Abstract
Malaria parasites contain a nonphotosynthetic plastid homologous to chloroplasts of plants. The parasite plastid synthesizes fatty acids, heme, iron sulfur clusters and isoprenoid precursors and is indispensable, making it an attractive target for antiparasite drugs. How parasite plastid biosynthetic pathways are fuelled in the absence of photosynthetic capture of energy and carbon was not clear. Here, we describe a pair of parasite transporter proteins, PfiTPT and PfoTPT, that are homologues of plant chloroplast innermost membrane transporters responsible for moving phosphorylated C3, C5, and C6 compounds across the plant chloroplast envelope. PfiTPT is shown to be localized in the innermost membrane of the parasite plastid courtesy of a cleavable N-terminal targeting sequence. PfoTPT lacks such a targeting sequence, but is shown to localize in the outermost parasite plastid membrane with its termini projecting into the cytosol. We have identified these membrane proteins in the parasite plastid and determined membrane orientation for PfoTPT. PfiTPT and PfoTPT are proposed to act in tandem to transport phosphorylated C3 compounds from the parasite cytosol into the plastid. Thus, the transporters could shunt glycolytic derivatives of glucose scavenged from the host into the plastid providing carbon, reducing equivalents and ATP to power the organelle.
Keywords: apicoplast, endosymbiosis, Plasmodium, translocator
Malaria is caused by parasites of the genus Plasmodium and remains one of the world’s major global diseases despite intensive efforts to control the mosquito vector, develop vaccines, and discover new drugs to replace existing drugs under pressure from parasite resistance. Malaria parasites harbor a vestigial plastid that is an evolutionary homologue of the chloroplasts of plant and algal cells (1). The parasite plastid arose by secondary endosymbiosis and occurs in most members of the Phylum Apicomplexa, to which malaria and numerous other obligate intracellular parasites belong (1). The plastid is known as the apicoplast (apicomplexan plastid) and is nonphotosynthetic but indispensable (1–3). Apicoplasts are emerging as excellent drug targets because they derive from endosymbiotic bacteria and thus have numerous bacterial-type pathways sensitive to compounds such as common antibiotics specifically directed against prokaryotes (4–7). Moreover, a number of herbicides are also parasiticidal because they perturb apicoplast function (8, 9).
Why parasites depend on their apicoplasts is not yet clear, but the Plasmodium plastid synthesizes several different compounds including fatty acids, isoprene subunits, heme and iron sulfur clusters, which might be exported from the apicoplast for use elsewhere in the parasite (10). The apicoplast is nonphotosynthetic, which begs the question of how it obtains energy, reducing power and components, particularly carbon, for anabolic syntheses. Plant and algal plastids typically generate energy and reducing power through photosynthesis, and they fix carbon from atmospheric carbon dioxide. However, not all plant tissues photosynthesize (roots and tubers, for instance, are underground), and these tissues still contain plastids that must be fuelled. Plants accomplish this by feeding sugars from the cytoplasm into their nonphotosynthetic plastids. These sugars are imported into the “dark” plastids using a set of membrane transporters (11). We previously hypothesized that the apicoplast’s carbon and energy system would resemble that of a “plant in darkness” (10). Our genome mining indicated to us that the apicoplast might also import sugars using transporters similar to those of plant chloroplasts (10). However, because apicoplasts are bounded by four membranes, whereas plant plastids have only two surrounding membranes, it was anticipated that apicoplasts might have extra transporters in their extra membranes. The parasite genome encodes two transporter-like proteins with clear homology to plant plastid transporters (12, 13), but their location and function are not yet known. Here we describe the localization of these two transporters to different membranes of the apicoplast, and present a model of how these transporters could fuel the organelle by shuttling the glycolytic products of glucose acquired from the host erythrocytes into the plastid.
Results and Discussion
PfiTPT and PfoTPT Are Related to Plastid Transporters.
