Abstract
Inositol 1,4,5-trisphosphate (IP3) receptors (IP3Rs) are IP3-gated Ca2+ channels that are located on intracellular Ca2+ stores. We previously identified an IP3R binding protein, termed IP3R binding protein released with IP3 (IRBIT). Because IRBIT is released from IP3R by physiological concentrations of IP3, we hypothesized that IRBIT is a signaling molecule that is released from IP3R and regulates downstream target molecules in response to the production of IP3. Therefore, in this study, we attempted to identify the target molecules of IRBIT, and we succeeded in identifying Na+/HCO3− cotransporter 1 (NBC1) as an IRBIT binding protein. Of the two major splicing variants of NBC1, pancreas-type NBC1 (pNBC1) and kidney-type NBC1 (kNBC1), IRBIT was found to bind specifically to pNBC1 and not to bind to kNBC1. IRBIT binds to the N-terminal pNBC1-specific domain, and its binding depends on the phosphorylation of multiple serine residues of IRBIT. Also, an electrophysiological analysis in Xenopus oocytes revealed that pNBC1 requires coexpression of IRBIT to manifest substantial activity comparable with that of kNBC1, which displays substantial activity independently of IRBIT. These results strongly suggest that pNBC1 is the target molecule of IRBIT and that IRBIT has an important role in pH regulation through pNBC1. Also, our findings raise the possibility that the regulation through IRBIT enables NBC1 variants to have different physiological roles.
Keywords: pH, acidosis, phosphorylation
Inositol 1,4,5-trisphosphate (IP3) receptors (IP3Rs) are intracellular Ca2+-release channels that are located on intracellular Ca2+-storage organelles, mainly the endoplasmic reticulum (ER) (1). IP3Rs release Ca2+ from the ER into the cytoplasm and increase the cytoplasmic concentration of Ca2+ in response to the binding of a second messenger, IP3. This IP3–Ca2+ pathway regulates many biological processes, including cell growth, cell differentiation, apoptosis, synaptic plasticity, secretion, and fertilization (1).
We identified (2) an IP3R binding protein, termed IP3R binding protein released with IP3 (IRBIT). IRBIT consists of an N-terminal domain (residues 1–104), which contains a serine/threonine-rich region, and a C-terminal domain (residues 105–530), which has homology with the methylation pathway enzyme S-adenosylhomocysteine hydrolase. We found (2) that the N-terminal amino acids 1–277 of IRBIT are sufficient for the interaction with the IP3R and that the interaction between IRBIT and the IP3R is inhibited by physiological concentrations of IP3, indicating that IRBIT interacts with the IP3R in the resting state and dissociates from the IP3R when IP3 production is induced by extracellular stimuli. Therefore, we speculated that IRBIT acts as a signaling molecule that dissociates from the IP3R and regulates target proteins in response to IP3 production, raising the possibility of the existence of an unidentified pathway, the IP3–IRBIT pathway.
The Na+/HCO3− cotransporter 1 (NBC1) is a membrane-integrated transporter protein that mediates the coupled movement of Na+ and HCO3− ions across the plasma membrane (3). Because NBC1 transports >1 HCO3− ion (2 or 3) per Na+ ion, the transport that is mediated by NBC1 is associated with a net movement of negative charge across the membrane. We and other researchers (4–8) have reported that inactivating mutations of the human NBC1 gene SLC4A4 cause proximal renal tubular acidosis associated with ocular abnormalities and stunted growth. Also, some patients with NBC1 mutations have elevated pancreatic enzyme levels and/or mental retardation (4, 5, 7, 8). These findings indicate that NBC1 has essential roles not only in whole-body acid–base balance but also in the maintenance of homeostasis in several tissues. There are two major splicing variants of NBC1, pancreas-type NBC1 (pNBC1) (9) and kidney-type NBC1 (kNBC1) (10–12). pNBC1 and kNBC1 are 93% identical, but the N-terminal 85 aa of pNBC1 are replaced by 41 different amino acids in kNBC1. Despite their high sequence similarity, these NBC1 variants are thought to have quite different physiological roles. pNBC1 is predominantly expressed in the pancreas, and lower levels are expressed in several organs, including the brain and eyes (9, 13). In pancreas, pNBC1 is thought to mediate HCO3− uptake into pancreatic duct cells in response to hormonal stimulation (3). However, expression of kNBC1 is almost completely limited to the kidney (9), where it mediates constitutive HCO3− exit from proximal tubules (3). However, the molecular mechanism underlying these different physiological roles of NBC1 variants has not been elucidated.
In this study, we demonstrated that IRBIT specifically binds to pNBC1 and does not bind to kNBC1 at all. Also, an electrophysiological analysis in Xenopus oocytes demonstrated that pNBC1 requires coexpression of IRBIT to manifest substantial activity comparable with that of kNBC1, which displays substantial activity independently of IRBIT. These results strongly suggest that pNBC1 is the target molecule of IRBIT, and raise the possibility that regulation through IRBIT explains the difference between the physiological roles of NBC1 variants.
Results
Identification of NBC1 as an IRBIT Binding Protein.
