Abstract
In eukaryotes, 40S ribosomal subunits move from their recruitment site on the mRNA to the initiation codon by an as yet poorly understood process. One postulated mechanism involves ribosomal shunting, in which ribosomal subunits completely bypass regions of the 5′ leader. For some mRNAs, shunting has been shown to require various mRNA elements, some of which are thought to base pair to 18S rRNA; however, the role of base pairing has not yet been tested directly. In earlier studies, we demonstrated that a short mRNA element in the 5′ leader of the Gtx homeodomain mRNA functioned as a ribosomal recruitment site by base pairing to the 18S rRNA. Using a model system to assess translation in transfected cells, we now show that this intermolecular interaction also facilitates ribosomal shunting across two types of obstacles: an upstream AUG codon in excellent context or a stable hairpin structure. Highly efficient shunting occurred when multiple Gtx elements were present upstream of the obstacles, and a single Gtx element was present downstream. Shunting was less efficient, however, when the multiple Gtx elements were present only upstream of the obstacles. In addition, control experiments with mRNAs lacking the upstream elements showed that these results could not be attributed to recruitment by the single downstream element. Experiments in yeast in which the mRNA elements and 18S rRNA sequences were both mutated indicated that shunting required an intact complementary match. The data obtained by this model system provide direct evidence that ribosomal shunting can be mediated by mRNA–rRNA base pairing, a finding that may have general implications for mechanisms of ribosome movement.
Keywords: mRNA, ribosome, translation, sequence complementarity, 5’ leader
In eukaryotes, the translation of mRNA into protein is a key site of gene regulation. For many mRNAs, this process is highly regulated at the initiation step, which begins with recruitment of the translation machinery at either the 5′ m7GpppN cap structure or at internal sequences (1–4). With few exceptions (e.g., ref. 4), 40S ribosomal subunits are recruited some distance upstream of the initiation codon, thus necessitating their movement to the initiation codon. Several mechanisms of subunit movement have been postulated. One postulated mechanism involves 5′ to 3′ linear scanning (5). Although this hypothesis is consistent with various experimental observations, it has not yet been technically possible to directly visualize scanning 40S ribosomal subunits, and the translation of a number of mRNAs appears to be inconsistent with this mechanism (e.g., refs. 6 and 7). In a few cases, it has been suggested that ribosomal subunits bypass or shunt segments of the 5′ leader of an mRNA, for example, in cauliflower mosaic virus and adenovirus mRNAs (8, 9). Highly efficient shunting across a hairpin structure introduced into the adenovirus mRNA was found to involve a protein that forms a complex with initiation factor eIF4G and poly(A)-binding protein (6). In addition, three cis-acting sequences in the adenovirus mRNA that are complementary to 18S rRNA have been proposed to facilitate shunting by base pairing to 40S ribosomal subunits (9). If shunting is promoted by mRNA–rRNA base-pairing interactions, it may be more widespread than previously thought inasmuch as complementary sequence matches to 18S rRNA are commonly found within cellular mRNAs (10, 11).
In the present studies, we developed a model system allowing analysis of the variables that affect shunting of ribosomal subunits within the 5′ leader during translation initiation. To this end, we generated synthetic mRNA constructs that enabled us to quantify shunting and to systematically evaluate parameters such as the nature and distribution of shunt sites. To test the notion that mRNA–rRNA base pairing mediates shunting, we used a sequence element from the 5′ leader of the Gtx homeodomain mRNA. In earlier studies, we showed that this element functioned as a binding site for 40S ribosomal subunits and that it enhanced translation by a mechanism that involved base pairing to a complementary segment of 18S rRNA (3, 12). We now show that 40S ribosomal subunits can shunt across an upstream AUG or stable hairpin structure using Gtx elements as shunt donor and acceptor sites (13). Furthermore, by altering both mRNA and rRNA sequences in yeast, we demonstrated that this shunting required base pairing to 18S rRNA.