PfiTPT (PFE1510c) and PfoTPT (PFE0410w) share maximum sequence identity (≈30%) with a family of plant plastid innermost membrane antiporters (Fig. 7, which is published as supporting information on the PNAS web site) that exchange inorganic phosphate (Pi) for phosphorylated C3, C5, and C6 compounds (14). The plant plastid transporters, known as triosephosphate/phosphate transporter (TPT), phosphoenolpyruvate/phosphate transporter (PPT), and glucose/phosphate transporter, have overlapping substrate preferences and move reduced carbon compounds either in to or out of the plant plastid. The plastid antiporters belong to a superfamily of drug metabolite transporters that also includes the malaria chloroquine resistance transporter protein located in the parasite’s food vacuole (15).
PfiTPT and PfoTPT resemble other members of the drug metabolite transporter superfamily (15) in having 10 putative α-helical transmembrane domains and two putative substrate binding sites: NKK in loop one and KR in transmembrane domain (TMD) 9 (Fig. 1). Direction of transport and substrate affinities cannot be determined for these transporters from the primary sequence (14). The characterized plant antiporters transport triose phosphates, 3-phosphoglycerate, 2-phosphoglycerate, dihydroxyacetonephosphate, phosphoenolpyruvate (PEP), and glucose-6-phosphate. The Plasmodium falciparum PfiTPT and PfoTPT could potentially transport any or all of these substrates into or out of the apicoplast. We attempted to express PfiTPT and PfoTPT in Xenopus laevis oocytes and assay transport of substrates, as has been done for other P. falciparum transporters (16), but were unsuccessful so substrate specificity remains unknown.
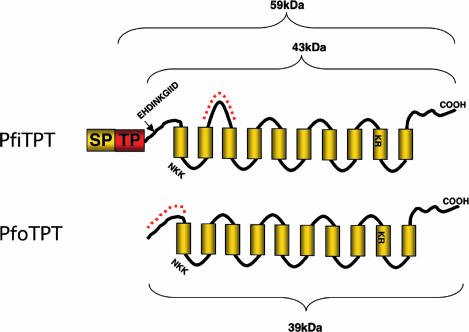
Fig. 1.
Schematics of the PfiTPT and PfoTPT proteins showing 10 transmembrane domains, two putative substrate binding sites, and a 140-aa leader comprising a signal peptide (SP) and a transit peptide (TP) in PfiTPT. Motifs selected from PfiTPT and PfoTPT for synthetic peptides used to generate antibodies are shown in dashed red lines. The N-terminal processing site for PfiTPT is marked by an arrow. Predicted molecular masses are shown.
PfiTPT bears an N-terminal extension that conforms to the general requirements of a bipartite apicoplast targeting signal with a signal peptide followed by a transit peptide (Fig. 1) and is predicted to be apicoplast targeted. PfoTPT bears no such targeting leader (Fig. 1), and it is predicted to be a multipass membrane protein with transmembrane domain 1 acting as an internal signal peptide.
PfiTPT and PfoTPT Are Apicoplast Membrane Proteins.
Immunofluoresence using antibodies directed against peptides from PfiTPT and PfoTPT (Fig. 1) localizes both proteins to the apicoplast (Fig. 2). PfiTPT and PfoTPT colocalize with apicoplast-resident acyl carrier protein (PfACP) (Fig. 2 A and B). Epitope tagged versions of PfiTPT and PfoTPT also localize to the apicoplast (Fig. 2 C--E). PfiTPT and PfoTPT partition into the detergent phase during Triton X-114 phase partitioning (17), indicating that they are membrane proteins (Fig. 2 A and B), congruent with bioinformatic predictions of 10 transmembrane domains (Fig. 1) (15). Epitope-tagged versions of PfiTPT and PfoTPT behave similarly to native versions, suggesting that tagging does not affect localization (Fig. 2 C–E). Conversely, epitope-tagged versions of mitochondrial and plasma membrane proteins do not target to the apicoplast (data not shown). Native PfiTPT has an apparent molecular mass of 43 kDa by SDS/PAGE (Fig. 2A), whereas PfoTPT has an apparent molecular mass of 34 kDa (Fig. 2B). We conclude that PfiTPT and PfoTPT are apicoplast membrane proteins.