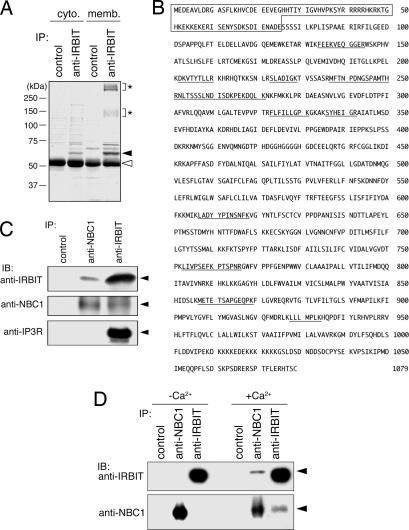
To identify the target molecules of IRBIT, we searched for IRBIT binding proteins in mouse cerebellar extracts, because IRBIT is highly expressed in the cerebellum (2). Because a previous report suggested that substantial amounts of IRBIT are tightly bound to the membrane fraction of the cerebellum (2), we fractionated cerebellar extracts into a cytoplasmic fraction and membrane fraction and analyzed them separately. IRBIT binding proteins were collected from the fractions by immunoprecipitation with anti-IRBIT Ab and separated by SDS/PAGE, and several proteins that were specifically immunoprecipitated by anti-IRBIT Ab were found in both fractions (Fig. 1A). We focused on the two major broad bands of anti-IRBIT immunoprecipitate from the membrane fraction (≈150 and ≈350 kDa, asterisks in Fig. 1A) and identified the proteins in the bands. Surprisingly, sequences of peptides obtained by digestion of both bands matched the sequence of a single protein, called NBC1 (Fig. 1B, underlined residues). NBC1 is a membrane-integrated transporter protein that mediates the coupled movement of Na+ and HCO3− ions across the plasma membrane (3). The predicted molecular mass of NBC1 is ≈120 kDa, and because NBC1 has several N-glycosylation sites, the broad band with an apparent molecular mass of 150 kDa should contain NBC1 monomer. We speculated that the broad band with an apparent molecular mass of 350 kDa contained the SDS-resistant multimer of NBC1.
Fig. 1.
Identification of NBC1 as an IRBIT binding protein. (A) Mouse cerebella were collected and fractionated into cytoplasmic (cyto.) and membrane (memb.) fractions, and each fraction was subjected to immunoprecipitation (IP) with control rabbit IgG and rabbit anti-IRBIT Ab. Immunoprecipitates were subjected to SDS/PAGE using a 10% acrylamide gel, and the gel was stained with Coomassie brilliant blue (CBB). Filled and open triangles indicate the migration of endogenous IRBIT and IgG proteins, respectively. Asterisks indicate the migration of membrane fraction-specific IRBIT binding proteins. (B) Amino acid sequence of pNBC1. The peptides obtained by digestion of membrane fraction-specific IRBIT binding proteins (asterisks in A) are underlined. The N-terminal pNBC1-specific domain is boxed. (C) Mouse cerebellar membrane fraction was subjected to immunoprecipitation with rabbit control IgG, rabbit anti-NBC1, and rabbit anti-IRBIT Ab. Immunoprecipitates were subjected to Western blotting (IB) with guinea pig anti-IRBIT, rabbit anti-NBC1, and rat anti-IP3R Ab. Filled triangles indicate the migration of endogenous IRBIT, NBC1, and IP3R proteins. (D) COS-7 cells were lysed by lysis buffer with (+Ca2+) or without (−Ca2+) 2 mM CaCl2. Each lysate was subjected to immunoprecipitation with rabbit control IgG, rabbit anti-NBC1, and rabbit anti-IRBIT Ab, and immunoprecipitates were subjected to Western blotting with guinea pig anti-IRBIT and guinea pig anti-NBC1 Ab. Filled triangles indicate the migration of endogenous IRBIT and NBC1 proteins.
We then confirmed the interaction between endogenous IRBIT and NBC1 by coimmunoprecipitation experiments. The cerebellar membrane fraction was immunoprecipitated with anti-IRBIT and anti-NBC1 Ab, and the immunoprecipitates were subjected to Western blotting with anti-IRBIT, anti-NBC1, and anti-IP3R Ab. Endogenous IRBIT was coimmunoprecipitated by anti-NBC1 Ab, and endogenous NBC1 was coimmunoprecipitated by anti-IRBIT Ab (Fig. 1C Top and Middle), confirming the interaction between endogenous IRBIT and NBC1. IP3R was coimmunoprecipitated by anti-IRBIT Ab, as described in ref. 2 but not by anti-NBC1 Ab (Fig. 1C Bottom), suggesting that IRBIT is incapable of interacting with both IP3R and NBC1 simultaneously to form a ternary complex. To investigate whether the interaction between endogenous IRBIT and NBC1 is specific for the cerebellum, we performed coimmunoprecipitation experiments with COS-7 cell lysate. We detected a considerable amount of endogenous IRBIT and NBC1 in each immunoprecipitate from the COS-7 cell lysate, but we did not detect coimmunoprecipitation of these proteins (Fig. 1D, −Ca2+), and we concluded that the interaction between endogenous IRBIT and NBC1 in COS-7 cells is regulated. Because IRBIT may mediate the signal downstream of IP3 production, we hypothesized that IP3R-induced Ca2+ release enhances the interaction between IRBIT and NBC1. We then added 2 mM CaCl2 to the lysis buffer containing 2 mM EDTA (estimated free Ca2+ concentration, ≈6.7 μM) and performed coimmunoprecipitation experiments. The results showed coimmunoprecipitation of endogenous IRBIT and NBC1 in the presence of CaCl2 (Fig. 1D, +Ca2+), suggesting that the interaction between endogenous IRBIT and NBC1 in COS-7 cells is regulated by the intracellular Ca2+ concentration. Because the interaction between endogenous IRBIT and NBC1 in the cerebellar membrane fraction did not require the addition of Ca2+ (Fig. 1C), the results suggested that the interaction between IRBIT and NBC1 is regulated in a cell-type-specific manner.
