Abstract
In this study, we report a serum-free culture system for primary neonatal pulmonary cells that can support the growth of octamer-binding transcription factor 4+ (Oct-4+) epithelial colonies with a surrounding mesenchymal stroma. In addition to Oct-4, these cells also express other stem cell markers such as stage-specific embryonic antigen 1 (SSEA-1), stem cell antigen 1 (Sca-1), and Clara cell secretion protein (CCSP) but not c-Kit, CD34, and p63, indicating that they represent a subpopulation of Clara cells that have been implicated as lung stem/progenitor cells in lung injury models. These colony cells can be kept for weeks in primary cultures and undergo terminal differentiation to alveolar type-2- and type-1-like pneumocytes sequentially when removed from the stroma. In addition, we have demonstrated the presence of Oct-4+ long-term BrdU label-retaining cells at the bronchoalveolar junction of neonatal lung, providing a link between the Oct-4+ cells in vivo and in vitro and strengthening their identity as putative neonatal lung stem/progenitor cells. Lastly, these Oct-4+ epithelial colony cells, which also express angiotensin-converting enzyme 2, are the target cells for severe acute respiratory syndrome coronavirus infection in primary cultures and support active virus replication leading to their own destruction. These observations imply the possible involvement of lung stem/progenitor cells, in addition to pneumocytes, in severe acute respiratory syndrome coronavirus infection, accounting for the continued deterioration of lung tissues and apparent loss of capacity for lung repair.
Keywords: differentiation, expression of Oct-4, lung stem/progenitor cells, slow-cycling cells
Stem cells exist in most adult organs, the umbilical cord, and bone marrow and have displayed a surprising ability for self-renewal and a multilineage differentiation capacity for the repair of damaged cells and tissues. Given a growing interest in the characterization of stem cells of lung tissues for regenerative therapy, attempts have been made to identify and enrich lung stem cells. The lung is an extremely complex, conditionally renewing organ composed of at least 40 differentiated cell types/lineages and can be divided into proximal cartilaginous airways (trachea and bronchi), distal bronchioles (bronchioles, terminal bronchioles, and respiratory bronchioles), and gas-exchanging airspace (alveoli). The lung is lined with functionally and structurally distinct epithelium that probably contains different and unique types of adult epithelial stem/progenitor cells. Because the epithelial surface is constantly open to potential injury, stem/progenitor cells serve as a primary protective lining armed with rapid response mechanisms for epithelial repair (1). The candidate stem/progenitor cells, which can repair the injured lungs and contribute to local needs in times of tissue damage, are the basal cells for mucosal gland development and renewal of the branched epithelium of the trachea (2, 3), the Clara cells of the bronchiole (4), and the type-2 pneumocytes of the alveolus (5). Lung injury models with naphthalene have suggested that there are cytochrome P450 negative (CyP450−)-variant Clara cells residing within neuroepithelial bodies or the bronchoalveolar duct junction that are spared from the toxicity of naphthalene and are responsible for the subsequent bronchiolar regeneration (6–8). In addition, the nonhematopoietic side population cells isolated from the lung airway have been shown to have the same molecular phenotype as the CyP450−-variant Clara cells (9, 10). Recently, pulmonary stem cells residing in the bronchoalveolar junction of adult lungs have been identified and characterized as CD34+ Sca-1+ CD45− PE-CAM− cells expressing both cytoplasmic Clara cell secretion protein (CCSP) and prosurfactant protein-C proteins, which are markers for Clara cells and type-2 pneumocytes, respectively (11). However, the developmental relationship among the above reported pulmonary stem and progenitor cells remains to be defined.
In 2003, a new atypical pneumonia, severe acute respiratory syndrome (SARS), spread across several countries with a high mortality rate resulting from acute lung failure (12). The SARS pathogen was then subsequently identified as a new variant coronavirus (SARS-CoV) based on its cytopathic effect on VeroE6 cells (13–15). A number of animal models have been used to study the pathogenesis of SARS-CoV infection. Although the monkey model mimics to certain degree the clinical course of SARS (16), the mouse model provides the first genetic evidence for angiotensin-converting enzyme 2 (ACE-2) as a crucial SARS-CoV receptor in vivo (17). Although type-1 pneumocytes, and to a lesser extent type-2 pneumocytes, have been shown to be the target cells of SARS-CoV infection in monkey studies (16, 18), the identity of mouse bronchiolar epithelial cells infected by SARS-CoV remains unclear (19, 20). To investigate the cellular tropism of SARS-CoV in the mouse lung, we set up primary cultures for the enzyme-released cells from lung tissues and attempted to establish pulmonary epithelial cell lines that may enable us to develop a more effective cell-based screening system for chemical inhibitors of virus entry and replication. In this article, we describe a serum-free culture system that can support the growth of octamer-binding transcription factor 4+ (Oct-4+) epithelial colonies with a surrounding stroma from neonatal lung tissues. In addition, we show that these Oct-4+ stage-specific embryonic antigen 1+ (SSEA-1+) stem cell antigen 1+ (Sca-1+) cytokeratin-7+ ACE-2+ lung stem/progenitor cells, but not the surrounding stroma, are susceptible to SARS-CoV infection. These observations suggest a potential role for lung stem/progenitor cells, in addition to the type-1 pneumocytes, in the continued deterioration of lung tissues and apparent loss of the capacity for lung repair after SARS-CoV infection.
Results
Primary Pulmonary Cell Cultures for SARS-CoV Infection.