Results
The scanning model (5) postulates that ribosomal subunits recruited by an mRNA scan along the 5′ leader in a 5′ to 3′ direction until they encounter an AUG codon in good context, which is then used as the initiation codon. In this model, an upstream AUG (uAUG) is predicted to block translation by diverting ribosomal subunits from the authentic initiation codon, whereas a hairpin structure is predicted to physically block scanning subunits. Some results that appear to be exceptions to this model have been explained as leaky scanning, whereby scanning ribosomal subunits sometimes bypass an AUG that resides in a suboptimal context (14, 15). Alternatively, reinitiation has been invoked, in which some of the ribosomes that terminate translation at a short upstream ORF remain associated with the mRNA, continue scanning and reinitiate translation at a downstream initiation codon (16, 17). In the present studies, an uAUG in optimal context and a hairpin structure were used as obstacles to scanning. We then assessed the ability of ribosomal subunits to bypass these obstacles in synthetic 5′ leaders using 8-nt Gtx elements (12) as potential shunt sites.
Shunting Across an Upstream Initiation Codon.
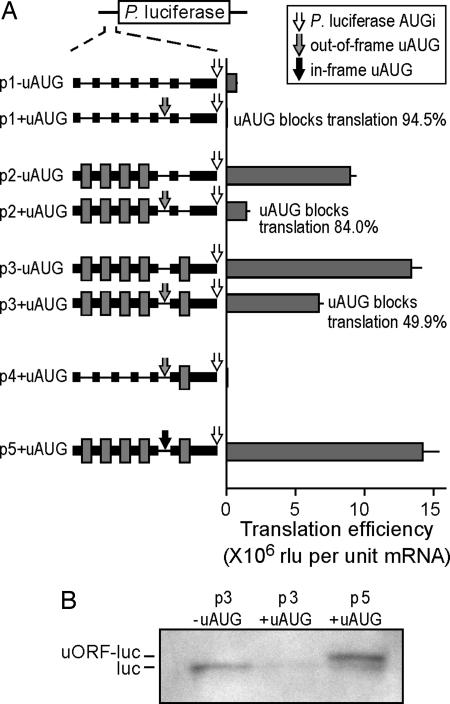
To evaluate the ability of ribosomal subunits to traverse an uAUG in excellent context, we used the Photinus luciferase reporter constructs shown in Fig. 1A. This uAUG generates an ORF that overlaps and is out-of-frame with the Photinus cistron and lies in a nucleotide context (ACCAUGGA) that should prevent leaky scanning (5). For these studies, we generated a reference construct containing poly(A) and β-globin mRNA sequences, which were shown in our previous studies not to enhance translation efficiency or facilitate internal initiation of translation (p1−uAUG) (18). The introduction of the uAUG into the 5′ leader reduced translation efficiency (raw light units per unit mRNA) by 94.5% (p1+uAUG). The residual 5.5% activity was ≈39,000 raw light units above the background luminescence of untransfected cells (≈200 raw lights units) and may represent ribosomes that bypassed the uAUG after recruitment at the cap.
Fig. 1.
Shunting across an uAUG can be mediated by Gtx-translational enhancer elements. (A) Photinus luciferase reporter mRNAs are indicated schematically. In the 5′ leaders, the Gtx elements are indicated by dark gray boxes, SI spacer sequences are indicated by black boxes, and poly(A) spacer sequences are indicated by a thin black line. The white arrow indicates the Photinus luciferase initiation codon (AUGi). The gray arrow indicates an uAUG, the resulting ORF overlaps the luciferase cistron in a different reading frame. The black arrow indicates an uAUG, the resulting ORF overlaps the luciferase cistron in the same reading frame. In the histogram, translation efficiencies (see Materials and Methods) in mouse N2a cells are expressed as raw light units (rlu) per unit mRNA, in the absence or presence of an uAUG, respectively. Error bars indicate SEM. The extent to which the uAUGs block translation is indicated as percent inhibition. In these experiments, the calculated translation efficiencies reflected the raw luciferase enzyme activities. (B) Western blot analysis for Photinus luciferase of lysates from N2a cells transfected with the indicated constructs.