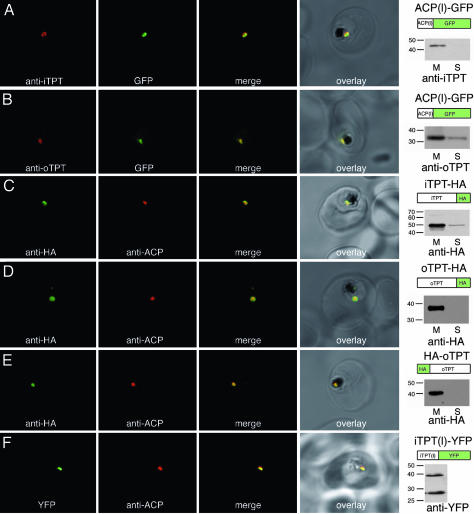
Fig. 2.
Immunolocalization of PfiTPT and PfoTPT to the apicoplast membranes. (A) Anti-PfiTPT (red) colocalizes with apicoplast-targeted GPF (green) in parasites expressing PfACP(leader)-GFP (18). PfiTPT is in the membrane fraction (M) not the soluble protein fraction (S) and has a molecular mass of 43 kDa (B) Anti-PfoTPT (red) colocalizes with apicoplast-targeted GFP (green). PfoTPT is in the membrane fraction (M) not the soluble protein fraction (S) and has a molecular mass of 34 kDa. (C) Anti-hemagglutinin (green) colocalizes with apicoplast-located protein PfACP (red) in parasites transfected with a C-terminal tagged copy of PfiTPT (iTPT-HA). PfiTPT-HA is in the membrane fraction (M) not the soluble protein fraction (S) and has a molecular mass of 49 kDa. (D) Anti-hemagglutinin (green) colocalizes with apicoplast-located protein PfACP (red) in parasites transfected with a C-terminal tagged copy of PfoTPT (oTPT-HA). PfoTPT-HA is in the membrane fraction (M) not the soluble protein fraction (S) and has a molecular mass of 38 kDa. (E) Anti-hemagglutinin (green) colocalizes with apicoplast-located protein PfACP (red) in parasites transfected with an N-terminal tagged copy of PfoTPT (HA-oTPT). HA-PfoTPT is in the membrane fraction (M) not the soluble protein fraction (S) and has a molecular mass of 40 kDa. (F) Fusion protein comprising the N terminus of PfiTPT and yellow fluorescent protein (green) colocalizes with apicoplast-located protein PfACP (red) in parasites transfected PfiTPT(l)-YFP. Mature YFP (27 kDa) and a precursor bearing the N-terminal transit peptide (39 kDa) are detected by Western blotting.
PfiTPT Is Targeted to the Innermost Apicoplast Membrane.
The ability of the bipartite leader sequence of PfiTPT to target the transporter to the apicoplast was explored by fusing the leader to the reporter protein yellow fluorescent protein (YFP). Fusion of the first 203 aa of the PfiTPT to the N terminus of YFP protein resulted in targeting of YFP to the apicoplast, demonstrating that the leader is sufficient for apicoplast targeting (Fig. 2F). Western blotting of reporter gene transfectants indicated that the bipartite leader was cleaved from YFP (Fig. 2F), as observed with other native apicoplast proteins and apicoplast leader/GFP fusions (18–22).
The apparent molecular mass of native PfiTPT in Western blots (Fig. 2A) suggested removal of its N-terminal leader (Fig. 1). We performed a pulse–chase experiment to examine processing of PfiTPT in vivo. Cells were labeled with radioactive methionine for 20 min, then chased for 2 h. Immunoprecipitation with anti PfiTPT antibody yielded several labeled proteins (Fig. 3A). A 43-kDa band exhibited a substantial increase after the 2-h chase, and N-terminal sequencing (see below) confirms that this band represents mature PfiTPT with the bipartite leader removed (Fig. 1). The accumulation of processed PfiTPT should be complemented by a concomitant decrease in a precursor, and indeed a 59-kDa band shows a marked decrease after the chase (Fig. 3A). Based on the gene sequence, the transit peptide-bearing form of PfiTPT has an estimated molecular mass of 59-kDa (Fig. 1), and we conclude that the 59-kDa radiolabeled band is a transit peptide-bearing precursor en route to the apicoplast, similar to that observed previously for the stromal apicoplast protein PfACP (18, 23).
