Abstract
The National Drug File Reference Terminology contains a novel reference hierarchy to describe physiologic effects (PE) of drugs. The PE reference hierarchy contains 1697 concepts arranged into two broad categories; organ specific and generalized systemic effects. This investigation evaluated the appropriateness of the PE concepts for classifying a random selection of commonly prescribed medications. Ten physician reviewers classified the physiologic effects of ten drugs and rated the accuracy of the selected term. Inter reviewer agreement, overall confidence, and concept frequencies were assessed and were correlated with the complexity of the drug’s known physiologic effects. In general, agreement between reviewers was fair to moderate (kappa range 0.08–0.49). The physiologic effects modeled became more disperse with drugs having and inducing multiple physiologic processes. Complete modeling of all physiologic effects was limited by reviewers focusing on different physiologic processes. The reviewers were generally comfortable with the accuracy of the concepts selected. Overall, the PE reference hierarchy was useful for physician reviewers classifying the physiologic effects of drugs. Ongoing evolution of the PE reference hierarchy as it evolves should take into account the experiences of our reviewers.
BACKGROUND AND INTRODUCTION
Reference terminology development is becoming an important aspect of health informatics.1, 2 The Department of Veterans Affairs, National Library of Medicine, Food and Drug Administration, National Institute of General Medical Sciences, the National Cancer Institute and several other organizations are collaborating to create a freely available reference terminology for medications. The National Drug File Reference Terminology (NDF RT) is defined by a semantic model containing several definitional roles, including physiologic effect, chemical structure, mechanism of action, and therapeutic intent.3, 4
Definitions for each active ingredient are being assigned from reference hierarchies that categorize each dimension (e.g. a hierarchy that categorizes mechanisms of action). It is anticipated that NDF RT will have a variety of uses, including patient care, research, data sharing, and pharmacogenomics investigation5–7.
As part of the process of creating the NDF RT, the development team (STR, JA, ME, JC, ML, SHB) recently modeled a reference hierarchy for physiologic effects of medications. The physiologic effects reference hierarchy categorizes the physiologic processes that drugs induce in bringing about both clinical effects and unintended consequences. Our working definition of physiologic effect is “cellular, tissue or organ processes or functions altered by drugs”. The physiologic effects reference hierarchy was initially seeded with candidate concepts extracted from the Chemical Actions subtree [D27.505+] of MeSH. These concepts were reviewed, were iteratively refined, and were supplemented through focus groups that included physicians, pharmacologists, informaticians, and terminology experts. The physiologic effect reference hierarchy subsequently evolved through testing against use case scenarios and face validity evaluation by subject matter experts.
The physiologic effects reference hierarchy is currently comprised of 1697 concepts arranged in a tree structure with some concepts residing in multiple branches of the hierarchy. The first level of branching segregates concepts used to describe organ specific effects from concepts that describe systemic effects. Subsequent branching segregates the individual organs and the organ specific physiologic processes (such as “bronchodilation”) from the individual classes of systemic physiologic processes (such as “decreased protein synthesis”). The PE reference hierarchy models intermediary processes that occur as a result of specific molecular interactions leading to intended therapeutic applications (or side effects).
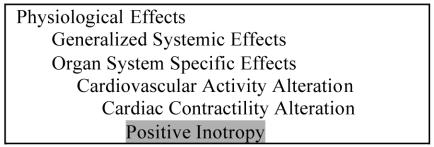
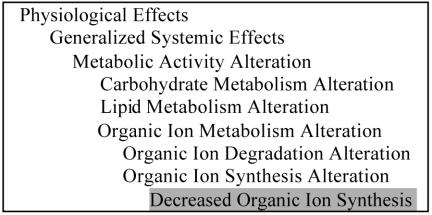
Mechanism of action and therapeutic intent are modeled in other hierarchies of the NDF RT. For example, the concept “Positive Inotropy” describes a physiologic effect of digoxin. Positive inotropy is a type of Cardiac Contractility Alteration, which is in turn a Cardiovascular Activity Alteration (Figure 1). The concept “Decreased Organic Ion Synthesis” could be used to classify the physiologic effects of both methotrexate and sulfa compounds (Figure 2). These physiologic effects are orthogonal to the therapeutic intent of the drugs, which may be treating congestive heart failure, neoplasia, or a bacterial infection, for digoxin, methotrexate, and sulfa derivatives, respectively.
