Abstract
The eukaryotic mRNA 3′ poly(A) tail and the 5′ cap cooperate to synergistically enhance translation. This interaction is mediated by the cap-binding protein eIF4E, the poly(A) binding protein (PABP), and eIF4G, a scaffolding protein that bridges between eIF4E and PABP to bring about the circularization of the mRNA. The translational repressor, Paip2 (PABP-interacting protein 2), inhibits translation by promoting the dissociation of PABP from poly(A). Here we report on the existence of an alternative mechanism by which Paip2 inhibits translation by competing with eIF4G for binding to PABP. We demonstrate that Paip2 can abrogate the translational activity of PABP, which is tethered to the 3′ end of the mRNA. Thus, Paip2 can inhibit translation by a previously unrecognized mechanism, which is independent of its ability to disrupt PABP–poly(A) interaction.
Keywords: circularization, translation initiation
Translational control is an important means by which cells govern gene expression. Initiation, the rate-limiting step of translation, is often the target of translational control (1). This control involves in many circumstances the cap structure at the mRNA 5′ end and the poly(A) tail at the mRNA 3′ end. Although both mRNA terminal structures stimulate translation on their own, their combined translational enhancement is synergistic. Such a mechanism was demonstrated in yeast, plant, and mammalian systems (2, 3) and plays a key role during development of Xenopus, Drosophila, and mouse (4). Translational synergy was also recapitulated in vitro (5–9). Hence, the cap–poly(A) tail synergy represents a common paradigm for translational control.
The mechanism by which the mRNA 3′ poly(A) tail synergizes with the 5′ cap to stimulate translation was first documented in yeast, where PABP was shown to interact directly with the eIF4G subunit of the cap-binding complex eIF4F (see below) (10, 11). In yeast, PABP is an essential protein because deletion of the PABP1 gene is lethal (12). However, because deletion of the eIF4G binding site in PABP causes only a mild effect on yeast cell growth (13), it is not clear that PABP–eIF4G interaction is the only mechanism by which PABP regulates translation and cell growth in yeast. In addition, Searfoss et al. (14) demonstrated that yeast Ski proteins inhibit the activity of eIF5 and eIF4B in 60S ribosomal subunit joining and that this inhibition is reversed by PABP. These results implicate an important role for PABP in 60S ribosomal subunit joining. In contrast to yeast, the expression of an eIF4G mutant that does not interact with PABP in Xenopus oocytes repressed translation of polyadenylated mRNAs and inhibited progesterone-induced oocyte maturation (15).
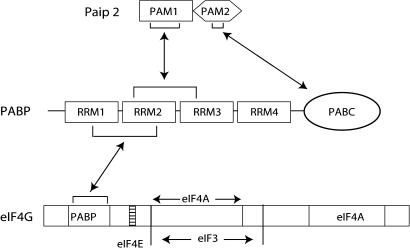
PABP contains four RNA-recognition motifs (RRMs) and a proline-rich C-terminal region, which is N-terminal to a highly evolutionarily conserved sequence termed PABC (see below) (16, 17). eIF4F is composed of the cap-binding subunit eIF4E, an RNA-dependent ATPase/ATP-dependent RNA helicase, eIF4A, and eIF4G (reviewed in ref. 18). The latter serves as a scaffolding for binding of PABP (19), eIF4E (20), eIF4A (21), and eIF3 (21), a 40S ribosome-associated initiation factor (22). Importantly, the N-terminal region of eIF4G harbors the binding site for PABP (10, 19), which brings about the circularization of the mRNA (23). The closed-loop model for mRNA circularization provides a structural basis for the synergistic enhancement of translation by the cap structure and the poly(A) tail (3, 24, 25). Depletion of PABP from mouse ascites cell-free translation extracts resulted in a dramatic reduction in translation initiation and 80S initiation complex formation, which could be rescued by the addition of recombinant PABP (26). These findings, therefore, ascribe a key role for PABP as a bona fide translation initiation factor and stress the importance of circularization for translation initiation.
Several mechanisms were proposed to explain how mRNA circularization enhances translation initiation. First, circularization is expected to increase the concentration of terminating ribosomes in the vicinity of the mRNA 5′ cap structure and thereby facilitate ribosome recycling. This notion is bolstered by the finding that PABP also interacts with the termination factor eRF3 (27). Thus, bridging of eIF4G and PABP by eRF3 engenders the looping out of the 3′ UTR and facilitates the shunting of the ribosomes to the 5′ end of mRNA. Second, the interaction between PABP and eIF4G may allosterically increase the affinity of eIF4E for the cap structure (26).