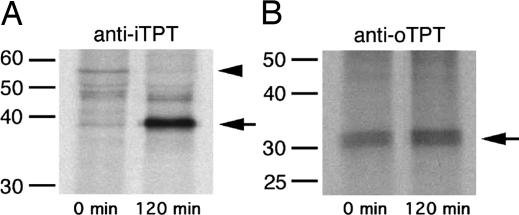
Fig. 3.
Pulse–chase analysis of PfiTPT and PfoTPT processing. (A) A 57-kDa PfiTPT precursor (arrowhead) is depleted during a 2-h chase, whereas a 43-kDa mature form of PfiTPT (arrow) increases in abundance. The 43-kDa immunoprecipitated protein was N-terminally sequenced and lacks the first 140 aa encoded by the PfiTPT gene. (B) The 34-kDa PfoTPT protein does not undergo any size change during a 2-h chase, suggesting no processing.
The N terminus of the mature, 43-kDa form of PfiTPT commenced with the following 10 aa (EHDINKGIID), indicating that the first 140 aa are removed (Fig. 1); this is only the third cleavage site to be determined for an apicoplast-targeted protein in Plasmodium (23, 24). No cleavage site consensus is yet evident. The mature form of PfiTPT has a predicted molecular mass of 43 kDa (Fig. 1), in good agreement with the Western blots (Fig. 2) and pulse–chase immunoprecipitation (Fig. 3). We conclude that the N-terminal 140 aa of PfiTPT act as an apicoplast targeting leader and are removed in the apicoplast stroma by the stromal processing peptidase (SPP) similar to other apicoplast targeted proteins (23). Because PfiTPT is clearly an apicoplast membrane protein, and its N terminus is apparently exposed to SPP, which is localized in the apicoplast stroma (23, 25), we conclude that PfiTPT is most likely located in the innermost apicoplast membrane. Plant homologues of PfiTPT are located in the innermost membrane of plant plastids and targeted there courtesy of a transit peptide that is removed by plant plastid SPP, but little is known about the mechanism (14). The possibility that the mechanism of targeting and insertion of plastid innermost membrane transporters may be conserved between plants and malaria parasites offers intriguing potential to investigate this process.
PfoTPT Is Targeted to the Outermost Apicoplast Membrane.
PfoTPT bears no obvious N-terminal extension (Fig. 1), yet it is clearly located in an apicoplast membrane (Fig. 2B). Moreover, both C- and N-terminal epitope-tagged versions of PfoTPT localize to the apicoplast membranes (Fig. 2 D and E), so the targeting information must reside within PfoTPT itself. The apparent PfoTPT molecular mass of 34 kDa (Fig. 2B) is slightly less than the predicted size for the entire ORF (39 kDa), which might suggest that processing of PfoTPT occurs. However, because our antibody is directed to the N terminus (Fig. 1), N-terminal processing can be ruled out. Similarly, C-terminal processing does not occur in the C-terminally tagged version of PfoTPT, which localizes faithfully to the apicoplast. Because tagged (i.e., unprocessed) versions of PfoTPT also migrate somewhat anomalously in SDS/PAGE (Fig. 2 D and E), we conclude that the extreme hydrophobicity of PfoTPT causes it to appear anomalously small. This conclusion is further supported by pulse–chase analysis of immunoprecipitated PfoTPT (Fig. 3B), which shows no evidence of processing, such as was observed for PfiTPT (Fig. 3A).