Figure 1.
A subset of the PE reference hierarchy of the NDF RT open to “Positive Inotropy”.
Figure 2.
A subset of the PE reference hierarchy open to “Decreased Organic Ion Synthesis”.
This investigation evaluates the usability of the physiologic effects reference hierarchy for the task of classifying a random selection of commonly prescribed medications from Tennessee Valley Healthcare System (TVHS), Vanderbilt University Medical Center, and rxlist.com.
METHODS
Lists of the most frequently prescribed drugs were obtained from the Vanderbilt University Hospital pharmacies, the Veterans Administration TVHS, and the 2001 top drug list from rxlist.com8. Entries from the three commonly prescribed medication lists were normalized with respect to spelling, case, generic name, and administration form. Eighteen drugs were retrieved at random from the aggregate list. Retrieved drugs were stratified on a three-point scale by the expected complexity of representing their physiologic effect using the PE reference hierarchy. Complexity was assigned by one of the authors (STR) based on the possible number of physiologic effects of the drug and on the complexity of the branches within the PE reference hierarchy that models the drugs’ physiologic effects. The retrieved drugs were randomly segregated into three groups. The first group, an example set, contained 3 drugs whose physiologic effects were modeled by one of the study authors (STR) and were shared with the evaluating clinicians. The second group, a practice set of 5 drugs, was given to the evaluating clinicians to familiarize them with the modeling task. The third group of 10 drugs comprised an evaluation set to be modeled.
Given the novelty of the physiologic effects axis, few experienced reviewers were available outside the group that had developed it. Instead, eight Internal Medicine physicians, one Pediatrician, and one Med/Peds physician were recruited as “naïve” reviewers to classify the selected drugs’ physiologic effects using the PE reference hierarchy. These physicians had no advanced training in pharmacology or prior experience using the PE reference hierarchy, and minimal exposure to other axes of the NDF RT. One additional “experienced” reviewer having familiarity with NDF RT and the PE reference hierarchy also completed the classification task to clarify ambiguous classification from the main reviewers and to provide additional use case feedback; these results were not included in the global analyses. To provide a level set of knowledge, each clinician was given a packet with the list of drugs in random sequence and reference material about the drugs from Goodman and Gilman’s “The Pharmacological Basis of Therapeutics”.9 Clinicians were advised that the exercise was “a test of the [terminology], not your knowledge”. The reviewers were given the following instructions for drug classification: “assign the most appropriate concepts that describe the physiologic effect used by the given drug X.” No specific therapeutic considerations were provided for the selected drugs.
In addition to classifying the drugs using the PE reference hierarchy, the reviewers also were asked to rate their comfort with the accuracy of each assigned PE concept for describing the physiologic effect. Reviewers were instructed to use a nine point confidence scale, according to the following guidelines: “The assigned physiologic effect concept X to categorize drug Y is accurate (7–9), ambiguous (4–6) or inaccurate (1–3).” All responses were entered into individualized research spreadsheets. The reviewers worked independently of each other.
We analyzed the results by generating descriptive statistics of the numbers of concepts identified for each drug, the number of raters who assigned each concept to the drugs, and the mean confidence score of the reviewers for the accuracy of the assigned terms. ANOVA testing with Bonferroni correction was used for multiple comparisons. Logistic regression was used to compare ordinal and categorical variables. Global inter-reviewer agreement was assessed by determining multiple reviewer Kappa statistics. The Kappa statistic tests inter rater independence where 0 is the agreement that would be expected by chance alone and 1 is complete agreement between raters. Landis and Koch define the following interpretation of inter rater agreement: 0.00–0.20 = slight; 0.21–0.40 = fair; 0.41–0.60 = moderate; 0.61–0.80 = substantial; and 0.81–1.00 = almost perfect.10 Confidence Intervals are reported as 95% level. All statistical evaluation was performed using SPSS v. 11.5 and Stata v. 7.0.