The introduction of four Gtx elements into the 5′ leader resulted in a large increase in translation efficiency for the construct lacking the uAUG (p2−uAUG). Such multiple Gtx elements were used to increase the signal/noise ratio as in our earlier studies (3, 12, 18). On comparing this construct with one containing an uAUG (p2+uAUG), it appears that ≈16% of ribosomes bypassed the uAUG. However, the uAUG was bypassed with an even higher efficiency, ≈50%, when flanked by Gtx elements (compare p3+/−uAUG constructs). The addition of a single Gtx element 3′ of the uAUG increased translation efficiency from ≈1.4 to ≈6.7 million. This increase could not be accounted for by the presence of the single Gtx element 3′ of the uAUG, which, when tested alone in construct p4+uAUG, yielded a translation efficiency of only 73,594. It was striking that the translation efficiencies from constructs containing the Gtx elements (p2+uAUG and p3+uAUG) was up to 9.4-fold higher than the activity of the reference construct (p1−uAUG) even though the Gtx constructs contained uAUGs that should preclude leaky scanning.
To confirm that the uAUG in the Gtx constructs was used as an initiation codon, we introduced an additional adenosine downstream of it to shift the uAUG into the same reading frame as the luciferase cistron. Translation initiating at the uAUG was expected to produce a luciferase protein with a 17-aa amino-terminal extension that increased the molecular mass by ≈2 kDa. The results showed that luciferase activity for the construct with the in-frame uAUG was ≈2-fold higher than that of the construct with the out-of-frame uAUG (compare p3+uAUG with p5+uAUG). This result is consistent with the prediction that the uAUG was efficiently used and that translation produced a luciferase fusion protein with enzymatic activity.
Western blot analysis using an anti-Photinus luciferase antibody provided physical evidence that the uAUG was used (Fig. 1B). A band of ≈60 kDa, corresponding to the luciferase protein (luc), was observed in cells transfected with the construct lacking the uAUG (p3−uAUG). The intensity of this band was markedly reduced in cells transfected with the construct containing the out-of-frame uAUG (p3+uAUG). However, in cells transfected with the construct containing the in-frame uAUG (p5+uAUG), two bands were observed: one corresponding to the luciferase protein and a more intense band of slightly increased molecular mass, corresponding to the fusion protein (uORF-luc).
Shunting Across a Stable Hairpin Structure.
To evaluate the ability of ribosomes to traverse a different obstacle, we used a stable hairpin structure with a predicted stability of −121.1 kcal/mol (Fig. 2A). In addition, this hairpin structure contained a second embedded obstacle, an AUG codon in an excellent context (ACAAUGGC) that overlapped the luciferase cistron in a different reading frame. If this hairpin structure was disrupted, for example by scanning ribosomal subunits, these subunits should be diverted from the luciferase cistron by the uAUG.
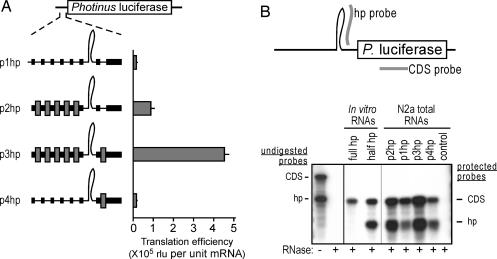
Fig. 2.
Shunting across a hairpin structure can be mediated by Gtx-translational enhancer elements. (A) Photinus luciferase reporter mRNAs are indicated schematically. Gray bars and lines are as in Fig. 1. The histogram represents translation efficiencies in mouse N2a cells. Error bars indicate SEM. In these experiments, the calculated translation efficiencies reflected the raw luciferase enzyme activities. (B) Ribonuclease protection assays. Riboprobes complementary to the 3′ stem of the hairpin (hp probe) and to the Photinus luciferase coding sequence (CDS probe) are indicated schematically as gray bars. Below is an autoradiograph of ribonuclease protection assays performed on in vitro transcribed RNAs containing the full hairpin (full hp) or 3′ stem of the hairpin (half hp) and on total RNAs isolated from N2a cells transfected with the constructs indicated. Full-length probes are indicated in the sample lacking RNase (−), and protected probes are indicated in the samples treated with RNase (+).
A construct containing only the cap structure as a ribosome recruitment site (Fig. 2A; p1hp) showed a low level of translation (14,549 raw light units) that was not due to the background luminescence of untransfected Neuro2a (N2a) cells. Translation efficiency was enhanced by ≈6-fold when multiple Gtx elements were introduced upstream of the hairpin structure (see construct p2hp). Significantly, translation efficiency was enhanced by ≈31-fold when a single Gtx element was also introduced downstream of the hairpin structure (see construct p3hp). Translational enhancement of an mRNA containing only a single Gtx element located downstream of the hairpin structure was ≈1.3-fold (see construct p4hp), indicating that this single element alone could not account for the observed increase in translation efficiency when Gtx elements flanked the hairpin structure.