IRBIT Specifically Binds to pNBC1.
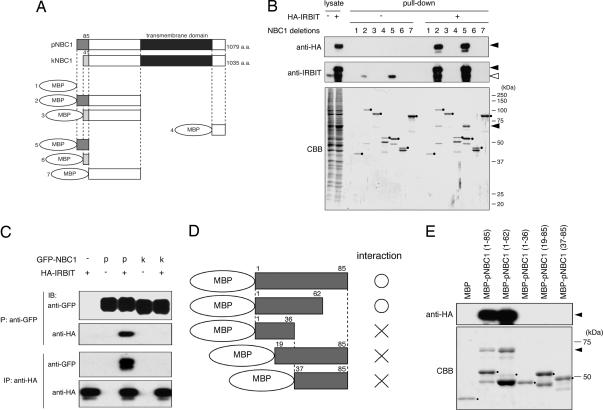
NBC1 is predicted to contain at least 10 transmembrane domains and cytoplasmic N and C termini (14). Because IRBIT is a cytoplasmic protein, either the N- or C-terminus of NBC1 may contain the region that interacts with IRBIT. NBC1 has two major splicing variants, pNBC1 (9) and kNBC1 (10, 11), and they differ only in their N-terminal sequence, with the first 85 aa in pNBC1 being replaced by 41 different amino acids in kNBC1 (Fig. 2A). To identify the region of NBC1 responsible for the interaction with IRBIT, we constructed the six deletion mutants of the N and C termini of NBC1 shown in Fig. 2A and analyzed their interaction with IRBIT by pull-down assay. Maltose-binding protein (MBP)-fused recombinant proteins of these deletion mutants were incubated with COS-7 cell lysate expressing hemagglutinin (HA)-tagged IRBIT (HA-IRBIT), and the proteins pulled down with recombinant proteins were subjected to Western blotting with anti-HA Ab. HA-IRBIT was specifically pulled down with the deletion mutant proteins containing pNBC1 specific 85 aa (Fig. 2B Top, pull-down, +, filled triangle). A pull-down assay with control COS-7 cell lysate demonstrated that endogenous IRBIT was also pulled down by the deletion mutant proteins containing the pNBC1-specific domain (Fig. 2B Middle, pull-down, −, open triangle). These results strongly suggest that the pNBC1-specific domain is required and sufficient for the interaction with IRBIT. Also, the only difference in Coomassie brilliant blue (CBB) staining pattern between samples containing and not containing HA-IRBIT were HA-IRBIT protein bands with substantial intensity (Fig. 2B Lower, filled triangle). If the interaction between HA-IRBIT and recombinant protein is mediated by another protein(s), such protein(s) should be present in substantial amounts and should be detected by CBB staining. Therefore, we considered that the interaction between IRBIT and pNBC1 is direct.
Fig. 2.
IRBIT specifically interacts with N-terminal pNBC1-specific domain. (A) Diagram of pNBC1, kNBC1, and NBC1 deletion mutants. MBP, maltose-binding protein. (B) COS-7 cells were transfected with empty vector (−) or hemagglutinin (HA)-tagged IRBIT (HA-IRBIT)-expressing plasmid (+). Cell lysates were incubated with each NBC1 deletion mutant protein, and bound proteins (pull-down) were subjected to Western blotting with anti-HA (Top) and rabbit anti-IRBIT Ab (Middle), and CBB staining (Bottom). Filled and open triangles indicate migrations of HA-IRBIT and endogenous IRBIT proteins, respectively. Filled circles indicate migrations of NBC1 deletion mutant proteins. (C) COS-7 cells were transfected with GFP-NBC1- and/or HA-IRBIT-expressing plasmid, cell lysates were subjected to immunoprecipitation (IP) with anti-GFP and anti-HA Ab, and immunoprecipitates were subjected to Western blotting (IB) with anti-GFP and anti-HA Ab. (D) Diagram of deletion mutants of pNBC1-specific domain. (E) HA-IRBIT-expressing COS-7 cell lysate was incubated with the indicated deletion mutant proteins of pNBC1-specific domain. Bound proteins were pulled down and subjected to Western blotting with anti-HA Ab (Upper) and CBB staining (Lower). Filled triangles indicate the migrations of HA-IRBIT proteins. Filled circles indicate migrations of deletion mutant proteins of pNBC1-specific domain.
We confirmed the interaction between IRBIT and the pNBC1-specific domain by coimmunoprecipitation assay. COS-7 cells were transfected with GFP-tagged NBC1 (GFP-NBC1)-expressing plasmid and/or HA-IRBIT-expressing plasmid, and cell lysates were subjected to immunoprecipitation with anti-GFP and anti-HA Ab. We found that GFP-pNBC1 and HA-IRBIT were coimmunoprecipitated with each other, whereas GFP-kNBC1 was not coimmunoprecipitated with HA-IRBIT (Fig. 2C). These results clearly demonstrated that IRBIT specifically interacts with pNBC1 and does not interact with kNBC1.