Several fragments of PABP stimulate translation in Xenopus oocytes independent of their ability to bind poly(A), as shown by tethering the fragments to the mRNA 3′ UTR. A fragment containing PABP RRMs 1–2 strongly stimulates translation (11), most probably through its interaction with eIF4G. poly(A)-stimulated translation is controlled by two PABP-interacting proteins (Paips), Paip1 and Paip2 (28, 29). Paip1 is an ≈70-kDa protein that stimulates translation in vivo (28). In contrast, Paip2, a much smaller protein (≈14 kDa), inhibits translation by (i) competing with Paip1 for binding to PABP and (ii) promoting the dissociation of the PABP–poly(A) complex, which in turn results in the destabilization of the mRNA closed loop (29). Furthermore, Paip2’s ability to impede 80S initiation complex formation demonstrates that Paip2 represses translation at the initiation level (29). Two distinct sequences in PABP, RRMs 2 and 3 and PABC, interact with two sites in Paip2 termed PABP-interacting motifs (PAMs). PAM1 spans amino acids 22–75 and binds RRMs 2 and 3 of PABP, whereas PAM2 spans amino acids 106–120 and binds to PABC (30).
The observation that eIF4G and the PAM1 motif of Paip2 interact with a shared sequence of PABP (RRM2) (see Fig. 1) prompted us to consider the possibility that eIF4G and Paip2 may compete for binding to PABP. In the present study we examined this idea. MS2 coat protein–PABP fusion protein (MS2–PABP), when tethered to the MS2 coat protein binding site in a reporter mRNA, stimulated translation in cis, but not in trans (11). Here we used this assay in a PABP-depleted Krebs-2 ascites cell extract to demonstrate that Paip2 and eIF4G compete for binding to PABP and that Paip2 inhibits translation through competition with eIF4G. Thus, our results demonstrate a mode of Paip2 action that disrupts the PABP–eIF4G interaction and thereby inhibits translation.
Fig. 1.
A possible mechanism of competition between Paip2 and eIF4G for PABP binding. Shown is PAM1 motif of Paip2 and the N-terminal segment of eIF4G interacting with an overlapping region in PABP.
Results
Paip2 Competes with eIF4G for Binding to PABP in Vitro.
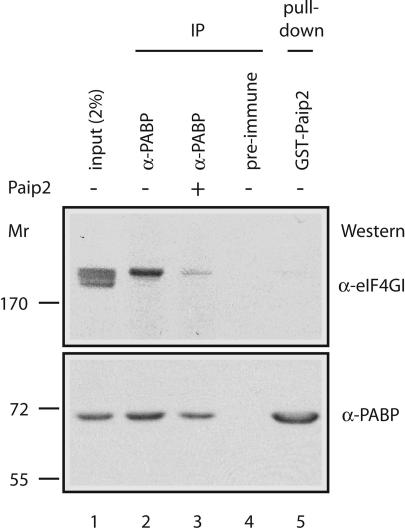
To determine whether Paip2 can compete with eIF4G for binding to PABP, we performed coimmunoprecipitation and pull-down assays using cytoplasmic extracts from Krebs-2 ascites cells. In agreement with our previous report (19), eIF4GI, which accounts for the majority of total eIF4G (there are two eIF4G forms, eIF4GI and eIF4GII) (31, 32), coimmunoprecipitated with PABP with an anti-PABP antibody, but not when a preimmune serum was used (Fig. 2, compare lanes 2 and 4). Strikingly, however, when extracts were preincubated with Paip2 (lane 3), coimmunoprecipitation of eIF4GI with PABP was reduced. In addition, eIF4GI failed to copurify with PABP that was pulled down with GST–Paip2-coupled beads (lane 5). These results indicate that eIF4GI and Paip2 binding to PABP is mutually exclusive. Interestingly, several closely migrating polypeptide species crossreacted with the eIF4GI antibody (lane 1), whereas only eIF4GI corresponding to the upper band coimmunoprecipitated with PABP. Thus, the use of some of the eIF4GI isoforms to initiate translation may be independent of PABP (see Discussion).
Fig. 2.
Coimmunoprecipitation of eIF4GI with anti-PABP is inhibited by Paip2. Extracts from Krebs-2 cells were incubated with anti-PABP (lanes 2 and 3) or preimmune serum (lane 4) coupled to beads in the absence (lane 2) or presence (lane 3) of Paip2. For the pull-down assay, extracts were incubated with GST–Paip2 (lane 5) that were immobilized on glutathione-Sepharose beads (33). Bound proteins were eluted in Laemmli sample buffer, resolved by SDS/7.5% PAGE, and subjected to Western blotting using anti-eIF4GI and anti-PABP antibodies. One-fiftieth of the extract used for immunoprecipitation or pull-down assay was loaded in lane 1.