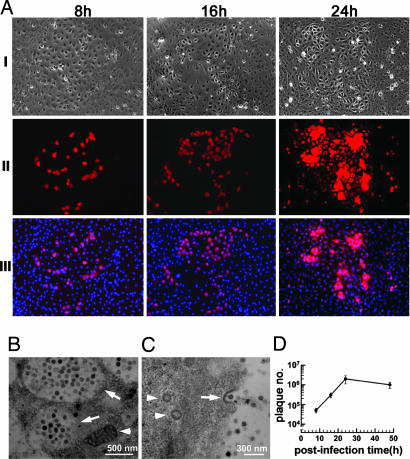
During our initial attempts to cultivate primary pulmonary cells for studies of SARS-CoV infection, serum-free monolayer cultures were prepared from nucleated pulmonary cells isolated from adult or neonatal mice, and the confluent cultures were then exposed to SARS-CoV at 1, 2, and 10 multiplicities of infection (moi). It was noted that infection occured only in the neonatal pulmonary cell culture in which a few tiny clusters with <30 cells stained positive for SARS-CoV nucleocapsid protein (data not shown). Because the incidence of the target cells was very low, we optimized the culture conditions by varying initial seeding cell density and the concentration of EGF in the culture so that epithelium-like colonies and clusters varying from a few tens to hundreds of cells appeared in the cultures after 10–14 days of incubation (see Fig. 6, which is published as supporting information on the PNAS web site). There were ≈106 ± 5 epithelium-like colonies with a surrounding stroma per culture when 3 × 105 nucleated neonatal lung cells were plated. The optimized primary pulmonary cultures were then exposed to SARS-CoV at 0.5 moi, and the kinetics of SARS-CoV infection was monitored. As shown in Fig. 1A, at 8 h postinfection ≈30% of cells within the pulmonary colonies displayed strong immunofluorescence for SARS-CoV nucleocapsid protein. The percentage of positive cells rose to ≈60% at 16 h, and by 24 h nearly every cell in the epithelial colonies was positive for SARS-CoV infection. Subsequently, the pulmonary colony cells exhibited cytopathic changes and detached by 48 h (data not shown). In contrast, none of the cells surrounding the epithelial colonies became infected at any time point examined (Fig. 1A) or on exposure to the SARS-CoV at a 20 times higher moi (data not shown). The susceptibility of epithelial colony cells, but not the surrounding stroma cells, to SARS-CoV infection was in line with the specific expression of ACE-2 on the former cell type (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
Identification of the pulmonary epithelial colony cells as the primary target for SARS-CoV infection. (A) Confluent primary pulmonary cultures were infected with SARS-CoV, and at 8, 16, and 24 h postinfection cultures were processed for immunostaining. Phase-contrast photographs of the pulmonary epithelial cells are shown in I. Immunostaining of infected cells within the colony are shown (red) by using antibodies directed against the nucleocapsid protein of SARS-CoV (II) and counterstained with DAPI (III). (B and C) Electron micrographs of infected cells at 16 h postinfection are shown. At this stage, the cytoplasm of infected cells was filled with swollen Golgi vesicles (arrows in B), which are shown to be full of mature SARS-CoV particles, and the mitochondria (arrowhead in B) are clearly illustrated. At a higher magnification, the virus vesicle was seen in the cell (arrowheads in C), and SARS-CoV particles with typical knob-like spikes were also observed outside the cell and attached to the plasma membrane via coated pits (arrow in C). (D) Kinetics of SARS-CoV replication in primary culture of pulmonary cells after virus infection at 0.5 moi.
To determine whether SARS-CoV replicated and produced infectious virus particles in the epithelial colony cells, we first used electron microscopy to examine the ultrastructure of the infected colony cells. At 16 h after infection, the cytoplasm of infected cells contained numerous vacuoles around the perinuclear region. At a higher magnification, these vacuoles were seen to be filled with virus particles (Fig. 1B), and mature virus particles were observed at the cell surface. Some of these extracellular virus particles were seen to associate with coated pits (Fig. 1C). We next collected the culture medium at indicated time points after infection to monitor the kinetics of virus replication. Fig. 1D shows that when the cultures were infected at a dose of 0.5 moi, the virus titers were 5 ± 1, 30 ± 8, 210 ± 75, and 104 ± 36 × 104 plaque-forming units/ml at 8, 16, 24, and 48 h postinfection, respectively. All these findings support the notion that the epithelial colony cells support active SARS-CoV replication with the virus titer peaked at 24 h postinfection.
Characterization of the Epithelial Colony Cells.
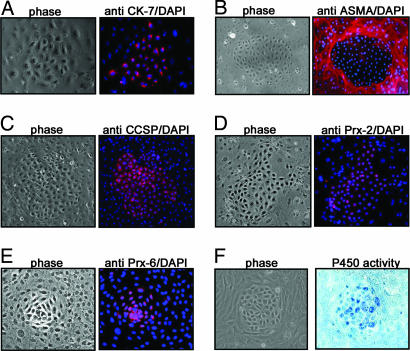
To characterize the type of cells grown in our serum-free medium cultures, we first performed immunocytochemistry by using a panel of epithelial and mesenchymal cell-specific antibodies. Immunostaining showed that the colony cells expressed cytokeratin-7 (Fig. 2A) but not cytokeratin-5/8, -18, and -19 (data not shown), whereas the cells surrounding the epithelial colonies stained positive for α-smooth muscle actin, suggesting that the surrounding stroma cells are mesenchymal in nature (Fig. 2B).
Fig. 2.
Characteristics of pulmonary epithelial colony cells. (A) Primary cell cultures were examined by using specific antibodies directed against cytokeratin-7 (CK-7). (B) To investigate the markers expressed in the surrounding cells, anti-α-smooth muscle actin (ASMA) was applied. (C) The cells were also examined for CCSP. Two other antibodies against peroxiredoxin 6 (Prx-6) (D) and peroxiredoxin 2 (Prx-2) (E), respectively, were applied. Both phase-contrast photographs and the respective immunostaining merged with DAPI are shown. The CyP450 enzyme activities were examined and are shown in F.