To more precisely identify the region of pNBC1 responsible for the interaction with IRBIT, we prepared the additional deletion mutant proteins shown in Fig. 2D and tested them for interactions with IRBIT by pull-down assay. The results showed that MBP-pNBC1 (1–62) interacted with HA-IRBIT (Fig. 2E). Further deletion of amino acids 1–18 or 37–62 interfered with the interaction with HA-IRBIT (Fig. 2E). These results indicated that the IRBIT-interacting region could not be narrowed down to a short primary amino acid sequence and that IRBIT may require the 3D structure of the pNBC1-specific domain for its interaction.
Interaction Between IRBIT and pNBC1 Depends on the Phosphorylation of IRBIT.
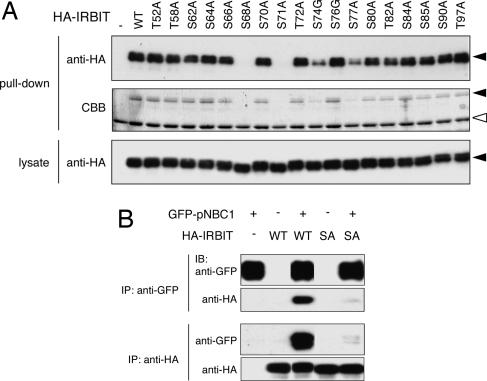
As reported in ref. 2, the interaction between IRBIT and IP3R depends on the phosphorylation of IRBIT. IRBIT has a serine/threonine-rich region in its N-terminal domain, and by testing alanine and glycine substitution mutants we demonstrated that phosphorylation of multiple serine residues in this region is important for the interaction with IP3R (20). To investigate the importance of phosphorylation of the serine/threonine-rich region for the interaction with pNBC1, we investigated the interaction between pNBC1 and these substitution mutants by pull-down assay. Of the 18 substitution mutants, 2 (the S68A and S71A mutants) had completely lost binding activity for pNBC1, and the S74G and S77A mutants had lost considerable binding activity for pNBC1 (Fig. 3A), suggesting that phosphorylation of the Ser-68, Ser-71, Ser-74, and Ser-77 residues is indispensable for the interaction with pNBC1. Next, we investigated the importance of phosphorylation of IRBIT by coimmunoprecipitation assay. COS-7 cells were transfected with HA-tagged WT or S68A IRBIT-expressing plasmid and/or GFP-pNBC1-expressing plasmid, and cell lysates were subjected to immunoprecipitation. S68A IRBIT hardly interacted with pNBC1 in the coimmunoprecipitation assay (Fig. 3B), and the results strongly suggested that phosphorylation of IRBIT is required for the interaction with pNBC1.
Fig. 3.
Phosphorylation of IRBIT is required for the interaction with pNBC1. (A) COS-7 cells were transfected with expression plasmids encoding indicated serine/threonine-substituted mutants of IRBIT. Cell lysates were incubated with recombinant protein of pNBC1-specific domain (no. 5 in Fig. 2A), and bound proteins were pulled down and subjected to Western blotting with anti-HA Ab (Top) and CBB staining (Middle). Cell lysates were subjected to Western blotting with anti-HA Ab (Bottom). Filled and open triangles indicate the migrations of HA-IRBIT proteins and recombinant protein of pNBC1-specific domain, respectively. (B) COS-7 cells were transfected with indicated plasmids, and cell lysates were subjected to immunoprecipitation (IP) with indicated Abs, and immunoprecipitates were subjected to Western blotting (IB) with indicated Abs.
Conformation of IRBIT Is Required for the Interaction with pNBC1.
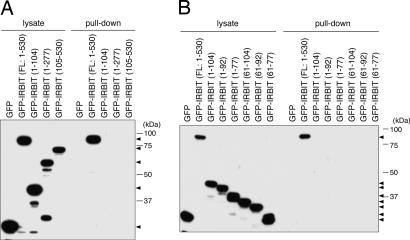
To identify the region of IRBIT that is responsible for the interaction with pNBC1, we investigated the interaction between pNBC1 and IRBIT deletion mutants by pull-down assay. Because serine residues required for the interaction with pNBC1 are present in the N-terminal domain of IRBIT, we speculated that the N-terminal domain of IRBIT contains the region responsible for the interaction with pNBC1. However, the results showed that neither the N-terminal nor the C-terminal domain of IRBIT interacted with pNBC1 (Fig. 4A). Because we doubted that the N-terminal domain of IRBIT contains an inhibitory region, we produced deletion mutants of the N-terminal domain of IRBIT and analyzed the interaction with pNBC1. As shown in Fig. 4B, none of the deletion mutants interacted with pNBC1. These results raise the possibility that the 3D structure of full-length IRBIT is required for the interaction with pNBC1. The N-terminal amino acids 1–277 of IRBIT, which are sufficient for the interaction with IP3R (2), did not interact with pNBC1 (Fig. 4A). Also, we found that the interaction between IRBIT and the pNBC1-specific domain was not inhibited by physiological concentrations of IP3 in pull-down assay (data not shown). These results demonstrated that the conformational requirements for the interaction with pNBC1 and IP3R are different.
Fig. 4.