RNA Analysis Suggests That the Hairpin Structure Is Cleaved in Vivo.
Because the hairpin is a long double-stranded RNA, it seemed possible that it could be a target of cleavage (19, 20). This hypothesis is an intriguing possibility because if true, it would mean that the observed shunting occurred across a noncovalently linked RNA. We assessed the integrity of the hairpin structure using ribonuclease protection assays (Fig. 2B). Two probes were used: one complementary to the 3′ stem of the hairpin structure (hp probe) and the other complementary to sequences contained within the luciferase coding sequence region (CDS probe). Control experiments performed using in vitro transcribed mRNAs showed that an mRNA containing the hairpin (full hp RNA) protected the CDS probe but did not protect the hp probe. To determine whether this result was due to the stability of the hairpin structure precluding hybridization of the probe, we tested an equivalent in vitro transcript lacking the 5′ half of the hairpin (half hp RNA). The results showed that this RNA could now protect the hp probe, suggesting that the intact hairpin inhibited hybridization to the probe.
RNase protection experiments were then performed by using DNase-treated total RNA extracted from cells transfected with the constructs in Fig. 2A. The results showed protection of both the CDS and hp probes by all of the reporter mRNAs. Ratios of the protected bands for three of the mRNAs (php2, php3, and php4) were the same as the ratio for the in vitro transcribed half hp mRNA, indicating that these hairpin structures were accessible to the hp probe, and consequently, it was likely that they were completely clipped in vivo. The hairpin in the fourth mRNA, p1hp, also appeared to be clipped but less extensively (≈60%).
The Ability of the Gtx Element to Mediate Shunting Requires Base Pairing to 18S rRNA.
To determine whether shunting is mediated by complementary base pairing between Gtx elements and 18S rRNA, we performed studies in Saccharomyces cerevisiae, which allowed us to experimentally alter both the mRNA and rRNA sequences. Yeast strain NOY908 lacks all chromosomal copies of the 35S rDNA but expresses the 35S rRNA from plasmid pNOY373, which is episomally maintained (21). The Gtx element has a poor complementary match to the yeast 18S rRNA (see Fig. 3A); in an earlier study, we showed that it was inactive in these cells but could function in yeast cells expressing a mouse–yeast hybrid 18S rRNA that contained mouse rRNA sequences that were complementary to the Gtx element (3).
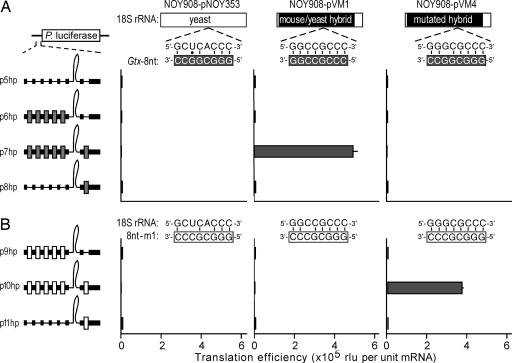
Fig. 3.
The ability of Gtx elements to mediate shunting requires base pairing to 18S rRNA. Schematic representations of the luciferase constructs are indicated to the left of the histogram. The 8-nt Gtx elements are indicated as dark gray boxes, and the mutated Gtx elements (8nt-m1) are indicated as white boxes. Constructs were tested in yeast strain NOY908 transformed with constructs expressing the 18S rRNAs indicated. Yeast rRNA sequences are indicated by the white bar and mouse sequences by the black bar. The histogram represents translation efficiencies in the indicated yeast cells expressed as raw light units per unit of mRNA. Error bars indicate SEM. (A) Sequence matches between the rRNA sequences (nucleotides 1132–1124 in the mouse), and the 8-nt Gtx element are indicated. Complementary nucleotides are indicated by vertical bars, and a black dot represents a potential G-U base pair. (B) Sequence matches between the rRNA sequences and the 8nt-m1 element are indicated as above. In both A and B, the calculated translation efficiencies reflected the raw luciferase enzyme activities.
