Abstract
The newly developed U.S. Common Medication Information Infrastructure was used as a basis to capture and formally express the properties of drugs relevant to research and the clinical application of pharmacogenomics. Two associated taxonomies within the model, Mechanism of Action and Physiologic Effect, were enriched to accommodate pharmacogenomic use-cases; the 4,000 active ingredients in the VA NDF-RT drug file were related to the enhanced taxonomies. Pharmacokinetics were independently modeled for pharmacogenomics and tested against thirty-one high-profile drugs to demonstrate our approach.
Introduction
The advent of the human genome has enabled or accelerated many medical disciplines. Not the least of these is the field of pharmacogenomics[1], or the role of inheritance in the individual variation in drug response. The promise of this field is an ability to identify the right drug and dose for each patient, despite demonstrable variability among individuals to activate, therapeutically utilize, or metabolize a given drug. Practical informatics application of pharmacogenomics data includes the reduction of adverse drug events [2], which has been demonstrated to have important clinical benefits[3].
The field of pharmacogenomics was introduced nearly 25 years ago, when it was shown that the activity of thiopurine S-methyltransferase (TPMT) in red blood cells could divide populations into at least three groups: persons with low, normal, and high levels of enzymatic functioning[4]. Furthermore, these traits are inherited in an autosomal codominant fashion. Subsequently, the clinical importance of these observations on the dosing of TPMT-metabolized drugs such as thiopurines was recognized, including an increased risk for life-threatening myelosuppression among patients with little or no TPMT functional activity.
Since that time, the field of pharmacogenomics has grown dramatically.The availability of high-throughput methods to discover underlying non-synonymous SNPs within a gene’s coding region has uncovered previously unappreciated variability in human metabolic functioning[5]. The NIGMS established the Pharmacogenomics Network in 2000 to coordinate the work of scientists on how genomic variations influence patients’ responses to drug therapy*. A specialized database of pharmacogenomic findings is being maintained as part of this network†[6].
Although appropriate ontologies for pharmacogenomics have been carefully considered[7], network members and NIGMS recognized that a publicly accessible ontology for drugs themselves was not available [8], never mind adopted for research or clinical applications in pharmacogenomics. Since drugs comprised “the third leg of the stool” together with genomics and phenotype, the critical nature of identifying or adopting an appropriate drug ontology was widely recognized. Fortunately, a multi-departmental initiative developed within the U.S. government, involving the Department of Veterans Affairs (VA), the Food and Drug Administration (FDA), and the National Library of Medicine (NLM), to establish a common medication information infrastructure[9].
This report outlines an NIGMS-funded initiative to adapt the emerging U.S. medication infrastructure to the clinical and research needs of pharmacogenomics.
Background
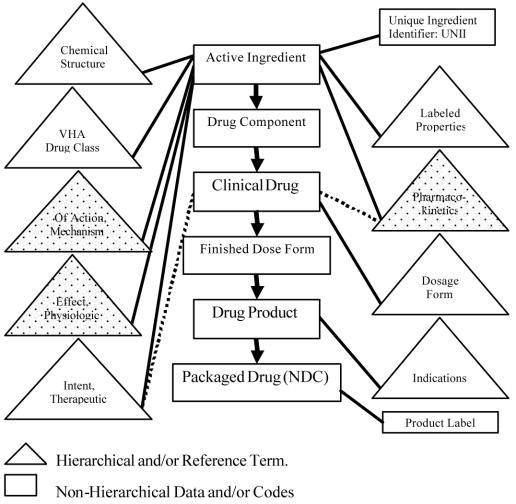
The U.S. medication infrastructure comprises an interlocking suite of projects, each emphasizing its own component, illustrated in Figure 1. These projects include the VA NDF-RT (National Drug File-Reference Terminology) core, the NLM’s RxNorm, and coordinated contributions from the FDA.
Figure 1.
Data model for the common medication infrastructure. The Pharmacogenomic extensions discussed involve extending or creating the “stippled” hierarchies and adding this information to the emerging federally funded medication initiative published by NLM. Dotted lines are secondary relationships. This content draws upon three Federal drug information initiatives: the VA NDF-RT (Veterans Administration National Drug File-Reference Terminology), FDA’s UNII project, and NLM’s RxNorm.
The VA NDF-RT Project
The legacy of the VA’s successful clinical information systems [10] has been widely admired. A component of the VA’s information environment has been a National Drug File, maintained to support VHA (Veterans’ Health Administration) applications. One of us (SHB) initiated the organization of this medication list into a formal representation of drugs, creating the NDF-RT. The process of transforming an informal catalog of drugs into a well-formed terminology is described elsewhere[11].
Many components of the common medication infrastructure, notably most of the associated taxonomies (triangle elements in Fig. 1) derive directly from the NDF-RT. Presently the catalog of drugs in this database exceeds 80,000 orderable compositions, associated with 3,997 active ingredients.