Conformation of IRBIT required for the interaction with pNBC1. (A and B) COS-7 cells were transfected with expression plasmids encoding GFP-fused IRBIT deletion mutants. Cell lysates were incubated with recombinant protein of pNBC1-specific domain (no. 5 in Fig. 2A), and bound proteins were pulled down and subjected to Western blotting with anti-GFP Ab. Filled triangles indicate the migrations of GFP-fused IRBIT deletion mutant proteins.
pNBC1 Requires Coexpression of IRBIT to Manifest Substantial Activity Comparable with That of kNBC1.
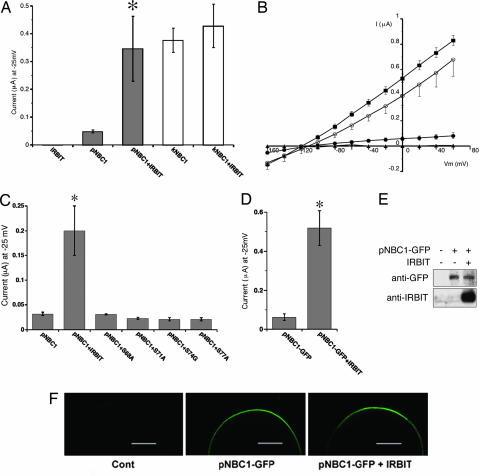
To examine the effect of IRBIT on pNBC1 function, we performed an electrophysiologic analysis in Xenopus oocytes. Oocytes were injected with NBC1-expressing cRNA and/or IRBIT-expressing cRNA and then perfused with HCO3−/CO2 solution, and the negative charge influx (current) originating from NBC1 was measured at a holding potential of −25 mV. Although the addition of HCO3−/CO2 solution induced a large current in oocytes expressing kNBC1, as reported in ref. 8, the current in oocytes expressing pNBC1 was much smaller (Fig. 5A). Surprisingly, however, coexpression of IRBIT increased the pNBC1 current by ≈7-fold, to a level comparable with the kNBC1 current (Fig. 5A). However, coexpression of IRBIT did not affect the kNBC1 current, and expression of IRBIT alone did not induce any NBC1 current (Fig. 5A). To examine the effect of IRBIT in greater detail, we determined the current–voltage (I–V) relationship by applying step pulses between Vm = −160 to 60 mV. As shown in Fig. 5B, coexpression of IRBIT markedly activated the pNBC1 current, and it became comparable with the kNBC1 current over the entire voltage range. However, IRBIT did not significantly modify the electrical properties of pNBC1, such as linearity and reversal potential (Fig. 5B). Next, to examine the role of the physical interaction between pNBC1 and IRBIT, we tested the effects of binding-deficient mutants of IRBIT (S68A, S71A, S74G, and S77A). As shown in Fig. 5C, none of the mutants increased the pNBC1 current, indicating that the direct interaction between IRBIT and pNBC1 is indispensable for stimulation of pNBC1 activity. To clarify the mechanism of pNBC1 activation by IRBIT further, we analyzed the effect of IRBIT on the amount and surface expression of pNBC1. To facilitate the detection of pNBC1, C-terminal GFP-tagged pNBC1 (pNBC1-GFP), which was similarly activated by IRBIT (Fig. 5D), was used in these experiments. Western blotting of the extracts of injected oocytes demonstrated that the amount of pNBC1-GFP was unaffected by coexpression of IRBIT (Fig. 5E), and examination with a confocal laser scanning microscope revealed that the surface expression of pNBC1-GFP was also unaffected by coexpression of IRBIT (Fig. 5F). These results indicate that IRBIT increases pNBC1 activity without changing its amount or surface expression.
Fig. 5.
pNBC1 requires coexpression of IRBIT to manifest substantial activity comparable with that of kNBC1. (A) NBC1-mediated currents in Xenopus oocyte. Influxes of anion charges, induced by solution change from ND-96 to HCO3−-containing solution, were measured at a holding potential of −25 mV. Numbers of observation were 5 (IRBIT), 9 (pNBC1), 11 (pNBC1 + IRBIT), 5 (kNBC1), and 5 (kNBC1 + IRBIT). ∗, P < 0.01 versus pNBC1. (B) Current–voltage (I–V) relationship of NBC1 currents in oocytes injected with cRNA of IRBIT (filled triangles), pNBC1 (filled circles), pNBC1 + IRBIT (open circles), and kNBC1 (filled squires). Step pulses between Vm = −160 and 60 mV were applied in the absence and presence of an NBC1 inhibitor, 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid (0.4 mM). Reversal potentials were 108 ± 6 mV (pNBC1), 115 ± 13 mV (pNBC1 + IRBIT), and 117 ± 3 mV (kNBC1). (C) Effects of WT IRBIT (IRBIT) or IRBIT mutants on pNBC1 currents measured at a holding potential of −25 mV. Numbers of observation were 5 for each construct. ∗, P < 0.05 versus pNBC1. (D) Effect of IRBIT on the currents mediated by C-terminal GFP-tagged pNBC1 (pNBC1-GFP). Numbers of observation were 4 for each construct. ∗, P < 0.05 versus pNBC1-GFP. (E) Xenopus oocytes injected with indicated cRNAs were extracted, and extracts were subjected to Western blotting with anti-GFP and guinea pig anti-IRBIT Ab. (F) Surface expression of pNBC1-GFP observed by a confocal laser scanning microscopy. (Scale bars, 300 μm.)