Yeast reporter mRNAs with 5′ leader sequences containing hairpin structures similar to those in Fig. 2A were expressed in cells that expressed either WT yeast (pNOY353) or mouse–yeast hybrid (pVM1) 18S rRNAs (Fig. 3A). The results showed that in cells expressing the WT yeast 18S rRNA all of the constructs were expressed only at background levels. In yeast cells that expressed the mouse–yeast hybrid 18S rRNA, however, the stable hairpin was efficiently bypassed when flanked by Gtx elements and was expressed ≈500-fold over the background (compare construct p7hp in NOY908 cells transformed with pNOY353 and pVM1). Shunting in yeast was also observed on mRNAs that contained an uAUG as an obstacle (data not shown).
To directly test base pairing between the Gtx element and 18S rRNA in mRNAs containing the hairpin, we mutated both sequences (see pVM4 in Fig. 3A and element 8nt-m1 in Fig. 3B) and assessed the effects of these mutations on translation. The results showed that constructs containing a point mutation within the Gtx elements were inactive in cells expressing either the WT yeast or mouse–yeast hybrid 18S rRNAs (Fig. 3B; NOY908-pNOY353 and NOY908-pVM1). However, when these constructs were tested in yeast expressing the mutated mouse–yeast hybrid 18S rRNA that restored complementarity to the mutated Gtx element (encoded by plasmid pVM4), the construct with mutated Gtx elements flanking the hairpin was expressed with high efficiency in these cells. Translation efficiency was ≈360-fold higher in NOY908 cells transformed with pVM4 compared with NOY908 cells transformed with pNOY353. This result demonstrates that base pairing between the Gtx elements and 18S rRNA was required for ribosomes to bypass the hairpin. Translation efficiency was not enhanced in the construct having Gtx elements only upstream of the hairpin (p6hp in Fig. 3A); this result differed from the result obtained in mammalian cells, which showed a 16% increase in translation efficiency (p2hp in Fig. 2A).
Discussion
The ability of ribosomal subunits to shunt may allow mRNAs that contain various obstacles in their 5′ leaders to be translated. For example, the occurrence of uAUG codons in natural mRNAs is not rare; it has been reported that up to 40% of human mRNAs contain uAUGs, approximately half of which are in similar or better contexts than the initiation codon (22, 23). Longer 5′ leaders contain more uAUGs, and the frequency of occurrence appears to be by chance, i.e., there does not appear to be any selective pressure during evolution to remove these elements. Although problems with some of these 5′ leaders have been identified (24), many mRNAs with uAUGs appear to be real. In some natural mRNAs, uAUGs can inhibit translation (25); however, our findings suggest that others may be bypassed.
The present studies were specifically designed to provide a model system to test mechanisms of shunting. The results provide direct experimental evidence that ribosomal shunting, which depends on mRNA–rRNA base pairing, can occur. In mammalian cells, we found that upstream obstacles were bypassed to various degrees in the mRNAs tested here. Translation efficiency increased when Gtx elements were present upstream of a hairpin structure or an uAUG and were increased to much higher levels when the Gtx elements flanked the obstacles. The failure of the uAUG and hairpin obstacles to block translation in some constructs strongly suggests that in these constructs they were shunted. In addition, the experiments in yeast using mutations to mRNA elements and to 18S rRNA demonstrated that shunting required an intact complementary match; shunting was facilitated by a mutated Gtx element only when a compensating mutation was introduced into the 18S rRNA.
In experiments using the hairpin structure as an obstacle, we found evidence that this structure was cleaved. One possible interpretation is that the observed translation occurred before the hairpin structures were cleaved. If so, ribosomal subunits would have been required either to shunt across the intact hairpins or to melt them. However, it was unlikely that scanning subunits melted the hairpin structure because of its high predicted stability and because it contained an uAUG that would block translation by diverting these ribosomal subunits from the luciferase cistron as in Fig. 1A.