We further examined the nature of the epithelial colony cells by using a panel of pulmonary cell-type-specific monoclonal antibodies. Immunostaining showed that the epithelial colony cells expressed CCSP (Fig. 2C) and peroxiredoxin 2 and 6 (Fig. 2 D and E) and displayed relatively high CyP450 activities (Fig. 2F). However, they lacked alkaline phosphatase activity and were negative for c-Kit, CD34, p63, surfactant protein C, and aquaporin 5 expression (data not shown).
Stem Cell Marker Expression in the Pulmonary Epithelial Colony Cells.
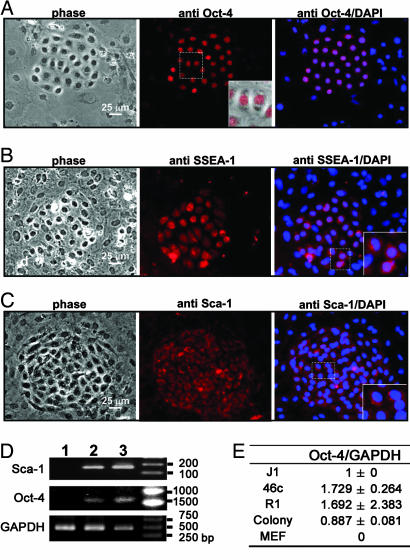
Because the epithelial colony cells lack markers for alveolar type-1 and type-2 pneumocytes but express CCSP and display detectable CyP450 activities, we next examined the expression of stem cell markers for embryonic and adult tissue stem cells. Immunostaining showed that the epithelial colony cells expressed Oct-4, SSEA-1, and Sca-1, whereas the surrounding cells did not (Fig. 3A–C). On close microscopic examination, Oct-4 expression was detected in the nuclei, whereas SSEA-1 and Sca-1 were mainly found in the cell surface and cytoplasm of the pulmonary colony cells (Fig. 3 A–C Inset). The presence of Oct-4 and Sca-1 mRNA was confirmed by RT-PCR analysis performed on the colony cells plucked from the cultures (Fig. 3D). The amplified products for Sca-1 and Oct-4 were consistent with the expected size of 160 and 1,121 bp, respectively. Because Oct-4 expression is relatively rare in adult cells and tissues, we next performed quantitative RT-PCR to evaluate the level of Oct-4 expression in the colony cells (see Fig. 8, which is published as supporting information on the PNAS web site). Fig. 3E shows that the Oct-4 expression level in the epithelial colony cells was quite high, approaching 51%, 52%, and 88% of mouse ES cell lines 46c, R1, and J1, respectively.
Fig. 3.
Stem cell characteristics of the pulmonary epithelial colony cells. Primary cultures were examined for expression of Oct-4 (A), SSEA-1 (B), and Sca-1 (C). Insets are magnifications of the white square dotted areas in respective photographs, showing the pattern of these cells expressing specific markers. Cells were also stained with DAPI and merged with their respective fluorescent images. (D) RT-PCR analysis for the cells of individually plucked pulmonary colonies. For Sca-1, the following cells were prepared and analyzed: lane 1, the A549 cell line (negative control); lane 2, the cells plucked from pulmonary epithelial colonies; and lane 3, BW5147 cells (positive control). For Oct-4, lane 1 was the TM4 cell line (negative control); lane 2 was the cells picked from pulmonary epithelial colonies; and lane 3 was the mouse R1 embryonic line (positive control). GAPDH was used as internal standard for both reactions. (E) Quantitative RT-PCR analysis of Oct-4 expression in pulmonary colony cells with respect to J1, 46c, and R1 embryonic stem cells; in contrast, mouse embryonic fibroblasts (MEF) were used as negative control. The results represent the average of three independent experiments with standard deviations. A graphic presentation of quantitative RT-PCR is given in Fig. 8.
Because the epithelial colony cells express both Oct-4 and ACE-2, we next investigated whether we could directly demonstrate the infection of SARS-CoV on Oct-4-expressing cells. As shown in Fig. 9, which is published as supporting information on the PNAS web site, 20–30% of the Oct-4+ cells showed cytoplasmic immunostaining for SARS-CoV nucleocapsid protein at 8 h after infection.
Differentiation Potential of the Epithelial Colony Cells.
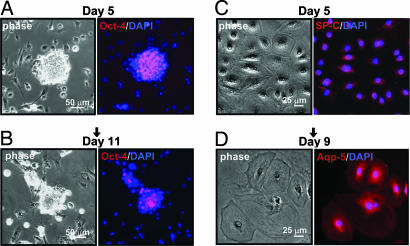
To address the self-renewal and differentiation potential of these pulmonary colony cells, individual colonies were plucked from the primary cultures and transferred to culture dishes either precoated with collagen I or preseeded with irradiated primary pulmonary cultured cells. The transferred colonies were then monitored for the expression of Oct-4, surfactant protein C, and aquaporin 5 protein. In the presence of preirradiated cultured pulmonary cells, the transferred colonies did not attach well and appeared as spherical aggregates in which Oct-4 expression was maintained in the majority of the colony cells up to day 5 (Fig. 4A); by day 11 the colony cells displayed diminished Oct-4 expression (Fig. 4B). No surfactant protein C or aquaporin 5 protein could be detected in the transferred colonies during the 11-day culture period. In contrast, the individually plucked colonies transferred into collagen I-coated plates attached well, and the cells continued to grow, migrated out, and appeared as thinly spread flattened cell clusters. Oct-4 expression decreased rapidly within 2 days, and by day 5 the expression of surfactant protein C was detected in the cytoplasm, especially in the perinuclear region of these flattened cells (Fig. 4C). These features are consistent with the type-2 pneumocytes. By day 9, the colony continued to expand and to spread thinly such that the average diameter of individual colony cells was ≈5-fold greater than that of the parental primary epithelial colony cells. Expression of surfactant protein C decreased, whereas expression of aquaporin 5 protein, a marker for type-1 pneumocytes, became pronounced (Fig. 4D). These observations suggest that Oct-4+ SSEA-1+ Sca-1+ pulmonary epithelial colony cells have the potential to differentiate to type-2 and -1 pneumocytes in a sequential fashion. In addressing whether these in vitro differentiated type-2 and -1 pneumocytes are susceptible to SARS-CoV infection, individual epithelial colonies were plucked and transferred to culture dishes precoated with collagen I. The cultures were then exposed to SARS-CoV at moi of 0.5 and 10 on days 5 and 9 after colony transfer. We found no evidence of SARS-CoV infection or virus replication on these differentiated type-2 and -1 pneumocytes by either immunostaining or virus titering (data not shown).