Discussion
We identified (2) an IP3R binding protein, termed IRBIT. Because IRBIT dissociates from IP3R in the presence of physiological concentrations of IP3 (2), we hypothesized that IRBIT is a signaling molecule that dissociates from IP3R and regulates downstream target molecules in response to the production of IP3. In this study, we demonstrated that IRBIT specifically and directly binds to pNBC1 (Fig. 2) and that IRBIT markedly potentiates the activity of pNBC1 in Xenopus oocytes (Fig. 5 A–D). These results indicate that pNBC1 is the target molecule of IRBIT and that IRBIT is a multifunctional molecule that regulates not only IP3R activity (20) but also pNBC1 activity. This finding demonstrates a function of IRBIT in the regulation of intracellular and extracellular pH through pNBC1. Also, the identification of another target molecule besides IP3R raises the possibility that IRBIT functions as a signaling molecule downstream of IP3 production. Further elucidation of IRBIT target molecules might reveal the full spectrum of biological processes regulated by IP3 production.
In this study, we showed that coexpression of IRBIT is required for pNBC1 to manifest substantial activity comparable with that of kNBC1, which displays substantial activity independently of IRBIT (Fig. 5 A and B). This marked and specific effect of IRBIT on pNBC1 may be of great relevance to the different physiological roles of NBC1 variants. kNBC1 is expressed predominantly in the kidney (9), where it mediates HCO3− reabsorption from renal proximal tubules (3), whereas pNBC1 is expressed predominantly in the pancreas (9), where it mediates HCO3− secretion from pancreatic duct cells (3). In contrast to the rather constant HCO3− reabsorption rates from renal proximal tubules, HCO3− secretion from pancreatic duct cells should be minimal in basal state and be rapidly enhanced by hormonal stimulation (3). Although HCO3− secretion from pancreatic duct cells is primarily regulated by cAMP production rather than by IP3 production, it is tempting to speculate that the interaction between pNBC1 and IRBIT may also contribute to this process. The importance of IRBIT in the regulation of pNBC1 activity in vivo remains to be tested by using an IRBIT knockdown system. Recently, we found that IP3R2 and IP3R3 double-knockout mice exhibit a defect in nutrient digestion caused by impaired pancreatic juice secretion (15). Because exocytosis of digestive enzymes from pancreatic acinar cells has been shown to require intracellular Ca2+ mobilization (16), we attributed the deficit of pancreatic juice secretion to a lack of exocytosis of digestive enzymes (15). However, the results of this study raise the possibility that HCO3− secretion from pancreatic duct cells may be also impaired in IP3R2 and IP3R3 double-knockout mice as a result of impairment of IRBIT mobilization in a cAMP-independent manner. Investigation of HCO3− secretion from pancreatic duct cells in the double-knockout mice would clarify the importance of IP3Rs and IRBIT in the regulation of pNBC1 activity in vivo.
Despite the marked pNBC1 activation by IRBIT, we were unable to detect any effect of IRBIT on the amount or the surface expression of pNBC1 in Xenopus oocytes (Fig. 5 E and F). Also, the cell surface biotinylation technique revealed that cell surface localization of pNBC1 was unaffected by coexpression of IRBIT in HEK293 cells (data not shown). These findings suggest that IRBIT activates pNBC1 through a molecular mechanism other than transcriptional regulation, translational regulation, or regulation of intracellular trafficking. One speculation is that the N-terminal-specific domain of pNBC1, but not of kNBC1, interferes with ionic flows through transmembrane domains in a manner analogous to the ball-and-chain model proposed for a voltage-dependent potassium channel (17) and that the binding of IRBIT may somehow free pNBC1 from this interference. Although this model nicely explains the much lower activity of pNBC1 than that of kNBC1 in the absence of IRBIT, its validity remains to be tested.
As shown in Fig. 1 C and D, addition of Ca2+ to the lysis buffer was necessary to detect any interaction between endogenous IRBIT and NBC1 in the COS-7 cell lysate, whereas the interaction between endogenous IRBIT and NBC1 in the cerebellar membrane fraction was detected in the absence of Ca2+, suggesting that the interaction between IRBIT and NBC1 is regulated in a cell-type-specific manner. We found that the apparent molecular mass of endogenous NBC1 of COS-7 cells was slightly higher than that of the cerebellar membrane fraction in SDS/PAGE (data not shown). This finding raises the possibility that NBC1 undergoes different posttranslational modification in COS-7 cells and in the cerebellar membrane fraction, and that the modification in COS-7 cells interferes with the interaction with IRBIT. Because exogenous pNBC1 interacted with IRBIT without Ca2+ addition even in COS-7 cells (Fig. 2C), this modification may be inefficient to modify all of pNBC1 molecules expressed exogenously.
We identified pNBC1 as a target molecule of IRBIT, indicating that IRBIT has an important role in the regulation of intracellular and extracellular pH through pNBC1. The regulatory mechanism through IRBIT may also be responsible for the characteristic difference in physiological roles of NBC1 variants.
Materials and Methods
Immunoprecipitations and MS Analysis.
