender, and category of infection – community versus hospital-acquired. The user is also given the opportunity to tailor the antibiotic output recommendations for important considerations such as patient allergies, age, renal function, and severity of illness (intravenous versus oral antibiotics).
The cultures from past patients that match the parameters of interest in the current patient are then selected and the best one- and two-drug antibiotic combinations are tallied against this list of queried pathogens and their actual measured and predicted antibiotic susceptibilities. The list of potential effective antibiotics is then trimmed based on the input from the user for considerations such as allergy, age, site of infection, and formulary restriction. The most effective antibiotics from the list are then resorted and ranked based on favorable costs, toxicities, and environmental impact profiles.
RESULTS
For the five years from 1997 through 2001, we found 26,196 unique patient culture specimens associated with 334,131 antibiotic susceptibility measurements. Using the bug-drug logic table, we amplified these results, generating an additional 804,809 predicted bug-drug susceptibility data points. From the resulting data set, a close examination of the tallied antibiotic susceptibility profile for Streptococcus pneumoniae shows that the list of potentially effective antibiotics is much larger than the seven antibiotics actively reported by the lab. Knowledge from these 6 measured antibiotic susceptibilities was amplified to predict susceptibility results for another 38 antibiotics, with 11 of them potentially efficacious for any given Streptococcus pneumoniae isolate (Table 2). Inspection shows that the summation for cefuroxime predicts it is only as effective as penicillin and that erythromycin is not predicted to be effective at all. In reality, the effectiveness of cefuroxime in labs that test this antibiotic against Streptococcus pneumoniae is usually in the range of 60–80%, and erythromycin is usually greater than 70%. The lower values in our dataset are due to the limitation of the technique, which uses pessimistic principles in generating the rule set - “never assign an antibiotic a surrogate that may be more effective, only allow surrogates that are less effective” and in the tallying of the results - “always assume that the isolate is resistant unless it or its surrogate antibiotic are found to be susceptible.” For the case of cefuroxime, the former principle dictates that the less effective antibiotic penicillin is assigned as a surrogate rather than the potentially more efficacious antibiotics cefotaxime or ceftriaxone. Potential methods to better match the predicted antibiograms with the national published antibiograms are discussed later.
Table 2.
“Extended” antibiotic susceptibility table for Streptococcus pneumoniae combining measured antibiotic susceptibilities (shown in bold) with predicted antibiotic susceptibilities (shown in italics). Not all 44 formulary antibiotics are shown.
| Drug | Percent Susceptible |
|---|---|
| vancomycin | 100% |
| chloramphenicol | 94% |
| cefotaxime | 73% |
| clindamycin | 71% |
| ceftriaxone | 68% |
| cefepime | 68% |
| penicillin | 45% |
| ampicillin | 45% |
| amoxicillin | 45% |
| cefuroxime | 45% |
| amoxicillin/clavulanate | 45% |
| oxacillin | 45% |
| erythromycin | 0% |
| ceftazidime | 0% |
Despite the above limitation, the end result of this method is a set of measured and predicted antibiotic susceptibilities that may be used as the foundation for a database-driven antimicrobial guidance program that calculates one- and two-drug antimicrobial recommendations that are clinically plausible to expert physicians and worthy of further evaluative testing (Figure 1). Although not as novel as the bug-drug logic tables, the method of generating antibiotic recommendations in our system merits mention and is more fully described in the caption of the figure.
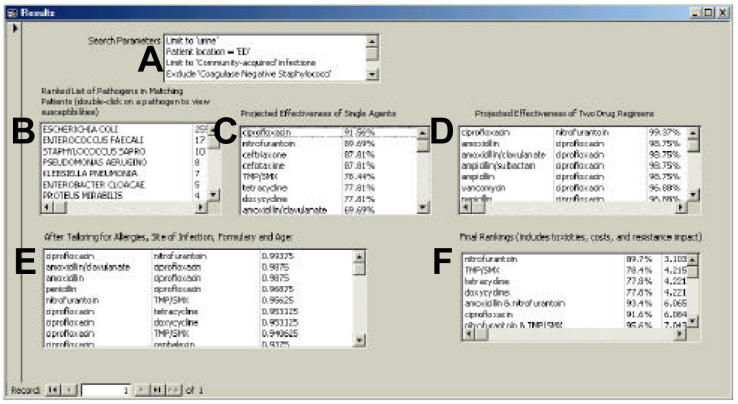
Figure 1.