Fig. 4.
Differentiation potential of pulmonary epithelial colonies. The expression of alveolar cell markers, surfactant protein C (SPC; a type-2 pneumocyte marker), and aquaporin 5 (Aqp-5; a type-1 pneumocyte marker), were examined in the cells after clone transfer at day 5 and day 9 or 11, respectively. In primary cell culture, cells (Oct-4+) in the pulmonary epithelial colonies were picked and subcultured onto irradiated 5-day-old primary lung cell cultures or collagen I-coated plates with conditioned media. (A) The picked colony cells were subcultured onto irradiated 5-day-old primary culture cells, and the spherical colonies were observed on day 5. The colonies stained Oct-4 positive. (B) The spherical colonies maintain Oct-4 expression but with a lower expression level on day 11. (C) The picked cells were subcultured onto collagen I-coated plates, and on day 5 the cells were positive for surfactant protein C protein. (D) On day 9, the phase-contrast photograph of the cells indicated that the morphology of individual cells changed; cells were flattened and stained positive with aquaporin 5 protein.
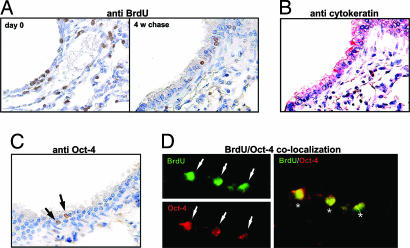
BrdU Label-Retaining Cells (LRC) in the Neonatal Lung Expressed Oct-4.
To address whether the Oct-4+ epithelial colony cells growing in our serum-free medium cultures arise de novo or from the Oct-4+ cells present in the neonatal lungs, we performed a pilot immunohistochemistry study and observed the presence of a small number of Oct-4-expressing cells at the bronchoalveolar junction of the neonatal lung (data not shown). To further ascertain whether these small number of Oct-4-expressing cells in neonatal lung can be the “putative” lung stem cells, we carried out BrdU pulse–chase experiments as described in Materials and Methods to label the long-term LRC. Immediately after five daily injections of BrdU, a large number of cells, including some alveolar cells (Fig. 5A Left), demonstrated nuclear staining for BrdU. The number of BrdU-positive cells in the lungs declined significantly with time such that by 4 weeks only a few cells still retained BrdU labeling (arrows in Fig. 5A Right). These LRC were found almost exclusively in the epithelial layer of terminal bronchioles adjacent to alveolar sacs, as revealed by staining with monoclonal antibodies against pan-cytokeratin (AE1/3), general markers for epithelial cells (Fig. 5B). We then performed immunohistochemical staining for Oct-4 and determined the number of Oct-4+ cells and BrdU LRC. The analysis revealed that there were 21 ± 8 dual Oct-4+ BrdU-retaining cells in each randomly selected lung section that contained bronchoalveolar junctions (Fig. 5D). Single Oct-4+ cells or BrdU LRC were not encountered. As estimated by histological grid analysis, there are ≈1.25 × 106 nucleated cells per slide, suggesting the presence of a small population (≈0.0016 ± 0.0006%) of slow-cycling, Oct-4-expressing stem cells in the neonatal lungs.
Fig. 5.
Immunohistochemical analysis of BrdU retention and Oct-4 expression in lung tissues. (A) Lung tissue sections of neonatal mouse were stained for BrdU immediately after a 5-day BrdU-labeling scheme (day 0) and a 4-week chase period (4 w chase). Cells with BrdU retention stained brown in the nuclei. (B) A successive tissue section of the 4-week chase period BrdU-retention experiment was stained with antibodies directed against cytokeratins. (C) The tissue section for the 4-week chase period experiment was stained for Oct-4 expression. (D) Immunofluorescence double-staining of the tissue section for the 4-week chase period experiment indicated that Oct-4 expression (Lower; red) was colocalized with BrdU retention (Upper; green). Double-stained cells are indicated by asterisks in the merged image.
Discussion
In this article, we report on a serum-free culture system that can support the growth of pulmonary Oct-4+ epithelial colonies in vitro. Culture of lung epithelial cells such as Clara cells and type-2 and -1 pneumocytes from different species had been reported in refs. 21–23. However, the expression of stem cell markers in these cultured lung cells has not been addressed. Our success in growing Oct-4+ SSEA-1+ Sca-1+ cytokeratin-7+ pulmonary epithelial stem/progenitor cells can be attributed to the use of collagen I as the attachment substrate, as well as the use of serum-free selection media, which minimizes the potential inconsistency problems associated with FCS.