RESULTS
A total of 18 drugs were retrieved from the aggregate list of most frequently prescribed drugs. Retrieved drugs included ten drugs for reviewer classification (Table 1). Each of the reviewers initially contacted successfully completed the task. Overall, reviewers provided 308 drug classifications using 127 unique concepts (ranging from 1 to 10 reviewers using each concept). The numbers of concepts assigned to a drug ranged from 2 for omeprazole to 34 for triamcinolone. The number of assigned concepts increased with advancing complexity rating, from 6.7 concepts for simple drugs and 11.3 concepts for moderately complex drugs to 22.0 concepts for complex drugs. There was an increase of 7.5 additional concepts for each increasing complexity rating (P=0.025).
Table 1. Overall Classification Rates.
The evaluation set of drugs, the expected complexity of classification, the individual number of concepts identified, and the mean confidence rating for all concepts classified by drug are listed. *The number of concepts identified by all 10 reviewers, by 8–9 reviewers, by 5–7 reviewers, and by only 1–4 reviewers.
| Reviewers identifying Terms* | |||||||
|---|---|---|---|---|---|---|---|
| Drug Name | Complexity | Retrieved | All 10 | 8–9 | 5–7 | 1–4 | Accuracy |
| Albuterol | simple | 9 | 1 | 0 | 1 | 7 | 7.3 |
| Ipratropium | simple | 8 | 1 | 0 | 1 | 6 | 8.0 |
| Omeprazole | simple | 2 | 1 | 0 | 0 | 1 | 8.2 |
| Ranitidine | simple | 8 | 1 | 0 | 0 | 9 | 7.8 |
| Meclizine | moderate | 12 | 0 | 0 | 2 | 10 | 7.3 |
| Nitroglycerine | moderate | 14 | 0 | 0 | 3 | 11 | 7.0 |
| Pravastatin | moderate | 8 | 0 | 0 | 1 | 7 | 7.5 |
| Doxycycline | complex | 8 | 0 | 1 | 1 | 6 | 6.8 |
| Ethinyl Estradiol | complex | 24 | 0 | 1 | 0 | 23 | 6.9 |
| Triamcinolone | complex | 34 | 0 | 0 | 1 | 33 | 6.9 |
The reviewers’ confidence rating for the accuracy of the selected physiologic effects terms’ representation of the identified physiologic process ranged from 1 to 9 with an overall mean confidence of 7.2 (95% CI, 7.0–7.4). This varied by the degree of complexity of the drug from 7.8 (7.4–8.2) for simple drugs, 7.2 (6.8–7.6) for moderately complex drugs, to 6.9 (6.6–7.2) for complex drugs. The reviewers assigned lower confidence ratings for complex drugs than they did for simple drugs (Corrected P=0.02). The number of PE concepts assigned to each drug was minimally inversely correlated with the mean confidence for that drug, decreasing by 0.03 for each additional concept (P=0.04).
Agreement measures were calculated for the first three levels of the PE reference hierarchy (Table 2). At the first branch, distinguishing physiologic effects into generalized systemic effects and organ specific effects, there was an overall kappa of 0.34. Within the generalized systemic effects branch, there was an overall kappa of 0.43. Within the organ specific effects, there was an overall kappa of 0.35. These results and the kappa statistics for the second level of the reference hierarchy are summarized in Table 2. In most cases, agreement was better than would be expected by chance alone.
Table 2. Inter Rater Independence relation to complexity of term.
The top three levels of the PE Ontology, with level of branching (depth), the number of branches beneath concepts (children) and the inter rater reliability for that concept (kappa). There is no relationship between the depth of the concept or the number of children and the inter rater reliability (P=0.668 and P=0.886, respectively).