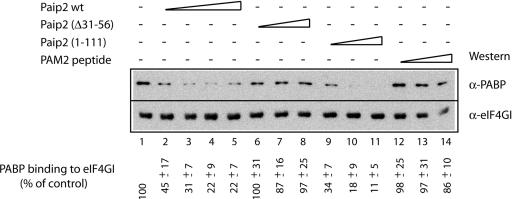
To corroborate the findings of competition between Paip2 and eIF4GI for PABP binding, a GST–eIF4GI (41–244) fragment, which binds PABP, was preadsorbed to glutathione-Sepharose beads and incubated with recombinant PABP in the absence or presence of increasing concentrations of recombinant full-length Paip2 or fragments thereof. The proteins were eluted with Laemmli sample buffer and analyzed by SDS/PAGE and Western blotting using anti-PABP and anti-eIF4GI antisera (Fig. 3). PABP copurified with full-length eIF4GI (lane 1). A gradual decrease in the amounts of PABP that copurified with eIF4GI was observed when increasing amounts of full-length Paip2 were used (22% of control at the highest concentration of Paip2; Fig. 3, compare lanes 1 and 5). The explanation for this residual activity is not immediately clear. Paip2 (Δ31–56), which can bind only to the C terminus, but not to the RRM region of PABP, failed to affect the amount of PABP that coprecipitated with eIF4GI (lanes 6–8). In contrast, incubation with increasing amounts of Paip2 (1–111), which can bind only to the RRM region, but not to PABC, progressively inhibited PABP binding to eIF4GI, similar to the effect obtained with full-length Paip2 (lanes 9–11). As expected, a peptide of Paip2 (PAM2 peptide), which can bind only to PABC, failed to affect the amount of PABP that coprecipitated with eIF4GI (lanes 12–14). These data suggest that Paip2 competes with eIF4GI for binding to PABP in vitro. Furthermore, the sequence responsible for this competition is the RRM-binding motif, PAM1, which is located in the middle region of Paip2 and represses translation (30). PAM2, which interacts with PABC, exerts no effect on the eIF4GI–PABP interaction and on translation (28).
Fig. 3.
Paip2 inhibits the interaction of PABP with eIF4GI. A solution of PABP was prepared with Paip2 WT or its mutants, Paip2 (Δ31–56) or Paip2 (1–111), and the PAM2 peptide (amino acids 112–127 of Paip2) using molar ratios of PABP to Paip2 of 1:1 (lanes 2, 6, 9, and 12), 1:2 (lane 3), 1:4 (lanes 4, 7, 10, and 13), or 1:8 (lanes 5, 8, 11, and 14). The mixtures were then incubated at 0°C for 30 min with recombinant GST-fused eIF4GI-Nt immobilized onto glutathione-Sepharose beads. After incubation, the resin was washed, and bound proteins were eluted with Laemmli buffer. After resolving on an SDS/10% polyacrylamide gel, proteins were analyzed by Western blotting using antibodies against eIF4GI or PABP, as indicated. The signals in different lanes were quantified by using nih image software (version 1.63). The value obtained in the absence of added competitor (lane 1) was set as 100%. Values representing the average from three independent experiments and standard deviations from the mean are shown.
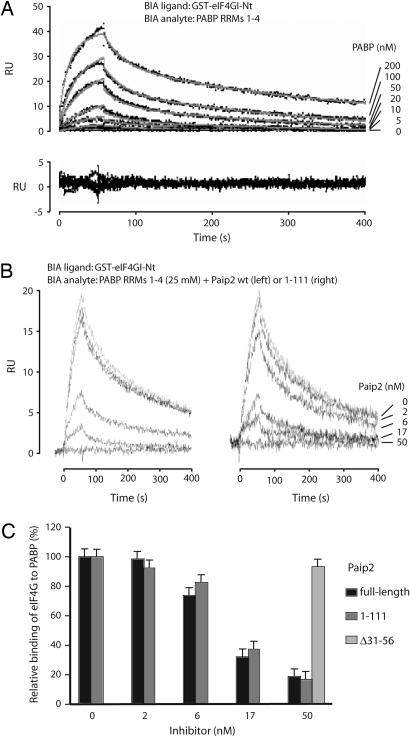
To obtain quantitative data for the eIF4GI–Paip2 competition, we first studied the thermodynamics of the eIF4GI–PABP interaction by using a surface plasmon resonance (SPR)-based biosensor, the Biacore. A recombinant GST–eIF4GI (41–244) fragment was coupled to sensor chip surfaces, and various concentrations of recombinant PABP RRMs 1–4 were randomly injected in duplicates over these surfaces. The resulting sensorgrams were globally analyzed by using a 1:1 stoichiometry model (Fig. 4A). The apparent kinetic and thermodynamic constants for the eIF4GI–PABP interaction are listed in Table 1. The constants for the Paip2–PABP RRM interactions (30) have been included for comparison. The eIF4GI–RRMs 1–4 interaction has an apparent on rate of 3.15 ± 0.7 × 105 M−1 s−1 (Table 1), which is significantly lower than those corresponding to the Paip2–RRMs 1–4 or Paip2–RRMs 2 and 3 interactions (Table 1; 19 ± 0.4 and 27 ± 2 × 105 M−1 s−1). The apparent “off rate” of the eIF4GI–PABP complex is 10 times higher than the apparent off rate corresponding to the Paip2–RRMs 1–4 complex (Table 1; 62 ± 6 × 10−4 and 6 ± 0.03 × 10−4 s−1, respectively), indicating that the eIF4GI–RRMs complex is significantly less stable than the Paip2–RRMs complex. These differences in association and dissociation rates result in a 60-fold weaker apparent affinity for the eIF4GI–RRMs 1–4 interaction versus the Paip2–RRMs 1–4 interaction (20 nM versus 0.31 nM, respectively). The significance of this difference is addressed in Discussion.
Fig. 4.