Oct-4, a member of the POU family of transcription factors, is now universally accepted as a robust marker of pluripotency for both mouse and human embryonic stem cells (24). Expression of Oct-4 is rarely seen in normal somatic tissues but can be detected in some human tumors (25–27). A recent study has shown that a few Oct-4+ cells can be found scattered in the basal layer of the epidermis; however, the identity of these cells has not been determined (28). In contrast, a growing list of cells have been shown to express Oct-4, including cells growing in cultures from mouse bone marrow CD34+ hematopoietic progenitors (29) and human umbilical cord blood (30), cultured epithelial cells lacking connexins and gap junctional intercellular communication (28), and mesenchymal stem cells (31, 32). In the current study, we have demonstrated that the neonatal lung-derived Oct-4+ epithelial colony cells also coexpress other stem cell markers such as SSEA-1 and Sca-1 but not c-Kit, CD34, and p63. In addition, by using real-time quantitative RT-PCR analysis, we have also demonstrated that the level of Oct-4 expression in these epithelial colony cells can be as high as 51–88% in three different mouse ES cell lines (see Fig. 3E). These Oct-4+ SSEA-1+ Sac-1+ colony cells also expressed markers such as cytokeratin-7, CCSP, and peroxiredoxin 2 and 6 and displayed relatively high CyP450 activities, most of which are known to be associated with Clara cells. It appears that these Oct-4+ colony cells represent a subpopulation of Clara cells, which have long been implicated in lung repair and regeneration after injury (6–8). Lastly, we have demonstrated that the LRC in neonatal lung coincide with Oct-4+ cells, thus providing a link between the Oct-4+ cells in vivo and in vitro that helps to argue against the notion that the Oct-4 expression in these cultured epithelial cells is due to induction after serum starvation, as had been reported for human mesenchymal stem cells (32). Importantly, these Oct-4+ cells were shown in colony transfer experiments to have the potential to differentiate into type-1- and -2-like pneumocytes sequentially. Our study thus demonstrated the expression of Oct-4 in adult stem cells. Recently, the bronchoalveolar stem cells in adult lung have been identified and characterized as CD34+ Sca-1+ CD45− PE-CAM− cells expressing both cytoplasmic CCSP and surfactant protein C, which are markers for Clara cells and type-2 pneumocytes, respectively (11).
A comparison of the reported adult bronchoalveolar stem cells and cultured Oct-4+ lung epithelial colony cells in our study reveals that they share common markers such as Sca-1 and CCSP but not others. For example, the reported adult bronchoalveolar lung stem cells express CD34 and surfactant protein C, whereas the Oct-4+ colony cells do not. The developmental relationship between the adult lung bronchoalveolar stem cells and neonatal lung Oct-4+ LRC remains to be elucidated. Although the Oct-4+ epithelial colony cells can be maintained for weeks, we have not been able to demonstrate extensive self-renewal of these cells in vitro except during the initial phase of culture. Our data further suggest that the mesenchymal stroma is important for the growth and maintenance of Oct-4+ pulmonary epithelial colony cells in the primary cultures. A better understanding and manipulation of the supportive mechanisms provided by the stroma may help us to address the self-renewal potential of these cells in vitro and their functional attributes in vivo.
SARS-CoV was the causative agent of the recent outbreak of SARS-induced atypical pneumonia (13, 14). The basis for continuous deterioration of lung function and the apparent loss of capacity for lung repair after viral load declines is not well understood (33). Clinical studies and in vivo studies with macaque monkeys revealed that the target cells of SARS-CoV infection are primarily type-1 pneumocytes, damage to which is closely associated with the subsequent diffuse alveolar damage (18). Our observations that the pulmonary Oct-4+ colony cells are susceptibility to SARS-CoV infection and allow virus replication leading to their own demise implies a potential role of the pulmonary stem/progenitor cells in the apparent loss of capacity for lung repair after SARS-CoV infection and later phases of lung failure. Currently, data to support the notion that Oct-4+ bronchoalveolar lung stem cells are the prime target cells for SARS-CoV infection in vivo are still lacking. However, the description of a highly restricted and patchy localization of SARS-CoV RNA in the epithelial lining of a subset of bronchi and terminal bronchioles of the day-3 infected mice (20) in part supports our in vitro observation.
In conclusion, we have demonstrated the presence of Oct-4-expressing LRC specifically located at the bronchoalveolar junction of 4-week-old mice. In addition, we have successfully developed a serum-free primary culture system for the growth of Oct-4+ SSEA-1+ Sca-1+ cytokeratin-7+ lung stem/progenitor cells, which are capable of differentiation into type-2 and -1 pneumocytes and are susceptible to SARS-CoV infection.
Materials and Methods
Pulmonary Primary Cell Culture.
Neonatal ICR mice were killed, and their lungs were removed and cut into small pieces. After washing in Hank’s buffer containing penicillin (100 units/ml) and streptomycin (100 μg/ml), the tissues were treated with 0.1% protease type-XIV (Sigma) in Joklik’s MEM (Sigma) at 4°C overnight. Afterward, tissues were transferred to 10% FCS/Joklik’s MEM, pipetted several times to release pulmonary cells, and then filtered through a 100-μm nylon cell strainer. The released cells were washed and resuspended in MCDB-201 medium containing insulin–transferrin–selenium supplements only (GIBCO). One neonatal mouse could yield ≈1.0–1.5 × 106 nucleated cells in this enzyme digestion procedure, which represents ≈5% to 8% of the total number of cells in lung tissues. These cells were cultivated at a density of 3 × 105 cells per milliliter in culture dishes coated with collagen I (10 μg/cm2; BD Bioscience). After 1 day of incubation, the primary cultures were washed with MCDB-201 medium to remove unattached cells, and fresh medium with insulin–transferrin–selenium supplement and epidermal growth factors (1 ng/ml; Invitrogen) was then added.
Virus Infection and Analysis.