A powerful architectural feature of the NDF-RT which made it particularly well suited for this project is the existence of description logic formalism to assert relationships between concepts, analogous to that used to build SNOMED -RT and CT [12]. For example, one can definitionally assert the mechanism of drug action, as illustrated in Figure 2.
Figure 2.
Description logic assertion in NDF-RT
RxNorm
The RxNorm project is an effort by NLM to define and enumerate a standard normal form in which a drug may be administered to a patient, as opposed to the form in which the manufacturer might supply it[13]. This emphasis derives from consensus within the HL7 Vocabulary Technical Committee on a desirable drug information model for clinical applications and messaging. In addition, a considerable amount of practical information, such as trade name mappings, ingredients, and dose-forms, is incorporated into the RxNorm database. Table 1 shows an enumeration of those relationships as of the 2003AA Metathesaurus..
Table 1.
Relationship types and counts within RxNorm as present within the 2003AA Meta-thesaurus
| Count | Relation Type |
|---|---|
| 25337 | consists_of |
| 25337 | constitutes |
| 813 | contained_in |
| 813 | contains |
| 18865 | dose_form_of |
| 997 | form_of |
| 18865 | has_dose_form |
| 997 | has_form |
| 19381 | has_ingredient |
| 1307 | has_tradename |
| 19381 | ingredient_of |
| 6 | inverse_isa |
| 6 | isa |
| 462 | mapped_from |
| 462 | mapped_to |
| 1307 | tradename_of |
To achieve the HL7 functional goals, RxNorm posits two types of semantic normal forms, which are illustrated in Tables 2 and 3. These forms distinguish Drug Components and Clinical Formulations, the latter often being an expression of components.
Table 2.
Semantic Normal Form for Drug Components
| Code | Short Name | Active Ingredient | Precise Ingredient | Strength | Units |
|---|---|---|---|---|---|
| 11111 | APAP | Acetaminophen | Acetaminophen | 325 | MG |
| 22222 | Codeine | Codeine | Codeine Phosphate | 30 | MG |
Table 3.
Semantic Normal Form for Clinical Formulations
| Code | Name | Components | Orderable Dose Form |
|---|---|---|---|
| 12345 | Acetaminophen 325MG/Codeine 30MG | 11111/22222 | oral capsule |
Tables adopted from RxNorm web page: http://umlsinfo.nlm.nih.gov/RxNorm.html [accessed 27 June 2003]
RxNorm in turn is an integral component of the medication information infrastructure, refining the content of Drug Component, Clinical Drug, and Dosage Form data types (Figure 1).
FDA Participation
The FDA is required to maintain a comprehensive database of drug ingredients. They are introducing a new electronic UNique Ingredient Identifier (UNII) repository, based on the Molecular Design Limited (MDL®) Molefile *. These identifiers are ASCII representations that enable the representation of molecular structure in many chemical drawing programs. The NLM is making the UNII identifiers publicly accessible. Additionally, NLM is creating UNIIs for internationally used ingredients outside the U.S. jurisdiction of the FDA. These identifiers correspond to the UNII and Active Ingredient data types in Figure 1.
Pharmacokinetics Extension
The mechanism by which most genomic variations manifest their effects on drug metabolism is by changing kinetics – either accelerating activation or degradation or significantly slowing it. Thus, drug ontology information relevant to pharmacogenomics would be expected to deal with metabolism and kinetics. The common medication model (Fig. 1) includes three associated taxonomies of pharmacogenomics interest, and was the focus of our attention.
Aims and Process
Taxonomy Extensions
We submitted a supplemental proposal to one of the PharmGKB cooperative agreements (Mayo) to extend the common medication infrastructure in a manner that would improve its application to pharmacogenomics. Our specific aims were:
To expand components of the reference terminology, the Mechanism of Action and Physiologic Effect hierarchies to accommodate more sophisticated elements pertinent to pharmacogenomics.
To attach elements of these extensions to the 80,000 orderable drugs in the VA NDF-RT.
To create a new component in the reference terminology that would capture pharmacokinetics and demonstrate the application of component elements to some high-profile drugs.
The common medication infrastructure has 9 major hierarchies associated within it; they are represented as triangles in Figure 1. The three directly pertinent to the present work are stippled. The list below repeats this information in tabular format. Bolded hierarchy names are those pertinent to this report.
Chemical Structure
VHA Drug Class (e.g., antibiotic)
Mechanism of Action (Specific Aim 1)
Physiologic Effect (Specific Aim 1)
Therapeutic Intent
Labeled Properties
Pharmacokinetics (Specific Aim 3)
Dosage Form
Indications
The initial taxonomies of interest, Mechanism of Action and Physiologic Effect, were populated by semantically relevant MeSH terms. Specific Aim 1 then comprised an intensive process of making these elements more complete, together with ensuring the correctness of their corresponding hierarchies. Because these data types take part in description logic relationship definitions, they are not themselves defined using description logics.