The possibility that cleavage per se increased translation by a mechanism other than shunting also appears unlikely inasmuch as the extent of cleavage did not correlate with the translation efficiencies (Fig. 2). For example, the hairpins appeared to be completely cleaved in constructs p3hp and p4hp, yet these mRNAs were translated with efficiencies that varied by >24-fold. This result indicates that the higher translation efficiency observed in construct p3hp was due to the presence of Gtx elements upstream of the hairpin. This finding suggests that the cleaved fragments remained base paired via the residual portions of the hairpin. These results also appear to be inconsistent with the notion that ribosomal subunits could scan through these 5′ leaders. Scanning subunits recruited by the capped 5′ fragment should never reach the initiation codon: either the clipped hairpin (linking the two fragments) would block scanning subunits, or if the clipped hairpin was melted, the two fragments would dissociate. In the latter case, the uncapped (luciferase) fragments should be rapidly degraded (26), but this did not appear to be the case in view of the high reporter activities obtained with some of these mRNAs (e.g., see Fig. 2; p3hp). The finding that shunting occurred on clipped mRNAs in which the fragments remained associated with each other suggests that the ribosomal subunits could move to the initiation codon by shunting across noncovalently linked nucleotides.
The Gtx element has only one functional binding site in the 18S rRNA (see ref. 3 and Fig. 3), suggesting that the ability of ribosomal subunits to bypass obstacles in the mRNAs used in our studies involved subunit recruitment at upstream Gtx elements, dissociation from these sites, and reassociation with the downstream element. We suggest that dissociation and reassociation of these subunits is not unidirectional, and thus, recruitment should be equally efficient from sites located 3′ of the initiation codon. This notion is supported by recent studies of the HIV-2 immunodeficiency virus RNA, which report ribosomal subunit recruitment from locations 3′ of the initiation codons (27). Our ongoing studies also support this notion (unpublished data).
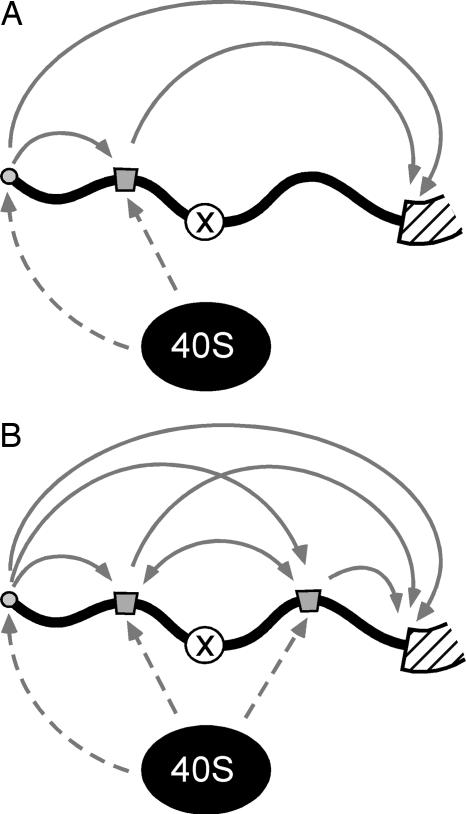
Based on the results of the present experiments, we propose a model of shunting in which ribosomal subunits are recruited to the mRNA via the eIF–4F complex at the cap structure or through internal mRNA elements (Fig. 4A). These elements may recruit the translation machinery by direct interactions (e.g., by base pairing to rRNA and by binding to ribosomal proteins) or indirectly (by binding to initiation factors or other proteins that can interact with the translation machinery). Although the extent to which these recruitment sites contribute to overall translation varies in different mRNAs, such recruitment sites would effectively increase the local concentration of 40S subunits and associated factors. This clustering of the translation machinery in the vicinity of the mRNA would enhance shunting by increasing the likelihood of interactions between ribosomal subunits and other accessible recruitment sites in the mRNA and may also increase the likelihood of interactions between the initiator tRNA-Met and the initiation codon itself. The efficiency of shunting would also depend on the relative positions of the mRNA elements. For example, obstacles in the mRNA that are flanked by recruitment sites may be effectively bypassed (Fig. 4B). This phenomenon may involve dissociation of ribosomal subunits from a site upstream of an obstacle and association with another site downstream of it. Alternatively, interactions at sites located upstream and downstream of the obstacle may be simultaneous, if these sites interact with the translation machinery by different mechanisms. This clustering of the translation machinery further predicts that translation efficiency will depend on the distance between the cluster and the initiation codon, e.g., translation should decline as this distance increases. In any event, although it is possible to invoke scanning between the cap, shunt sites, and the initiation codon, it does not appear necessary to do so.