SPR analysis of eIF4GI interactions with PABP RRMs 1–4: effect of Paip2 and related proteins on the interactions. (A) Control-corrected sensorgrams corresponding to the interaction of RRMs 1–4 with biosensor surface-immobilized eIF4G. RRMs 1–4 from PABP was injected in duplicate for 60 s at different concentrations over surface-immobilized eIF4GI (400 resonance units). The dissociation of the complexes was followed for up to 400 s. The resulting control-corrected sensorgrams (Upper) were fitted by using a 1-to-1 stoichiometry model. Dotted lines show experimental sensorgrams, and solid lines show calculated fits. The residuals (difference between calculated and experimental data points) are shown in Lower, and the related kinetic and thermodynamic constants determined from the fit are listed in Table 1. (B) SPR in-solution competition experiments. RRMs 1–4 from PABP (25 nM) was injected alone or after preincubation with Paip2 (Left), Paip2 (1–111) (Right), or Paip2 (Δ31–56) (at 50 nM; sensorgram not shown). (C) Control-corrected SPR signal recorded after 55 s of injection (shown in B; triplicates not shown) were expressed as percentages (100% corresponds to RRMs 1–4 binding in the absence of Paip2) and plotted as a function of Paip2 or Paip2 mutant concentrations.
Table 1.
Kinetic and thermodynamic parameters related to the interaction of various PABP regions with Paip2 and eIF4G
| Interactions | kass, ×10−5 M−1 s−1 | kdiss, ×104 s−1 | Kd app, nM |
|---|---|---|---|
| RRMs 1–4/eIF4G | 3.15 ± 0.7 | 62 ± 6 | 20 ± 2.5 |
| RRMs 1–4/Paip2 | 19 ± 0.4 | 6.00 ± 0.03 | 0.31 ± 0.01 |
| RRMs 2 and 3/Paip2 | 27 ± 2 | 23 ± 1 | 0.85 ± 0.09 |
Next, an SPR in-solution competition experiment was carried out by injecting recombinant RRMs 1–4 (25 nM) preincubated with increasing amounts of recombinant full-length Paip2 or Paip2 fragments, Paip2 (1–111) or Paip2 (Δ31–56), over an eIF4GI surface. Increasing concentrations of both Paip2 and Paip2 (1–111) preincubated with RRMs 1–4 resulted in a decrease of net binding to surface-captured GST–eIF4GI–Nt, with an EC50 of ≈15 nM for Paip2 and Paip2 (1–111) (Fig. 4 B and C). These values are in agreement with the affinity of Paip2 for RRMs 1–4 that was determined to be in the low nanomolar range (Table 1) (30). In contrast, preincubation of RRMs 1–4 with Paip2 (Δ31–56) had no effect on the net binding to GST–eIF4GI–Nt even at Paip2 (Δ31–56) concentrations as high as 50 nM (data not shown and Fig. 4C).
Paip2 Inhibits Translation by Disrupting the PABP/eIF4G Interaction.
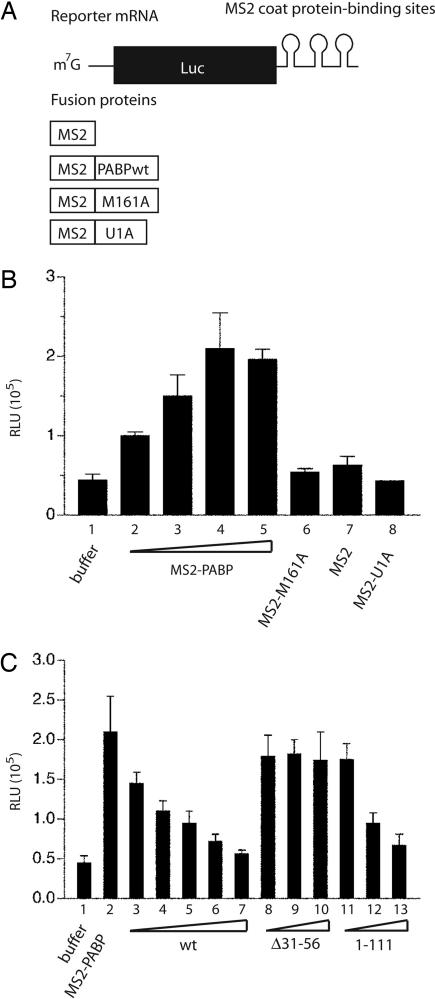
Paip2 was reported to inhibit translation by selective disruption of PABP–poly(A) interaction (29). To determine whether the Paip2 can also inhibit translation in a poly(A)-independent manner, we used a capped firefly luciferase mRNA that lacks a poly(A) tail but instead contained several MS2 coat protein binding sites at its 3′ end (Fig. 5A). In vitro translation experiments were carried out by using PABP-depleted Krebs-2 cell extracts (33) that were supplemented with increasing amounts of recombinant MS2-coat protein fused to PABP (MS2–PABP). As expected, translation was progressively stimulated with the addition of up to 82 ng (1 pmol) of MS2–PABP (Fig. 5B). The PABP portion of the fusion protein was responsible for the stimulatory effect of MS2–PABP on translation, because MS2 alone or MS2–U1A, a splicing RNA binding protein fused to MS2, did not stimulate translation (Fig. 5B) (11). In agreement with the data of Gray et al. (11), PABP lacking MS2 coat protein was inefficient in stimulating translation (data not shown). Importantly, a point mutant, MS2–PABP M161A, which is unable to bind to eIF4GI, failed to stimulate translation (Fig. 5B), consistent with earlier data showing that PABP–eIF4GI interaction is necessary for translational enhancement (26). To determine whether Paip2 negates the PABP-induced translational stimulation, extracts were supplemented with MS2–PABP (1 pmol) and increasing amounts of Paip2. Translation was progressively inhibited by increasing amounts of Paip2 (Fig. 5C). This effect was specific, as Paip2 (Δ31–56), which cannot bind to RRMs 1–2 of PABP (30), failed to inhibit translation. Paip2 (1–111), which binds only to the RRM region of PABP, inhibited translation (Fig. 5C). These findings clearly demonstrate that PABP stimulation of in vitro translation is inhibited by Paip2, which acts by specific disruption of PABP–eIF4G interaction.