All experiments for SARS-CoV infection were carried out at a P4 facility in the Institute of Preventive Medicine (Taipei, Taiwan). Confluent primary cultures of pulmonary cells were incubated with SARS-CoV (strain Tw7) (34) at various moi (0.5, 1, 2, and 10) for 1 h at room temperature. Cells were washed once with PBS and incubated in MCDB-201 medium containing supplements. At 8, 16, 24, and 48 h postinfection, the supernatants of the cultures were collected for viral titration and the cells were washed with PBS and fixed in ethanol/acetone (1:1) for immunofluorescence analysis. The SARS-CoV was detected by using a mouse monoclonal antibody (1:2,000) generated against the recombinant SARS-CoV nucleocapsid protein for this study. The epitope of this monoclonal antibody was localized to the N-terminal region of the nucleocapsid (M.D.K., unpublished data).
Virus titers were determined by plaque-forming assay with modifications using VeroE6 cells. Briefly, serial dilutions of the harvested supernatant were added into confluent culture of VeroE6 cells. After incubation for 30 min at 37°C, the unabsorbed virus was washed off and the culture was overlaid with 1% agar in DMEM and 2% FBS. After plaques were formed, the agar overlay was removed, the cells were stained with crystal violet solution (10%) for 10 min, and the plaques were counted.
Electron Microscopy.
Transmission electron microscopy was performed as described, with modification (35). The pulmonary epithelial cells were cultivated on collagen I-coated ACLAR–Fluoropolymer films (Structure Probe). The cells were infected with SARS-CoV at 0.5 moi as described. At 16 h postinfection, cells were fixed with 2% glutaraldehyde/4% paraformaldehyde/PBS for 2 h, followed by 1% osmium tetroxide for 1 h, and then embedded in Spurr’s resin. Ultrathin sections (60 nm) of the embedded cells were prepared, stained with 2% uranyl acetate and 1% lead citrate, and analyzed with an H-7000 electron microscope (Hitachi, Tokyo).
Immunocytochemistry.
Cells in primary cultures were fixed in methanol/acetone (1:1) for 3 min at room temperature. For analysis of Sca-1 expression, cells were fixed in 4% paraformaldehyde/PBS for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 5 min, and then blocked with 3% BSA/PBS for 30 min. Cells were incubated at 4°C with primary antibodies against the following antigens: peroxiredoxin 2 [a kind gift from J. H. Chen (National Defense Medical Center)]; peroxiredoxin 6 (ab16824; Abcam); CD34 (clone RAM34; BD Biosciences); aquaporin 5 (AB3069), cytokeratin-5/8 (MAB3228), cytokeratin-7 (MAB3226), and cytokeratin-18 (MAB3234), p63 (MAB4135), pan-cytokeratin (clone AE1/3), surfactant protein C (AB3786), and SSEA-1 (MAB4301) (all from Chemicon); α-smooth muscle actin (clone 1A4; DAKO); cytokeratin 19 (IF15; Oncogene); c-Kit (MAB1356) and Sca-1 (AF1229) (both from R & D Systems); and ACE-2 (sc20998), CCSP (sc9773) and Oct-4 (sc9081) (all from Santa Cruz Biotechnology). After overnight incubation, cells were washed and incubated for 1 h at room temperature with the following respective Cy3-labeled secondary antibodies: donkey anti-goat IgG, goat anti-rabbit IgG, goat anti-mouse IgG, goat anti-mouse IgM (μ-chain-specific), and goat anti-rat IgG (Jackson ImmunoResearch). Cells were then counterstained with DAPI. Primary cultures were also analyzed for enzymatic activity of alkaline phosphatase according to standard protocols using an AP detection kit (Chemicon) and CyP450, as described in ref. 36.
RT-PCR and Quantitative RT-PCR.
Pulmonary epithelial colony cells were collected for analysis. Under a microscope, a 26-gauge needle was used to delineate the boundary of pulmonary epithelial colonies, and the colonies were gently plucked from the cultures by using a finely drawn Pasteur pipette. Three mouse ES cell lines, 46c (37), R1, and J1, were used for a positive control for Oct-4 expression, and the TM4 cell line (mouse testis Sertoli cells; American Type Culture Collection) and MEF (mouse embryonic fibroblast) were used for a negative control. For Sca-1, the BW5147 cell line (T lymphocyte cell) was used for a positive control, and the A549 cell line (type-2 pneumocyte) was used for a negative control (both from American Type Culture Collection). Total RNA was prepared with the RNeasy Micro Kit (Qiagen, Valencia, CA), and reverse transcription was carried out with random primers by using the SuperScript first-strand synthesis system (Invitrogen) according to the manufacturer’s instructions. For PCR, the forward and reverse primers were as follows: (i) 5′-ATGGCTGGACACCTGGCTTC-3′ and 5′-CCAGGTTCTCTTGTCTACCTC-3′ for Oct-4 expression (38), (ii) 5′-GGACACTTCTCACACTACAAAG-3′ and 5′-TAACACAGACTCCATCAGGGTAG-3′ for Sca-1 expression (39), and (iii) 5′-ACCACAGTCCATGCCATCAC-3′ and TCCACCACCCTGTTGCTGTA-3′ for GAPDH as the internal control in both reactions (38). Quantitative RT-PCR was performed by using the ABI Prism 7000 sequence detection system (Applied Biosystems) following the manufacturer’s instructions. The primer/probe sets for mouse Oct-4 (TaqMan gene expression assay no. Mm00658129_gH; Applied Biosystems) and mouse GAPDH (TaqMan gene expression assay no. Mm99999915_g1) were used. Quantitative RT-PCR was carried out for 45 cycles, and raw data were analyzed by ABI Prism 7000 sds software (Applied Biosystems). The cycle threshold, Ct, of each sample was generated with the default setting. The Oct-4 expression level of each sample was normalized to the expression level of GAPDH in the same sample by the following formula: Oct-4/GAPDH = 2−(Ct of Oct-4 − Ct of GAPDH). The Oct-4/GAPDH ratio of the 46c cell line was set to 1.0, and the values of all others were recalculated accordingly. The result represents the average of three independent experiments, with standard deviations.