Fig. 4.
A model of ribosome shunting. 5′ leader sequences are represented schematically as bold black lines, the coding regions as a hatched bar, the cap as gray circles, and the key mRNA elements that can affect shunting by interacting with ribosomal subunits directly or indirectly as gray boxes. The “X” in the 5′ leaders represents an obstacle, for example, hairpin or uAUG. The 40S ribosomal subunits and associated factors (ribosomal complexes) are indicated as black circles. Ribosomal subunits are recruited to the mRNA via the cap structure and mRNA elements (indicated by the dashed arrows). For clarity, we indicate individual ribosomal complexes only. Likewise, we indicate one mRNA element upstream of the obstacle in A and two mRNA elements flanking the obstacle in B. These recruitment events increase the local concentration of the translation machinery and the likelihood of ribosomes interacting with other accessible sites in the mRNA (indicated by the solid arrows). The presence of one or more mRNA elements downstream of the obstacle in B increases the efficiency of shunting. One assumption of the model is that movement of the translational machinery between mRNA elements need not necessarily be unidirectional.
The model of ribosomal shunting explains how obstacles in a 5′ leader can be bypassed in mRNAs containing elements that function as shunt sites. However, such nonlinear ribosomal subunit movement does not depend on the presence of these obstacles, as seen for example, in the Cauliflower mosaic virus (8). We therefore expect that the nonlinear movement of ribosomal subunits may also occur in mRNAs lacking obstacles. Our findings, together with various observations (22, 23) of the frequent occurrence of uAUGs, raise the possibility that ribosomal shunting may be more general than previously thought.
Materials and Methods
DNA Constructs.
Photinus luciferase reporter constructs were based on plasmids pGL3c 5′ multiple cloning site and pYESFFlucH (3, 12). Plasmids encoding yeast and mouse–yeast hybrid 18S rRNAs (pNOY353, pVM1, and pVM4, respectively) were identical to those used in our previous studies (3). The 5′ leader sequences used in this study are presented in Table 1, which is published as supporting information on the PNAS web site. Mammalian reporter constructs were all generated by cloning inserts (PCR amplified from oligonucleotide templates using Pfu DNA polymerase). Hairpin constructs were generated by cloning three inserts into pGL3c 5′ multiple cloning site. The 5′ inserts correspond to sequences 5′ of the hairpin structure (with SpeI as the 5′ site and MfeI or EcoRI as the 3′ sites), the middle insert contained the 5′ half of the stable secondary structure (with EcoRI and AatII sites), and the 3′ insert contained the 3′ half of the stable secondary structure followed by 5′ leader sequences to the initiation codon (with AatII and NcoI sites). For yeast constructs, the 5′ leader sequences from mammalian reporter constructs were amplified by PCR using Pfu polymerase and cloned into the pYESFFlucH yeast expression construct (using MfeI and NarI sites). Site-directed mutagenesis was performed with primers containing mutated nucleotides using Phusion DNA polymerase (New England Biolabs).
Analyses of Reporter Gene Activity.
Transfection of mouse N2a cells and transformation of yeast strain NOY908, reporter gene assays, and total RNA isolations were performed as described in our studies (3, 12). Translation efficiencies are expressed as luciferase activities in raw light units normalized for luciferase mRNA levels. Reporter mRNA levels in mammalian cells were determined by using ribonuclease protection assays (RPAIII kit; Ambion) with 1 μg of DNase-treated total RNA. Protected fragments were size-fractionated on 6% polyacrylamide–urea gels, visualized on a Storm 860 PhosphorImager (Molecular Dynamics), and quantified by using alphaeasefc stand-alone software (Alpha Innotech, San Leandro, CA). Reporter mRNA levels in yeast cells were determined by using Northern blots hybridized with a riboprobe corresponding to the luciferase CDSs.
Protein expression was determined by Western analysis using 7% Tris-acetate gels in Tris-acetate buffer (Invitrogen). Proteins were transferred to poly(vinylidene difluoride) membranes and probed with goat anti-Photinus luciferase polyclonal IgG 1° antibody and donkey anti-goat IgG 2° antibody conjugated to alkaline phosphatase (Promega) by using the Western Breeze chemiluminescent Western blot immunodetection kit (Invitrogen).