The process by which these taxonomies were extended included validation of the target drug information model against some use-cases actually encountered in the Pharmacogenomics Network. Intensive, face-to-face modeling sessions were conducted in Washington and Nashville with pharmacologists. The initial MeSH hierarchies were used as the starting template, greatly expanded, and moderately restructured to represent what is judged to be an optimal representation of Mechanism of Action and Physiologic Effect for general application, while also accommodating the pharmacogenomic use-cases. The final re-assignment of each active ingredient to an appropriate, potentially new, node within these pharmacogenomically enhanced taxonomies (Specific Aim 2) presents a straightforward, albeit tedious, task.
Pharmacokinetics
Creating a useful representation of kinetics exceeds the expressive capacity of term hierarchies, and raises the utility or desirability of representing descriptions as complex objects within an ontology. Our process for working out how this should be done emulated that employed for taxonomies, in that we convened several face-to-face sessions with pharmacologists, supplemented with electronic correspondence and teleconferences. Several alternatives were examined as a basis for kinetics objects, ranging from adoption of NCI’s caBIO* suite of biomedical objects about molecular functions and physiologic pathways to complex XML substructures. Our taxonomic strategy of adopting MeSH concepts as a starting point had less utility for capturing kinetics.
Results
Taxonomy Extensions
Mechanism of Action
The Mechanism of Action hierarchy began its pre-enrichment life with 123 concepts appropriated from the MeSH hierarchies. Its final size is 210 concepts, suggesting the relevance of Mechanism of Action to the medical literature as indexed by MeSH. The excerpt below shows the top of this hierarchy. The bar-delimited (|) numbers to the right of some concepts reflect the original MeSH concept number, if one existed before expansion.
-
Cellular or Molecular Interactions
-
Biological Macromolecular Agents
-
Enzymatic Agents
Hydrolases|D006867
Pancrelipases|D020799
Tissue Plasminogen Activators|D010959
-
Lipoproteins
Surfactants|D013501
Bile Acids
Structural Macromolecules
-
-
-
Receptor Interactions
-
Ion Channel Interactions
-
Sodium Channel Interactions
-
Cholinergic Nicotinic Interactions
Cholinergic Nicotinic Agonists|D018722
Cholinergic Nicotinic Antagonists|D018733
-
-
-
Physiological Effects
The pre-enrichment version of the Physiologic Effects hierarchy had only 21 loosely-structured entries, extracted from MeSH. This impoverished representation was dramatically expanded to 1,638 entries, and represented in a formally structured hierarchy. The excerpt below illustrates the first few lines of this hierarchy, and illustrates its basic structure. The strict hierarchy is a limitation of the overarching information model for the ontology.
The absence of compositional expressions or description logics within this taxonomy of relationships is immediately evident, as the permuted use of dichotomized modifiers such as Increase or Decrease with respect to Production or the triad of Production, Activity and Degradation across families of effect classes. These permutations are responsible for inflating the entries from a score to thousands. This limitation of course does not pertain to instantiating drug-centric descriptions composed with elements from these taxonomies.
-
Organ System Nonspecific Activity
-
Immunologic Activity
-
Production Of Immunologically Active Molecules
-
Increased Production Of Immunologically Active Molecules
Increased Production Of Adhesion Factors
Increased Production Of Complement
Increased Production Of Cytokines
Increased Production Of Antibodies
-
Increased Production Of Immunologically Active Biogenic Amines
Increased Production Of Histamine
Increased Production Of Serotonin
-
Increased Production Of Kinins
Increased Production Of Bradykinin
Increased Production Of Kallidins
-
Increased Production Of Lipid-Derived
Immunologically Active Molecules
Increased Production Of Platelet-Activating Factors
-
Increased Production Of Eicosanoids
Increased Production Of Prostaglandins
Increased Production Of Thromboxanes
Increased Production Of Leukotrienes
-
-
-
Attachment to Orderable Drugs
Trained personnel edited the attachments of taxonomy elements among the 4,000 unique active ingredients to reflect the expanded hierarchies for Mechanism of Action and Physiologic effects. As depicted in Figure 1, this as the effect of relating these association to all 80,000 orderable drugs presently in the ontology.
Pharmacokinetics Model
We concluded that kinetic information must have the capacity to invoke object behaviors. To accommodate the reality that our kinetic representations exist within an ontology, we adopted a frame-like representation expressed as XML. This affords the opportunity to have slots within a kinetic object frame assume values of URLs to complex objects such as caBIO component. We are invoking Protégé[14]* to author these slots.