The results screen of the prototype of the antibiotic guidance program. The scenario entered in this example is of a urinary tract infection in a woman presenting to the emergency department. The requested output is oral antibiotics only. In this figure, the data boxes are labeled with letters for clarity. The search parameters are repeated back to the user in the top box, labeled A. Next, the ranked list of pathogens from patients matching the query parameters are shown in the box labeled B. Escherichia coli is the most common isolate. Box C displays the projected effectiveness of single antibiotics, based on the tally of the measured and predicted antibiotic susceptibilities of the matching pathogens. These antimicrobials are ranked in descending order of projected effectiveness. Box D shows a similar tally for the 990 two-drug combinations of our formulary antibiotics. Box E displays the list of agents after patient allergies, site of infection, antibiotic restrictions, age-based toxicities, and requested formulation (intravenous versus oral) have been considered. The final recommendation, nitrofurantoin, is presented in box F. This suggestion is calculated by taking the most effective antibiotics on the left and then re-ranking based on drug cost, toxicities, and environmental impact scores. In this system, each drug is assigned a score from one to three in each of these categories. Nitrofurantoin is inexpensive (cost score = 1), relatively safe (toxicity score = 1), and has a low risk or impact on environmental resistance (resistance impact = 1). The final score is calculated by subtracting the predicted effectiveness, 89.7%, from 100%, dividing by 100, and adding the cost, toxicity, and resistance impact scores (((100% − 89.7%)/100) + 1+ 1 + 1 = 3.103 for nitrofurantoin). For the final score, lower is better. In the production version of this system, only the search parameters, the matching pathogens, and the final antimicrobial rankings will be shown to the users.
DISCUSSION
The above methodology has rendered a working antibiotic guidance tool that potentially brings better information to the bedside than is routinely available. A unique advantage of this type of program is that as bacteria develop resistance and susceptibility profiles evolve, the calculated antibiotic recommendations will likewise evolve. This provision of calculated empiric antibiotic therapy can therefore potentially impact critical clinical decisions and improve patient outcomes. Additionally, the program may positively influence associated antibiotic costs, toxicities, and environmental impact of unnecessary use of broad-spectrum antibiotics.
As described above, the method of extending antibiotic susceptibility predictions does not generate antibiotic susceptibility profiles that fully mirror known nationally published antibiograms. The simplest solution would be to implore the microbiology lab to test all pathogens against all antibiotics. However, that is unlikely to occur, given cost considerations. As designed presently, use of surrogate antibiotics are limited to drugs of the same or similar class. However, in the future, one could enhance the bug-drug logic tables to take advantage of increasing scientific knowledge regarding mechanisms of antibiotic resistance. For instance, susceptibility of a pathogen to drugs in class I may predict susceptibility to drugs in class II secondary to an absence of a common antibiotic efflux mechanism found in this species but not seen in this isolate. Conversely, rather than measuring antibiotic susceptibilities in the lab, the genetic material of an isolate may be analyzed by PCR techniques and a range of antibiotic susceptibilities be inferred by the presence or absence of known susceptibility genes.
Lastly, this antibiotic program matches patients by using simple parameters such as age and location. As hospital information systems evolve, improvements in matching algorithms and antibiotic targeting may be demonstrated with systems that match on such factors as recent antibiotic use, presence of neutropenia, innate genetic susceptibilities of the patient, etc. The methodologies of antibiotic guidance programs will need to evolve as both medical knowledge and the depth of clinical information systems advance.
CONCLUSION
Bug-drug logic tables can be used to successfully derive antibiotic susceptibility predictions, which may then serve as a foundation for a computerized antibiotic guidance program. Our computer program merits further offline testing to assess its safety and evaluate its effectiveness.
REFERENCES
- 1.Kollef MH, Sherman G, Ward S, Fraser VJ. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest. 1999;115(2):462–74. doi: 10.1378/chest.115.2.462. [DOI] [PubMed] [Google Scholar]
- 2.Leibovici L, Shraga I, Drucker M, Konigsberger H, Samra Z, Pitlik SD. The benefit of appropriate empirical antibiotic treatment in patients with bloodstream infection. J Intern Med. 1998;244(5):379–86. doi: 10.1046/j.1365-2796.1998.00379.x. [DOI] [PubMed] [Google Scholar]
- 3.Leibovici L, Gitelman V, Yehezkelli Y, et al. Improving empirical antibiotic treatment: prospective, nonintervention testing of a decision support system. J Intern Med. 1997;242(5):395–400. doi: 10.1046/j.1365-2796.1997.00232.x. [DOI] [PubMed] [Google Scholar]
- 4.Evans RS, Classen DC, Pestotnik SL, Lundsgaarde HP, Burke JP. Improving empiric antibiotic selection using computer decision support. Arch Intern Med. 1994;154(8):878–84. [PubMed] [Google Scholar]
- 5.Evans RS, Pestotnik SL, Classen DC, et al. A computer-assisted management program for antibiotics and other antiinfective agents. N Engl J Med. 1998;338(4):232–8. doi: 10.1056/NEJM199801223380406. [DOI] [PubMed] [Google Scholar]
- 6.Personal communication with RS Evans; 2001.