Supplementary Material
Acknowledgments
We thank Luke Burman for excellent technical assistance and Dr. Masayasu Nomura (University of California, Irvine) for providing yeast strain NOY908 and plasmid pNOY353. Funding was provided by National Institutes of Health Grant GM61725 (to V.P.M.), the G. Harold and Leila Y. Mathers Charitable Foundation (V.P.M.), and the Skaggs Institute for Chemical Biology (S.A.C. and J.D.).
Abbreviations
- uAUG
upstream AUG
- CDS
coding sequence
- N2a
Neuro2a.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Kapp L. D., Lorsch J. R. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 2.Komar A. A., Hatzoglou M. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 3.Dresios J., Chappell S. A., Zhou W., Mauro V. P. Nat. Struct. Mol. Biol. 2006;13:30–34. doi: 10.1038/nsmb1031. [DOI] [PubMed] [Google Scholar]
- 4.Pisarev A. V., Shirokikh N. E., Hellen C. U. C. R. Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Kozak M. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xi Q., Cuesta R., Schneider R. J. Genes Dev. 2004;18:1997–2009. doi: 10.1101/gad.1212504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers G. W., Jr., Edelman G. M., Mauro V. P. Proc. Natl. Acad. Sci. USA. 2004;101:2794–2799. doi: 10.1073/pnas.0308576101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryabova L. A., Pooggin M. M., Hohn T. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:1–39. doi: 10.1016/S0079-6603(02)72066-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yueh A., Schneider R. J. Genes Dev. 2000;14:414–421. [PMC free article] [PubMed] [Google Scholar]
- 10.Mignone F., Pesole G. Appl. Bioinformatics. 2002;1:145–154. [PubMed] [Google Scholar]
- 11.Mauro V. P., Edelman G. M. Proc. Natl. Acad. Sci. USA. 1997;94:422–427. doi: 10.1073/pnas.94.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chappell S. A., Edelman G. M., Mauro V. P. Proc. Natl. Acad. Sci. USA. 2004;101:9590–9594. doi: 10.1073/pnas.0308759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson R. J. In: Translational Control of Gene Expression. Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Woodbury, NY: Cold Spring Harbor Lab. Press; 2000. pp. 127–183. [Google Scholar]
- 14.Herzog E., Guilley H., Fritsch C. Virology. 1995;208:215–225. doi: 10.1006/viro.1995.1145. [DOI] [PubMed] [Google Scholar]
- 15.Kos M., Denger S., Reid G., Gannon F. J. Biol. Chem. 2002;277:37131–37138. doi: 10.1074/jbc.M206325200. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch A. G., Wek R. C., Dever T. E., Cigan A. M., Feng L., Donahue T. F. Translational Regulation of Gene Expression. Vol. 2. New York: Plenum; 1993. pp. 87–116. [Google Scholar]
- 17.Gaba A., Wang Z., Krishnamoorthy T., Hinnebusch A. G., Sachs M. S. EMBO J. 2001;20:6453–6463. doi: 10.1093/emboj/20.22.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chappell S. A., Edelman G. M., Mauro V. P. Proc. Natl. Acad. Sci. USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond S. M. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 20.Doma M. K., Parker R. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wai H. H., Vu L., Oakes M., Nomura M. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peri S., Pandey A. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki Y., Ishihara D., Sasaki M., Nakagawa H., Hata H., Tsunoda T., Watanabe M., Komatsu T., Ota T., Isogai T., et al. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- 24.Kozak M. Genomics. 2000;70:396–406. doi: 10.1006/geno.2000.6412. [DOI] [PubMed] [Google Scholar]
- 25.Meijer H. A., Dictus W. J., Keuning E. D., Thomas A. A. J. Biol. Chem. 2000;275:30787–30793. doi: 10.1074/jbc.M005531200. [DOI] [PubMed] [Google Scholar]
- 26.Wilusz C. J., Wormington M., Peltz S. W. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 27.Herbreteau C. H., Weill L., Decimo D., Prevot D., Darlix J. L., Sargueil B., Ohlmann T. Nat. Struct. Mol. Biol. 2005;12:1001–1007. doi: 10.1038/nsmb1011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.