| Concept | Depth | Children | Kappa |
|---|---|---|---|
| Physiological Effects | 1 | 2 | 0.34 |
| Generalized Systemic Effects | 2 | 3 | 0.43 |
| Cellular Activity Alteration | 3 | 6 | 0.65 |
| Immunologic Activity Alteration | 3 | 3 | 0.39 |
| Metabolic Activity Alteration | 3 | 4 | 0.32 |
| Organ System Specific Effects | 2 | 10 | 0.35 |
| Cardiovascular Activity Alteration | 3 | 5 | 0.49 |
| Dermatologic Activity Alteration | 3 | 7 | 0.13 |
| Digestive/GI System Activity Alteration | 3 | 11 | 0.48 |
| Endocrine Activity Alteration | 3 | 12 | 0.14 |
| Hemic/Lymphatic Activity Alteration | 3 | 1 | 0.08 |
| Musculoskeletal Activity Alteration | 3 | 4 | 0.15 |
| Neurological & Neuromuscular System Activity Alteration | 3 | 2 | 0.51 |
| Renal/Urological Activity Alteration | 3 | 6 | 0.10 |
| Reproductive System Activity Alteration | 3 | 2 | 0.17 |
| Respiratory/Pulmonary Activity Alteration | 3 | 7 | 0.41 |
Manual review of the assigned concepts revealed that much of the reviewer independence resulted from identification of different physiologic processes for the given drugs, rather than different physiologic effects to model single processes. Focusing on disparate processes appeared to result from inconsistent recognition of all therapeutic uses and side effects. For example, ten reviewers identified “bronchodilation” and five identified “positive chronotropy” as physiologic effects of albuterol. Only one reviewer identified “cellular activity alteration”, “emesis”, “neurological and neuromuscular system alteration”. Two other reviewers each identified an additional term, “increased renal K+ secretion” and “increased smooth muscle epinephrine activity”.
Dispersion due to incomplete recognition of all physiologic processes was more pronounced for complex drugs. For example, ethinyl estradiol induces physiologic processes related ovulation prevention, bone density alteration, adverse cardiovascular events, circulating lipids, and reduction of the symptoms of estrogen deficiency, among others. All reviewers classified the physiologic processes bone density regulation and preventing ovulation but only six addressed lipid alteration, five addressed cardiovascular events, and four addressed menopausal symptoms. After completing the task, many reviewers stated that they had difficulty with the complex drugs due to the intricacy of identifying all possible physiologic effects.
The reviewers assigned drugs to five physiologic effect concepts that do not exist in the PE reference hierarchy falling into thee categories: confusion with other NDF RT branches, synonyms of existing terms, and true content deficiencies. The most common problem was distinguishing physiologic effects from molecular mechanism of action. An example was “HMG CoA reductase inhibitor” assigned to pravastatin. An example of missed synonymy is “tachycardia” assigned to albuterol and ipratropium. Tachycardia is represented by the existing concept “positive chronotropy”. This investigation only revealed one example of a true deficiency in the PE reference hierarchy: “decreased LH secretion” was absent in the tested version of the reference hierarchy.
The reviewers made additional suggestions about concepts they believed should be represented. These too fell into the three categories of wrong branch, synonymy, and true deficiency. Examples of “headache” and “stops dizziness” belong in the therapeutic intent hierarchy. “Increased clotting tendency” is a reasonable synonym of the modeled concept “hemostasis alteration”. True missing concepts that were suggested include processes such as “WBC demargination’”, although “cellular locomotion alteration” overlaps with this concept.
The reviewer with previous experience using the PE reference terminology also successfully completed the classification task. Among the 42 physiologic effects identified, thirty matched concepts selected by the naïve reviewers. Ten of the remaining concepts represented distinct physiologic processes that would not have been expected to match. Only two physiologic processes were modeled using different concepts from the naïve reviewers’ results: “RNA replication alteration” for doxycycline, instead of “decreased protein synthesis” and “glucose metabolism alteration” for triamcinolone, instead of “decreased glycolosis”, “increased gluconeogenesis”, or “increased glycogenesis” as other reviewers selected. Both examples of divergence in classification involved complex drugs.
DISCUSSION
Our results suggest that the physiologic effects reference hierarchy is appropriate for modeling the physiologic effects of medications. A group of physicians without advanced training in pharmacology was able to use the hierarchy to classify a random selection of commonly used medications. Drugs that induce many physiologic processes were more likely to have more concepts selected than simple drugs. The frequency with which the reviewers selected the same concepts to classify drugs was inversely correlated with the complexity of the drug.