Several relationship types were recognized as necessary to express kinetics associated with drugs. At present, these new relationships, expressed as frame slots, include activated_by, degraded_by, coverts_to, and the inverse relationships associated with drug effects on enzymes. This last point requires at least one level of indirection since a given drug may have a spectrum of effects on more than one enzyme, such as induction, competitive binding, or suppression. In all instances, these slots may have more than one value, though a determination of which metabolic pathway among multiple options exceeds the capacity of our model at present.
We encountered some philosophic issues surrounding thresholds of evidence needed to assert kinetic relationships. On the one hand, weak but intriguing evidence has considerable value in the pharmacogenomics research community, though arguably it has no place for clinical applications. This requires the capacity to assign levels of confidence to kinetic information, for which for the time being we have adopted the four-point scale of unconfirmed, possible, probable, and confirmed. These ordinal confidence values populate slots in our frame structure, potentially associated with each fact.
We have successfully outlined the relevant kinetic parameters for 31 drugs that are strongly influenced by pharmacogenomic effects. This result is preliminary, and represented in Excel spreadsheets outline parameters and dosing implications. Transfer of these results into the object-based kinetics model remains incomplete work.
Discussion
We describe our efforts to enrich the already fertile base afforded by the common medication infrastructure, to support drug-related knowledge pertinent to pharmacogenomic use-cases. The effort entailed two complementary activities: the extension of taxonomies for mechanism of action and physiologic effect already extant in the drug ontology; and the definition and refinement of a frame-based XML structures to accommodate the complex elements associated with pharmacokinetics.
The nested model for pharmacokinetics raises two interesting challenges. On the one hand, we embedded knowledge into an ontology in a manner typically outside the domain of ontologies. The second aspect involves the overt creation of yet additional taxonomies, such as coverts_to, which recursively call components of the main model, introducing potential cyclic structures. Resolving such challenges comprises our future work.
Acknowledgments
We thank Peter Covitz, Frank Hartel, and Sherri De Coronado at NCI; Mike Lincoln from the VA, Dave Flockhart at Indiana University; and Joseph Awad at Vanderbilt. This work was sponsored by U01 GM61388-03S1 from NIGMS (RM Weinshilboum, PI).
Footnotes
http://www.nigms.nih.gov/pharmacogenetics/[accessed 27 June 2003]
http://www.pharmgkb.org/[accessed 27 June 2003]
www.mdli.com/downloads/ctfile/ctfile_subs.html [accessed 27 June 2003]
http://ncicb.nci.nih.gov/core/caBIO [accessed 27 June 2003]
http://protege.stanford.edu/[accessed 27 June 2003]
REFERENCE
- 1.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348(6):529–37. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 2.O’Kane DJ, Weinshilboum RM, Moyer TP. Pharmacogenomics and reducing the frequency of adverse drug events. Pharmacogenomics. 2003;4(1):1–4. doi: 10.1517/phgs.4.1.1.22588. [DOI] [PubMed] [Google Scholar]
- 3.Phillips KA, et al. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. Jama. 2001;286(18):2270–9. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 4.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet. 1980;32(5):651–62. [PMC free article] [PubMed] [Google Scholar]
- 5.Altman RB, et al. Indexing pharmacogenetic knowledge on the World Wide Web. Pharmacogenetics. 2003;13(1):3–5. doi: 10.1097/00008571-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Hewett M, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30(1):163–5. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliver DE, et al. Ontology development for a pharmacogenetics knowledge base. Pac Symp Biocomput. 2002:65–76. [PubMed] [Google Scholar]
- 8.Chute CG. Drugs, Codes, Standards, and Other Incompatible Things in the Dark. MD Computing. 2001;18(1):45–46. [PubMed] [Google Scholar]
- 9.Brown, S., et al., United States Government Progress Towards a Common Medication Information Infrastructure. submitted, 2003.
- 10.Kolodner, R.M. and J.V. Douglas, Computerizing large integrated health networks: the VA success 1997, New York: Springer. xxv, 515 p.
- 11.Carter JS, et al. Initializing the VA Medication Reference Terminology Using UMLS Metathesaurus Co-Occurrences. Proc AMIA Symp. 2002:116–20. [PMC free article] [PubMed] [Google Scholar]
- 12.Spackman KA, et al. Role Grouping as an Extension to the Description Logic of Ontylog, Motivated by Concept Modeling in SNOMED. Proc AMIA Symp. 2002:712–6. [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson SJ, et al. A Semantic Normal Form for Clinical Drugs in the UMLS: Early Experiences with the VANDF. Proc AMIA Symp. 2002:557–61. [PMC free article] [PubMed] [Google Scholar]
- 14.Musen M. Domain ontologies in software engineering: Use of Protege with the EON architecture. Methods of Information in Medicine. 1998;37(4–5):540–550. [PubMed] [Google Scholar]