Fig. 5.
Paip2 negates the stimulatory activity of tethered PABP. (A) Diagram of the components used for the assay of the tethered PABP function. The capped reporter luciferase mRNA (Luc-MS2) was not polyadenylated but contained binding sites for MS2 coat protein within its 3′ UTR. The fusion proteins contained the MS2 coat protein sequence at their N terminus. (B) Tethered functional analysis of PABP. The assay was conducted in a PABP-depleted Krebs-2 cell extract. Translation reactions (10 μl) containing increasing concentrations of MS2–PABP (2.8, 5.5, 8.2, and 11 μg/ml in lanes 2, 3, 4, and 5, respectively) were incubated at 30°C for 1 h with 25 ng of Luc-MS2 mRNA. Translation was measured by luciferase assay and is expressed in relative light units (RLU). As negative controls, MS2 protein, either alone or fused to U1A, or PABP mutant M161A were used (at 82 μg/ml). (C) Inhibition of the tethered PABP function by Paip2. Luc–MS2 mRNA translation was assayed as in B in the presence of 82 ng (1 pmol) of MS2–PABP and increasing amounts of recombinant Paip2 WT or its mutants, Δ31–56 or 1–111 [used at the MS2–PABP to Paip2 molar ratios of 1:1 (lanes 3, 8, and 11), 1:2 (lanes 4, 9, and 12), 1:4 (lanes 5, 10, and 13), 1:8 (lane 6), or 1:16 (lane 7)]. In B and C the levels of luciferase in the presence of buffer (25 mM Hepes-KOH, pH 7.3/50 mM KCl/1.5 mM MgCl2/1 mM DTT) are shown in lane 1. The data are the average from three experiments with standard deviations from the mean.
Discussion
The importance of the eIF4G–PABP interaction for efficient translation initiation has been established in many systems (34), although in yeast it might not have a critical role (3). Here we demonstrate that this interaction is a direct target for translational inhibition by Paip2, an inhibitor of translation initiation (29). We show that Paip2 competes with eIF4GI for binding to PABP. Our observation that the eIF4G–RRMs 1–4 interaction is not as strong as the Paip2–PABP and Paip1–PABP interactions (Table 1) (35) is consistent with our inability to detect PABP in association with the eIF4F complex by Western blotting, whereas eIF4E, eIF4G, and eIF4A were readily detected (H. Imataka and N.S., unpublished data). We estimated by quantitative Western immunoblotting that the content of PABP in the HeLa and Krebs-2 cell S10 fraction is ≈0.2% (or 29 fmol/μg of total protein) (data not shown and ref. 29). The molar concentration of Paip2 is 5- to 7-fold lower than that of PABP (29). The content of eIF4GI is 0.08% or 4.5 fmol/μg total HeLa cytoplasmic protein based on our previous measurements (30), which is ≈6-fold less than that of PABP. It could therefore be argued that PABP is not limiting for translation and that Paip2 and eIF4GI interact with PABP in a noncompetitive manner. However, because a significant fraction of PABP is likely to be sequestered in complexes with other proteins (36), the size of the PABP pool that is accessible to Paip2 and eIF4Gs is smaller than total PABP. Therefore, competition between Paip2 and eIF4GI for PABP binding may occur under physiological conditions.