In Vitro Differentiation.
To analyze differentiation potential, cells were plucked from individual colonies as described above and transferred to either new culture dishes free of the surrounding stromal cells, or new culture dishes with the irradiated primary culture cells as a feeder layer. The primary culture cells used for the feeder layer were grown to near confluence and preirradiated with 1,500 rad in a 137Cs source (Atomic Energy, Ottawa). In this assay, conditioned media were applied to both conditions. The conditioned media were harvested from confluent pulmonary primary cultures and filtered with 0.2-μm filters.
BrdU Labeling, Oct-4 Expression, and Immunohistochemical Analysis.
Neonatal ICR mice were injected i.p. with 50 mg/kg BrdU (Sigma) in PBS twice a day for 5 days. Mice were maintained without further BrdU injection and killed on day 0 or after 1, 2, 3, or 4 weeks of chase for BrdU labeling. Lungs were removed, fixed in 10% formalin fixative, and embedded in paraffin. Afterward, 5-μm sections of lung tissues were obtained and stained for BrdU as described in ref. 40. For Oct-4 expression, the general staining protocol was the same as described in ref. 41, with the following modifications: antigen retrieval was carried out by heating for 8 min in sodium citrate buffer (10 mM; pH 6.0), followed by a 15-min incubation at room temperature. Mouse monoclonal anti-BrdU (DAKO) and rabbit anti-Oct-4 antibodies (Santa Cruz Biotechnology) were added, respectively. Afterward, immunohistochemical analysis was performed with a peroxidase detection kit (Vector Laboratories) by using diaminobenzene as substrate according to the manufacturer’s instructions. Furthermore, anti-pan-cytokeratin antibodies (AE1/3 clone; Chemicon) and an alkaline phosphatase detection kit with Fast-Red as a substrate were used to delineate the location of bronchoalveolar junctions (42). All sections were counterstained with Mayer’s hematoxylin.
For double staining of immunofluorescence with anti-BrdU and Oct-4 primary antibodies, the FITC-conjugated goat anti-mouse and Cy3-conjugated goat anti-rabbit antibodies (Jackson ImmunoResearch) were used, respectively.
Supplementary Material
Acknowledgments
We greatly appreciate the guidance provided by the late Dr. Nai-San Wang. Mouse ES cell lines 46c, R1, and J1 were a generous gift from Meng Li (University of Edinburgh) and Dr. Chia-Ning Shen (Academia Sinica). We thank Y. K. Huang, W. W. Chen, and M. L. Chiang for their excellent technical assistance and the Transmission Electron Microscopy Core Facility (Institute of Cellular and Organismic Biology, Academia Sinica) for expert assistance. This work was supported by National Science Council Grant NSC93-2751-B-001-001-Y.
Abbreviations
- CyP450
cytochrome P450
- CCSP
Clara cell secretion protein
- SARS
severe acute respiratory syndrome
- SARS-CoV
SARS coronavirus
- ACE-2
angiotensin-converting enzyme 2
- moi
multiplicity of infection
- SSEA-1
stage-specific embryonic antigen 1
- Oct-4
octamer-binding transcription factor 4
- Sca-1
stem cell antigen 1
- LRC
label-retaining cells.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Gail D. B., Lenfant C. J. Am. Rev. Respir. Dis. 1983;127:366–387. doi: 10.1164/arrd.1983.127.3.366. [DOI] [PubMed] [Google Scholar]
- 2.Engelhardt J. F., Schlossberg H., Yankaskas J. R., Dudus L. Development (Cambridge, U.K.) 1995;121:2031–2046. doi: 10.1242/dev.121.7.2031. [DOI] [PubMed] [Google Scholar]
- 3.Hong K. U., Reynolds S. D., Watkins S., Fuchs E., Stripp B. R. Am. J. Pathol. 2004;164:577–588. doi: 10.1016/S0002-9440(10)63147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans M. J., Cabral-Anderson L. J., Freeman G. Lab. Invest. 1978;38:648–653. [PubMed] [Google Scholar]
- 5.Fehrenbach H. Respir. Res. 2001;2:33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hong K. U., Reynolds S. D., Giangreco A., Hurley C. M., Stripp B. R. Am. J. Respir. Cell Mol. Biol. 2001;24:671–681. doi: 10.1165/ajrcmb.24.6.4498. [DOI] [PubMed] [Google Scholar]
- 7.Giangreco A., Reynolds S. D., Stripp B. R. Am. J. Pathol. 2002;161:173–182. doi: 10.1016/S0002-9440(10)64169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds S. D., Giangreco A., Power J. H., Stripp B. R. Am. J. Pathol. 2000;156:269–278. doi: 10.1016/S0002-9440(10)64727-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majka S. M., Beutz M. A., Hagen M., Izzo A. A., Voelkel N., Helm K. M. Stem Cells. 2005;23:1073–1081. doi: 10.1634/stemcells.2005-0039. [DOI] [PubMed] [Google Scholar]
- 10.Summer R., Kotton D. N., Sun X., Ma B., Fitzsimmons K., Fine A. Am. J. Physiol. Lung Cell Mol. Physiol. 2003;285:L97–104. doi: 10.1152/ajplung.00009.2003. [DOI] [PubMed] [Google Scholar]
- 11.Kim C. F., Jackson E. L., Woolfenden A. E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R. T., Jacks T. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. Weekly Epidemiol. Rec. 2003;78:81–83. [Google Scholar]