The reviewers were generally comfortable with their classifications, leading to an overall perceived confidence score for accuracy of physiologic effects concept assignments of 7.2 (accurate), although the accuracy rating decreased as the drugs’ complexity increased.
This study has limitations that merit discussion. The inter-reviewer agreement, although better than would be expected from chance alone, was lower than had been anticipated. Terminology content coverage and agreement studies of this type are typically performed by non-naïve reviewers to minimize bias introduced by the novelty of the terminology itself. The selection of naïve reviewers for this study likely led to the relatively low inter-rater agreement, as patterns of selected physiologic concepts suggest that incomplete concept identification was related to inconsistent recognition of all physiologic processes of the evaluation drugs. Within identified physiologic processes, agreement was much greater. Future studies involving the PE reference hierarchy should utilize reviewers more experienced with the physiologic effects of drugs. Furthermore, indicating therapeutic uses and adverse effect of drugs, or providing accompanying clinical case scenarios prior to classifying physiologic effects would likely prompt users to be complete and would improve agreement among reviewers.
This investigation provides valuable knowledge for the continued evolution and development of the PE reference hierarchy. Feedback from use case scenarios and actual classification tasks will likely continue to reveal the need to model additional terms, and to embed concept synonymy. Ongoing performance will need to be monitored to take these factors into account and the style guides given to modelers will be iteratively refined to maximize accuracy and reliability. Additionally, our experience that clinicians incompletely classified physiologic effects of drugs when presented without clinical information, may generalize to others building or investigating novel pharmacology reference terminologies. As reference terminology development plays an increasing role in informatics projects, inter-rater evaluation of the application of concepts may improve the quality of the reference terminologies and also help end users set appropriate expectations for consistency and completeness.11
Acknowledgments
This work was supported in part by NIGMS, NIH 3 U01 GM61388-03S1 (Weinshilboum) and by NLM LM6918-A102 (Elkin). We also appreciate the feedback from Stuart Nelson, MD, Soaring Bear, PhD, and Mark Tuttle, FACMI, during the development of the terminology.
REFERENCES
- 1.Chute CG. Clinical classification and terminology: some history and current observations. J Am Med Inform Assoc. 2000;7:298–303. doi: 10.1136/jamia.2000.0070298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chute CG, Cohn SP, Campbell JR. A framework for comprehensive health terminology systems in the United States. J Am Med Inform Assoc. 1998;5:503–10. doi: 10.1136/jamia.1998.0050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson SJ, Brown SH, Erlbaum MS, et al. A Semantic Normal Form for Clinical Drugs in the UMLS: Early Experiences with the VANDF. Proc AMIA Symp. 2002:557–61. [PMC free article] [PubMed] [Google Scholar]
- 4.Carter JS, Brown SH, Erlbaum MS, et al. Initializing the VA Medication Reference Terminology Using UMLS Metathesaurus Co-Occurrences. Proc AMIA Symp. 2002:116–20. [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver DE, Rubin DL, Stuart JM, Hewett M, Klein TE, Altman RB. Ontology development for a pharmacogenetics knowledge base. Pac Symp Biocomput. 2002:65–76. [PubMed] [Google Scholar]
- 6.Hewett M, Oliver DE, Rubin DL, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30:163–5. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinshilboum RM. The genomic revolution and medicine. Mayo Clin Proc. 2002;77:745–6. doi: 10.4065/77.8.745. [DOI] [PubMed] [Google Scholar]
- 8.http://rxlist.com/top200.htm, 2001.
- 9.Goodman LS, Hardman JG, Limbird LE, Gilman AG. Goodman & Gilman’s the pharmacological basis of therapeutics. New York: McGraw-Hill, 2001:xxvii, 2148 p.
- 10.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- 11.Elkin PL, Brown SH, Carter J, et al. Guideline and quality indicators for development, purchase and use of controlled health vocabularies. Int J Med Inf. 2002;68:175–86. doi: 10.1016/s1386-5056(02)00075-8. [DOI] [PubMed] [Google Scholar]