One important aspect of translation initiation that might not be accommodated by the closed-loop model is the ability of PABP to stimulate the translation of nonpolyadenylated mRNAs. This regulation was first reported for a yeast system in vitro (37). Sachs and colleagues (37) defined this phenomenon as transactivation, suspecting that it is mediated by PABP that is not directly bound to mRNA. Supporting the possibility of PABP acting in trans, PABP mutants with reduced RNA binding activity stimulated translation (37). Given that an A stretch is required for the interaction between eIF4G and PABP in yeast (10), it was suggested that a factor(s) other than eIF4G is targeted by PABP within the translation initiation complex (37). However, the PABP binding sites in yeast and mammalian eIF4G markedly differ, and the presence of poly(A) is not necessary for the human eIF4G–PABP interaction. Association of human eIF4GI and PABP was detected without addition of poly(A), and treatment of proteins with micrococcal nuclease did not decrease the interaction (19). We showed previously that, in a Krebs-2 cell extract, either PABP depletion or PABP inactivation by Paip2 inhibited the translation of poly(A) mRNA (29, 33, 38). Consistent with these observations, our current results demonstrate that PABP can mediate in cis translational stimulation of mRNAs that are devoid of a poly(A) tail. This stimulation can be abrogated by specific inhibition of the PABP–eIF4GI interaction and further supports the conclusion that translational stimulation by PABP requires its interaction with eIF4GI. It is therefore conceivable that, in mammalian systems, the PABP–eIF4GI complex, whose formation is prevented by Paip2, mediates transactivation of translation by PABP. It would be of interest to determine whether this mechanism preferentially affects naturally mRNAs, which recruit PABP through DAZL proteins (39), or mRNAs whose poly(A) tail become shortened as they age.
Generation of multiple eIF4GI isoforms (a, b, c, d, and e) because of alternative translation initiation at different in-frame AUGs was demonstrated in HeLa cells (40). Interestingly, the smallest eIF4GI isoform (e) lacks the PABP binding domain and failed to interact with PABP. Data for Krebs-2 cells also reveal several eIF4GI forms and argue for nonequivalent regulation of individual isoforms of eIF4GI by PABP (Fig. 2). However, we detected only the largest eIF4GI isoform in the eIF4GI–PABP coimmunoprecipitate, indicating that a larger fraction of eIF4GI molecules could be refractory to PABP-mediated regulation than was previously thought (40).
Materials and Methods
Plasmids.
The constructs for expression of Paip2 and its fragments in pGEX-6P2 vector were reported previously (29, 30). The pGEX-eIF4GI construct (originally 1–204, but renamed 41–244, according to ref. 40) was described earlier (19). The construct pET3b-PABP-His was described earlier (29). By using the latter plasmid as a template, the RRMs 1–4 fragment of PABP was PCR-amplified with the forward primer 5′-CGGGATCCATGAACCCCAGTGCCC and the reverse primer 5′-GCCGCTCGAGTTTGCGCTGAGCTAAAGC. The resulting fragment was digested with BamHI/XhoI and ligated in-frame into BamHI/XhoI-cleaved vector of pGEX-6P2 (Amersham Pharmacia Biotech). The point mutation M161A in the PABP coding region was generated by using the plasmid pET3b-PABP-His as a template that was annealed with the primers 5′-A/ATG/AAT/GGA/UUGCG/UUCTC/CTA/AAT/GAT/CGC and 5′-GCG/ATC/ATT/TAG/GAG/UUCGC/UUTCC/ATT/CAT/T by using the QuikChange Site-Directed Mutagenesis kit (Stratagene). The point mutations are underlined, and codons are separated by slashes. The pET-MS2, pET-MS2-U1A construct and the firefly luciferase containing three MS2 binding sites reporter were generous gifts from Marvin Wickens (University of Wisconsin, Madison). Human PABP from pET3b-PABP-His was subcloned as a BamHI and NheI fragment into the pET-MS2 vector to construct pET-MS2-PABP-His plasmid that expresses an MS2-fused version of human PABP-His upon isopropyl β-d-thiogalactopyranoside induction. Likewise, the point mutant MS2-PABP-His M161A was generated in the pET vector.
Production and Purification of Recombinant Proteins.
Production and purification of C-terminal His6-tagged PABP by using BD Talon metal affinity resin (BD Biosciences) were previously described (29). The same procedure was used to prepare MS2, MS2–U1A, and PABP WT and mutant (M161A) proteins that were fused to the MS2 coat protein at their N terminus. The expression and purification of GST-tagged proteins, Paip2, and its mutants and the subsequent cleavage of the GST tag from the protein with PreScission protease were carried out according to the manufacturer’s protocol (Amersham Pharmacia Biotech). All of the purified proteins were dialyzed in reaction buffers indicated in the figure legends. Paip2 (112–127) fragment that consists of the PAM2 peptide was synthesized by the Sheldon Biotechnology Center at McGill University.
GST Pull-Down Assay and Western Blotting.
Five micrograms of purified GST-eIF4G-Nt fragment or a mutant that is unable to bind to PABP (amino acids 134KRERK138 in the PABP-binding site were converted to alanines) (19) were incubated end-over-end with a 50% slurry of glutathione 4B-Sepharose (20 μl; Amersham Pharmacia Biotech) for 30 min at 4°C with 300 μl of buffer A (20 mM Tris·HCl, pH 7.5/100 mM KCl/2.5 mM MgCl2/0.1 mM EDTA/10% glycerol/0.1% Triton X-100). Next, 1 μg of His-tagged PABP was mixed with Paip2 WT or its mutants, Paip2 (Δ31–56) or Paip2 (1–111), or the PAM2 peptide (112–127 of Paip2) at molar ratios ranging from 1:1 to 1:8 in 300 μl of buffer A, and the mixture was incubated at 4°C for 30 min. The resin was washed three times with 500 μl of buffer A to remove unbound protein before the PABP–Paip2 protein complexes were added to the resin. After a further incubation of 30 min at 4°C, the resin was washed as before, and the proteins were eluted with 50 μl of Laemmli sample buffer, boiled for 5 min, centrifuged at top speed to collect the supernatant, resolved by SDS/PAGE, and transferred to a nitrocellulose membrane (pore size 0.2 μm; Protran Bioscience). Western blotting analysis using eIF4GI and PABP antibodies was performed as previously described (19).