- 13.Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Penaranda S., Bankamp B., Maher K., Chen M. H., et al. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 14.Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., et al. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 15.Gillim-Ross L., Taylor J., Scholl D. R., Ridenour J., Masters P. S., Wentworth D. E. J. Clin. Microbiol. 2004;42:3196–3206. doi: 10.1128/JCM.42.7.3196-3206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuiken T., Fouchier R. A., Schutten M., Rimmelzwaan G. F., van Amerongen G., van Riel D., Laman J. D., de Jong T., van Doornum G., Lim W., et al. Lancet. 2003;362:263–270. doi: 10.1016/S0140-6736(03)13967-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W., et al. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haagmans B. L., Kuiken T., Martina B. E., Fouchier R. A., Rimmelzwaan G. F., van Amerongen G., van Riel D., de Jong T., Itamura S., Chan K. H., et al. Nat. Med. 2004;10:290–293. doi: 10.1038/nm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subbarao K., McAuliffe J., Vogel L., Fahle G., Fischer S., Tatti K., Packard M., Shieh W. J., Zaki S., Murphy B. J. Virol. 2004;78:3572–3577. doi: 10.1128/JVI.78.7.3572-3577.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glass W. G., Subbarao K., Murphy B., Murphy P. M. J. Immunol. 2004;173:4030–4039. doi: 10.4049/jimmunol.173.6.4030. [DOI] [PubMed] [Google Scholar]
- 21.Richards R. J., Oreffo V. I., Lewis R. W. Environ. Health Perspect. 1990;85:119–127. doi: 10.1289/ehp.85-1568339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isakson B. E., Seedorf G. J., Lubman R. L., Boitano S. In Vitro Cell Dev. Biol. Anim. 2002;38:443–449. doi: 10.1290/1071-2690(2002)038<0443:HCOPAE>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 23.Murphy S. A., Dinsdale D., Hoet P., Nemery B., Richards R. J. Methods Cell Sci. 1999;21:31–38. doi: 10.1023/a:1009825008023. [DOI] [PubMed] [Google Scholar]
- 24.Saito S., Liu B., Yokoyama K. Hum. Cell. 2004;17:107–115. doi: 10.1111/j.1749-0774.2004.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 25.Monk M., Holding C. Oncogene. 2001;20:8085–8091. doi: 10.1038/sj.onc.1205088. [DOI] [PubMed] [Google Scholar]
- 26.Abate-Shen C. Cancer Cell. 2003;4:329–330. doi: 10.1016/s1535-6108(03)00277-0. [DOI] [PubMed] [Google Scholar]
- 27.Gidekel S., Pizov G., Bergman Y., Pikarsky E. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 28.Tai M. H., Chang C. C., Kiupel M., Webster J. D., Olson L. K., Trosko J. E. Carcinogenesis. 2005;26:495–502. doi: 10.1093/carcin/bgh321. [DOI] [PubMed] [Google Scholar]
- 29.Goolsby J., Marty M. C., Heletz D., Chiappelli J., Tashko G., Yarnell D., Fishman P. S., Dhib-Jalbut S., Bever C. T., Jr., Pessac B., et al. Proc. Natl. Acad. Sci. USA. 2003;100:14926–14931. doi: 10.1073/pnas.2434383100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuckin C. P., Forraz N., Baradez M. O., Navran S., Zhao J., Urban R., Tilton R., Denner L. Cell Prolif. 2005;38:245–255. doi: 10.1111/j.1365-2184.2005.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izadpanah R., Joswig T., Tsien F., Dufour J., Kirijan J. C., Bunnell B. A. Stem Cells Dev. 2005;14:440–451. doi: 10.1089/scd.2005.14.440. [DOI] [PubMed] [Google Scholar]
- 32.Pochampally R. R., Smith J. R., Ylostalo J., Prockop D. J. Blood. 2004;103:1647–1652. doi: 10.1182/blood-2003-06-1967. [DOI] [PubMed] [Google Scholar]
- 33.Peiris J. S., Chu C. M., Cheng V. C., Chan K. S., Hung I. F., Poon L. L., Law K. I., Tang B. S., Hon T. Y., Chan C. S., et al. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeh S. H., Wang H. Y., Tsai C. Y., Kao C. L., Yang J. Y., Liu H. W., Su I. J., Tsai S. F., Chen D. S., Chen P. J. Proc. Natl. Acad. Sci. USA. 2004;101:2542–2547. doi: 10.1073/pnas.0307904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng M. L., Tan S. H., See E. E., Ooi E. E., Ling A. E. J. Gen. Virol. 2003;84:3291–3303. doi: 10.1099/vir.0.19505-0. [DOI] [PubMed] [Google Scholar]
- 36.Oreffo V. I., Morgan A., Richards R. J. Environ. Health Perspect. 1990;85:51–64. doi: 10.1289/ehp.85-1568317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. Nat. Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 38.Shimozaki K., Nakashima K., Niwa H., Taga T. Development (Cambridge, U.K.) 2003;130:2505–2512. doi: 10.1242/dev.00476. [DOI] [PubMed] [Google Scholar]
- 39.Bonyadi M., Waldman S. D., Liu D., Aubin J. E., Grynpas M. D., Stanford W. L. Proc. Natl. Acad. Sci. USA. 2003;100:5840–5845. doi: 10.1073/pnas.1036475100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borthwick D. W., Shahbazian M., Krantz Q. T., Dorin J. R., Randell S. H. Am. J. Respir. Cell Mol. Biol. 2001;24:662–670. doi: 10.1165/ajrcmb.24.6.4217. [DOI] [PubMed] [Google Scholar]
- 41.Rajpert-De M. E., Hanstein R., Jorgensen N., Graem N., Vogt P. H., Skakkebaek N. E. Hum. Reprod. 2004;19:1338–1344. doi: 10.1093/humrep/deh265. [DOI] [PubMed] [Google Scholar]
- 42.Krause D. S., Theise N. D., Collector M. I., Henegariu O., Hwang S., Gardner R., Neutzel S., Sharkis S. J. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.