The cerebella of 8-week-old mice were homogenized in a buffer containing 10 mM Hepes (pH 7.4), 320 mM sucrose, 2 mM EDTA, and protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride/10 μM leupeptin/10 μM pepstatin A/10 μM E-64) with a glass–Teflon homogenizer (1,000 rpm, 10 strokes), and the homogenate was centrifuged at 1,000 × g for 10 min. The supernatant was centrifuged at 100,000 × g for 60 min to obtain the cytoplasmic fraction (the supernatant) and the crude microsome fraction (the pellet). The crude microsome fraction was suspended in a buffer containing 50 mM Hepes (pH 7.4), 2 mM EDTA, 1% Nonidet P-40, and protease inhibitors, and incubated for 30 min at 4°C. After centrifugation at 20,000 × g for 30 min, the supernatant was diluted two times with a buffer containing 200 mM NaCl, 2 mM EDTA, 1% Nonidet P-40, and protease inhibitors to obtain the membrane fraction. COS-7 cells were lysed with lysis buffer (10 mM Hepes, pH 7.4/150 mM NaCl/2 mM EDTA/1% Nonidet P-40) containing Complete protease inhibitor mixture (Roche Molecular Biochemicals). The cerebellar fractions and COS-7 cell lysate were precleaned by incubation with Protein G Sepharose 4 Fast Flow (GE Healthcare Bio-Science) and incubated with Ab coupled to Protein G Sepharose 4 Fast Flow. Immunoprecipitates were washed three to five times with lysis buffer and subjected to Western blotting and CBB staining. IRBIT binding proteins were identified by MS analysis as described (18).
Plasmids, Recombinant Proteins, and Transfections.
Deletion mutants of NBC1s were constructed by PCR and subcloned into pMALc plasmid (New England Biolabs). MBP-fused recombinant proteins were expressed in BL21-CodonPlus (DE3) Escherichia coli (Stratagene) and purified according to the supplier’s protocol (New England Biolabs). Full-length IRBIT cDNA was subcloned into pHM6 plasmid (Roche Molecular Biochemicals) to generate HA-tagged IRBIT expression plasmid. Site-directed mutants of IRBIT were generated by using the QuickChange II site-directed mutagenesis kit (Stratagene). Deletion mutants of IRBIT were constructed by PCR and subcloned into pEGFP-C plasmid (BD Biosciences) the same as full-length IRBIT (2). Coding sequences of human pNBC1 and kNBC1 were subcloned into pEGFP-C plasmid to make GFP-tagged pNBC1 and kNBC1 expression plasmids. Transfections were performed with TransIT-LT1 (Mirus, Madison, WI). For expression in Xenopus oocytes, coding sequences of human pNBC1 and kNBC1 were subcloned into pcDNA3.1(+) plasmid (Invitrogen). For C-terminal GFP-tagged pNBC1 expression in Xenopus oocytes, coding sequence of pNBC1 is conjugated with GFP sequence by subcloning into pEGFP-N plasmid (BD Biosciences) and conjugated sequence (pNBC1-GFP) is subcloned into pcDNA3.1(+) plasmid.
Abs.
Rabbit anti-IRBIT Ab was described in ref. 2. Guinea pig anti-IRBIT antiserum was raised against purified recombinant protein of N-terminal domain of IRBIT (1–104). Guinea pig anti-NBC1 antiserum was raised against the recombinant protein of C-terminal cytoplasmic domain of NBC1. These antisera were affinity-purified by a column of CNBr-activated Sepharose 4B (GE Healthcare Bio-Science) covalently coupled with each antigen. Rabbit anti-NBC1 Ab (AB3204, Chemicon), mouse anti-HA Ab (16B12, Abcam, Inc., Cambridge, MA), and mouse anti-GFP Ab (sc-9996, Santa Cruz Biotechnology) were purchased. Rat anti-IP3R Ab (18A10) was described in ref. 19.
Pull-Down Assays.
COS-7 cells were lysed with binding buffer (20 mM Tris·HCl, pH 7.5/120 mM NaCl/2 mM EDTA/20 mM NaF/10 mM sodium pyrophosphate/0.1% Triton X-100/1 mM DTT) containing Complete protease inhibitor mixture. Cell lysates were incubated with recombinant protein, and binding proteins were pulled down by amylose (New England Biolabs). Pulled-down proteins were washed three times by binding buffer and subjected to Western blotting and CBB staining.
Xenopus Oocyte Manipulations.
Oocytes collection, cRNA injection, and the NBC1 current detection were performed as described in ref. 8. The total injection volume was adjusted to 50 nl (5 ng of each cRNA). Oocytes were initially perfused with nominally HCO3−-free ND96 solution (5 mM Hepes, pH 7.4/96 mM NaCl/2 mM KCl/1 mM MgCl2/1.8 mM CaCl2), and solution was changed to HCO3−-containing solution (5 mM Hepes, pH 7.4/66 mM NaCl/30 mM NaHCO3/2 mM KCl/1 mM MgCl2/1.8 mM CaCl2), which was equilibrated with 5% CO2 in oxygen (pH 7.4). The NBC1 currents induced by solution changes from ND96 to HCO3−-containing solution were measured by the two-electrode voltage-clamp method (8). The data were presented as mean ± SEM. For Western blotting, oocytes were extracted by extraction buffer (20 mM Tris·HCl, pH 7.5/150 mM NaCl/2 mM EGTA/5 mM MgCl2/1% Triton X-100). A TCS SL confocal laser scanning microscope (Leica, Deerfield, IL) was used to detect the surface expression of C-terminal GFP-tagged pNBC1.