PABP Depletion from Krebs-2 Cell Extract.
PABP was depleted from nuclease-treated Krebs-2 cell extract by affinity chromatography using recombinant GST–Paip2 immobilized on glutathione-Sepharose beads as described previously (33).
In Vitro Transcription and Translation Assay.
The MS2 plasmid was linearized with BglII and used as a template in an in vitro transcription reaction catalyzed by T7 RNA polymerase to synthesize the capped luciferase RNA without a poly(A) tail. A 10-μl translation reaction that contained 1 μl of the recombinant proteins was added to 7 μl of PABP-depleted Krebs-2 cell extract and 1 μl of water and was briefly incubated on ice for 5 min. Recombinant proteins were resuspended in translation buffer (25 mM Hepes-KOH, pH 7.3/50 mM KCl/1.5 mM MgCl2/1 mM DTT). Firefly luciferase mRNA containing MS2 coat protein binding sites was then added, and the mixture was incubated for 10 min on ice before being transferred to a 30°C water bath for a 1-h incubation. The tubes were chilled on ice, and luciferase activity was measured in duplicates by using Promega’s luciferase assay system and the Lumat LB 9507 bioluminometer (Berthold Technologies).
SPR-Based Biosensor Analysis.
SPR experiments were carried out at 25°C on a Biacore 3000 optical biosensor (Biacore, Piscataway, NJ). The data collection rate was set to 10 Hz for every assay. Hepes-buffered saline (20 mM Hepes-KOH, pH 7.4/100 mM KCl/2.5 mM MgCl2/0.005% Tween 20) was used as running buffer for SPR experiments as well as to dialyze and dilute the injected proteins. The GST-eIF4G-Nt fragment (250 nM, in 10 mM acetic acid, pH 4.5) was covalently immobilized onto CM4 biosensor chips by using standard amine coupling chemistry (≈400 resonance units) (41). For every eIF4G-Nt surface a separate flow cell was similarly activated and blocked to be used as control.
Kinetic Analysis of the GST-eIF4G-Nt/RRMs 1–4 Interactions.
Kinetic experiments were carried out in duplicate at a flow rate of 50 ml/min. The interaction between GST-eIF4G-Nt with the RRMs 1–4 domains of PABC was monitored as follows: RRMs 1–4 (5–200 nM), in buffer were randomly injected for 60 s, followed by a 360-s buffer injection over both GST-eIF4G-Nt and mock surfaces. Surface regeneration between each RRMs 1–4 injection was performed with a 24-s pulse of 1% acetic acid solution (50 μl/min) followed by an EXTRACLEAN procedure and a RINSE procedure (Biacore instrument handbook). Before any data collection, the GST-eIF4G-Nt and mock surfaces were conditioned by five consecutive regeneration protocols as described here to increase reproducibility.
Biacore Data Preparation and Analysis.
The data preparation was done as described elsewhere by double referencing (42). All of the corrected sensorgrams were reduced to 700 evenly spaced sampling points. Global analysis of the set of sensorgrams was performed by using kinetic models available in the sprevolution software package (43).
SPR Competition Experiments.
Competition experiments were performed by injecting solutions of PABP RRMs 1–4 (final concentration of 25 nM) preincubated with 2–50 nM of full-length Paip2, Paip2 (1–111), or Paip2 (Δ31–56) over a mock surface and more than ≈800 resonance units of GST-eIF4G-Nt immobilized on a CM4 surface, as described above. Regeneration between injections was performed as described above.
Acknowledgments
We thank Marvin Wickens for plasmids and Kfir Madjar for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01 GM66157 (to N.S.). N.S. is a Canadian Institute of Health Research Distinguished Scientist and a Howard Hughes Medical Institute International Scholar. M.C.-M. is supported by a Canadian Institute of Health Research postdoctoral fellowship.