Acknowledgments
We thank K. Suzuki for excellent technical assistance. We also thank the Research Resource Center of Brain Science Institute for MS analysis of IRBIT-binding proteins. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Abbreviations
- IP3
inositol 1,4,5-trisphosphate
- IP3R
IP3 receptor
- IRBIT
IP3R binding protein released with IP3
- NBC1
Na+/HCO3− cotransporter 1
- MBP
maltose-binding protein
- HA
hemagglutinin
- CBB
Coomassie brilliant blue
- pNBC1
pancreas-type NBC1
- kNBC1
kidney-type NBC1.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Berridge M. J. Nature. 1993;361:315–325. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 2.Ando H., Mizutani A., Matsu-ura T., Mikoshiba K. J. Biol. Chem. 2003;278:10602–10612. doi: 10.1074/jbc.M210119200. [DOI] [PubMed] [Google Scholar]
- 3.Soleimani M., Burnham C. E. J. Membr. Biol. 2001;183:71–84. doi: 10.1007/s00232-001-0055-8. [DOI] [PubMed] [Google Scholar]
- 4.Igarashi T., Inatomi J., Sekine T., Cha S. H., Kanai Y., Kunimi M., Tsukamoto K., Satoh H., Shimadzu M., Tozawa F., et al. Nat. Genet. 1999;23:264–266. doi: 10.1038/15440. [DOI] [PubMed] [Google Scholar]
- 5.Igarashi T., Inatomi J., Sekine T., Seki G., Shimadzu M., Tozawa F., Takeshima Y., Takumi T., Takahashi T., Yoshikawa N., et al. J. Am. Soc. Nephrol. 2001;12:713–718. doi: 10.1681/ASN.V124713. [DOI] [PubMed] [Google Scholar]
- 6.Dinour D., Chang M. H., Satoh J., Smith B. L., Angle N., Knecht A., Serban I., Holtzman E. J., Romero M. F. J. Biol. Chem. 2004;279:52238–52246. doi: 10.1074/jbc.M406591200. [DOI] [PubMed] [Google Scholar]
- 7.Inatomi J., Horita S., Braverman N., Sekine T., Yamada H., Suzuki Y., Kawahara K., Moriyama N., Kudo A., Kawakami H., et al. Pflügers Arch. 2004;448:438–444. doi: 10.1007/s00424-004-1278-1. [DOI] [PubMed] [Google Scholar]
- 8.Horita S., Yamada H., Inatomi J., Moriyama N., Sekine T., Igarashi T., Endo Y., Dasouki M., Ekim M., Al-Gazali L., et al. J. Am. Soc. Nephrol. 2005;16:2270–2278. doi: 10.1681/ASN.2004080667. [DOI] [PubMed] [Google Scholar]
- 9.Abuladze N., Lee I., Newman D., Hwang J., Boorer K., Pushkin A., Kurtz I. J. Biol. Chem. 1998;273:17689–17695. doi: 10.1074/jbc.273.28.17689. [DOI] [PubMed] [Google Scholar]
- 10.Romero M. F., Hediger M. A., Boulpaep E. L., Boron W. F. Nature. 1997;387:409–413. doi: 10.1038/387409a0. [DOI] [PubMed] [Google Scholar]
- 11.Burnham C. E., Amlal H., Wang Z., Shull G. E., Soleimani M. J. Biol. Chem. 1997;272:19111–19114. doi: 10.1074/jbc.272.31.19111. [DOI] [PubMed] [Google Scholar]
- 12.Amlal H., Wang Z., Burnham C., Soleimani M. J. Biol. Chem. 1998;273:16810–16815. doi: 10.1074/jbc.273.27.16810. [DOI] [PubMed] [Google Scholar]
- 13.Usui T., Hara M., Satoh H., Moriyama N., Kagaya H., Amano S., Oshika T., Ishii Y., Ibaraki N., Hara C., et al. J. Clin. Invest. 2001;108:107–115. doi: 10.1172/JCI11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatishchev S., Abuladze N., Pushkin A., Newman D., Liu W., Weeks D., Sachs G., Kurtz I. Biochemistry. 2003;42:755–765. doi: 10.1021/bi026826q. [DOI] [PubMed] [Google Scholar]
- 15.Futatsugi A., Nakamura T., Yamada M. K., Ebisui E., Nakamura K., Uchida K., Kitaguchi T., Takahashi-Iwanaga H., Noda T., Aruga J., Mikoshiba K. Science. 2005;309:2232–2234. doi: 10.1126/science.1114110. [DOI] [PubMed] [Google Scholar]
- 16.Williams J. A. Annu. Rev. Physiol. 2001;63:77–97. doi: 10.1146/annurev.physiol.63.1.77. [DOI] [PubMed] [Google Scholar]
- 17.Zhou M., Morais-Cabral J. H., Mann S., MacKinnon R. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 18.Doi H., Mitsui K., Kurosawa M., Machida Y., Kuroiwa Y., Nukina N. FEBS Lett. 2004;571:171–176. doi: 10.1016/j.febslet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 19.Maeda N., Niinobe M., Nakahira K., Mikoshiba K. J. Neurochem. 1988;51:1724–1730. doi: 10.1111/j.1471-4159.1988.tb01151.x. [DOI] [PubMed] [Google Scholar]
- 20.Ando H., Mizutani A., Kiefer H., Tsuzurugi D., Michikawa T., Mikoshiba K. Mol. Cell. 2006 doi: 10.1016/j.molcel.2006.05.017. in press. [DOI] [PubMed] [Google Scholar]