Abbreviations
- PABP
poly(A) binding protein
- RRM
RNA-recognition motif
- PAM
PABP-interacting motif
- SPR
surface plasmon resonance.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Mathews M. B., Sonenberg N., Hershey J. W. B. In: Translational Control of Gene Expression. Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2000. pp. 1–31. [Google Scholar]
- 2.Gallie D. R. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 3.Sachs A. In: Translational Control of Gene Expression. Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2000. pp. 447–465. [Google Scholar]
- 4.Wickens M., Goodwin E., Kimble J., Strickland S., Hentze M. W. In: Translational Control of Gene Expression. Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2000. pp. 295–370. [Google Scholar]
- 5.Iizuka N., Najita L., Franzusoff A., Sarnow P. Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarun S. Z., Jr., Sachs A. B. Mol. Cell. Biol. 1997;17:6876–6886. doi: 10.1128/mcb.17.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preiss T., Hentze M. W. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 8.Gebauer F., Corona D. F., Preiss T., Becker P. B., Hentze M. W. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel Y. M., Poncet D., Piron M., Kean K. M., Borman A. M. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 10.Tarun S. Z., Sachs A. B. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 11.Gray N. K., Coller J. M., Dickson K. S., Wickens M. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachs A. B., Davis R. W., Kornberg R. D. Mol. Cell. Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kessler S. H., Sachs A. B. Mol. Cell. Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Searfoss A., Dever T. E., Wickner R. Mol. Cell. Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakiyama M., Imataka H., Sonenberg N. Curr. Biol. 2000;10:1147–1150. doi: 10.1016/s0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- 16.Adam S. A., Nakagawa T., Swanson M. S., Woodruff T. K., Dreyfuss G. Mol. Cell. Biol. 1986;6:2932–2943. doi: 10.1128/mcb.6.8.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sachs A. B., Bond M. W., Kornberg R. D. Cell. 1986;45:827–835. doi: 10.1016/0092-8674(86)90557-x. [DOI] [PubMed] [Google Scholar]
- 18.Gingras A. C., Raught B., Sonenberg N. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 19.Imataka H., Gradi A., Sonenberg N. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mader S., Lee H., Pause A., Sonenberg N. Mol. Cell. Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morino S., Imataka H., Svitkin Y. V., Pestova T. V., Sonenberg N. Mol. Cell. Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershey J. W. B., Merrick W. C. In: Translational Control of Gene Expression. Sonenberg N., Hershey J. W. B., Mathews M. B., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2000. pp. 33–88. [Google Scholar]
- 23.Wells S. E., Hillner P. E., Vale R. D., Sachs A. B. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson A. In: Translational Control. Hershey J. W. B., Mathews M. B., Sonenberg N., editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 451–480. [Google Scholar]
- 25.Kahvejian A., Roy G., Sonenberg N. Cold Spring Harbor Symp. Quant. Biol. 2001;66:293–300. doi: 10.1101/sqb.2001.66.293. [DOI] [PubMed] [Google Scholar]
- 26.Kahvejian A., Svitkin Y. V., Sukarieh R., M’Boutchou M. N., Sonenberg N. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uchida N., Hoshino S., Imataka H., Sonenberg N., Katada T. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- 28.Craig A. W., Haghighat A., Yu A. T., Sonenberg N. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 29.Khaleghpour K., Svitkin Y. V., Craig A. W., DeMaria C. T., Deo R. C., Burley S. K., Sonenberg N. Mol. Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 30.Khaleghpour K., Kahvejian A., De Crescenzo G., Roy G., Svitkin Y. V., Imataka H., O’Connor-McCourt M., Sonenberg N. Mol. Cell. Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gradi A., Imataka H., Svitkin Y. V., Rom E., Raught B., Morino S., Sonenberg N. Mol. Cell. Biol. 1998;18:334–342. doi: 10.1128/mcb.18.1.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svitkin Y. V., Gradi A., Imataka H., Morino S., Sonenberg N. J. Virol. 1999;73:3467–3472. doi: 10.1128/jvi.73.4.3467-3472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Svitkin Y. V., Sonenberg N. Methods Mol. Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- 34.Mangus D. A., Evans M. C., Jacobson A. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy G., De Crescenzo G., Khaleghpour K., Kahvejian A., O’Connor-McCourt M., Sonenberg N. Mol. Cell. Biol. 2002;22:3769–3782. doi: 10.1128/MCB.22.11.3769-3782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim N. S., Kozlov G., Chang T. C., Groover O., Siddiqui N., Volpon L., De Crescenzo G., Shyu A. B., Gehring K. J. Biol. Chem. 2006;281:14376–14382. doi: 10.1074/jbc.M600307200. [DOI] [PubMed] [Google Scholar]
- 37.Otero L. J., Ashe M. P., Sachs A. B. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svitkin Y. V., Imataka H., Khaleghpour K., Kahvejian A., Liebig H. D., Sonenberg N. RNA. 2001;7:1743–1752. [PMC free article] [PubMed] [Google Scholar]
- 39.Collier B., Gorgoni B., Loveridge C., Cooke H. J., Gray N. K. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byrd M. P., Zamora M., Lloyd R. E. Mol. Cell. Biol. 2002;22:4499–4511. doi: 10.1128/MCB.22.13.4499-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnsson B., Lofas S., Lindquist G. Anal. Biochem. 1991;198:268–277. doi: 10.1016/0003-2697(91)90424-r. [DOI] [PubMed] [Google Scholar]
- 42.Rich R. L., Myszka D. G. Curr. Opin. Biotechnol. 2000;11:54–61. doi: 10.1016/s0958-1669(99)00054-3. [DOI] [PubMed] [Google Scholar]
- 43.De Crescenzo G., Grothe S., Lortie R., Debanne M. T., O’Connor-McCourt M. Biochemistry. 2000;39:9466–9476. doi: 10.1021/bi992987r. [DOI] [PubMed] [Google Scholar]