Protein structure predictions suggest that PfoTPT is a multipass membrane protein utilizing the first transmembrane domain as an internal, noncleavable signal peptide (Fig. 1), which would result in PfoTPT residing in a membrane derived from the endomembrane system. The outermost membrane of the apicoplast is part of the endomembrane system (18, 26, 27), so we tested the hypothesis that PfoTPT resides in the outermost apicoplast membrane. We liberated intact apicoplasts from cells and incubated them with antibodies to apicoplast proteins without fixation so that the apicoplast would remain intact and only antigens on the apicoplast surface could be labeled. Anti-PfoTPT peptide antibodies clearly decorate intact apicoplasts, and in immunofluorescence assays, the PfoTPT label can be resolved as peripheral when compared to GFP targeted to the apicoplast stroma by the bipartite leader of PfACP (Fig. 4A). Stromal proteins such as GFP, ACP, and the innermost membrane protein PfiTPT were not labeled by antibodies when intact apicoplasts were used, and were only detectable after fixation and permeabilization (data not shown). The fluorescence labeling of intact apicoplasts is consistent with PfoTPT being located in the outermost membrane and suggests that the N terminus (from which the peptide used to generate the antibody was derived) projects into the cytoplasm. Epitope-tagged versions (both C- and N-terminal) of PfoTPT were also detected by anti-hemagglutinin antibody reacted with intact apicoplasts (data not shown), which further indicates that the N terminus projects into the cytoplasm and corroborates the predictions that the C terminus also projects into the cytoplasm based on an even number of transmembrane domains (Fig. 1) (11, 15).
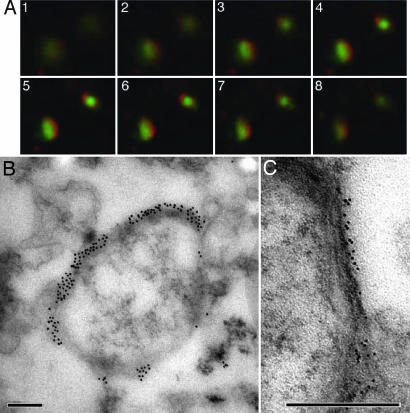
Fig. 4.
Immunolocalization of PfoTPT to the outside of free, intact apicoplasts. (A) Z-stack of immunofluorescent images (1–8) showing localization of anti-PfoTPT (red) around apicoplast-targeted GPF (green) in parasites expressing PfACP(leader)-GFP (18). (B) Electron micrograph of intact apicoplast labeled with anti-PfoTPT antibodies and 20-nm colloidal gold. (C) Tannic acid fixation showing four bounding membranes around isolated apicoplast labeled with anti-PfoTPT antibodies and 10-nm colloidal gold. (Scale bars, 200 nm.)
To confirm that PfoTPT is indeed located in the outermost membrane, we labeled intact apicoplasts with either anti-PfoTPT or anti-hemagglutinin and colloidal gold before fixing and embedding the apicoplasts for electron microscopy. Apicoplasts were heavily decorated with gold (Fig. 4B), and four bounding membranes were clearly visible in these labeled apicoplasts (Fig. 4C), suggesting that all four bounding membranes were intact in these released apicoplasts and confirming that PfoTPT is located in the outermost membrane with the N and C termini facing the cytosol.
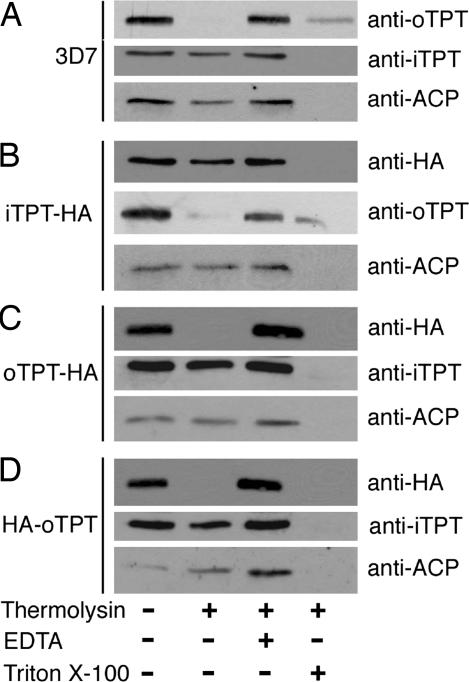
Because PfoTPT in intact apicoplasts is accessible to antibodies, we reasoned that it should also be accessible to proteases. Intact apicoplasts were incubated with thermolysin, then examined by Western blotting (28). Thermolysin treatment of intact apicoplasts digested PfoTPT but left stromal (ACP) and innermost membrane proteins (PfiTPT) intact, further confirming that PfoTPT is located in the outermost membrane (Fig. 5). Inhibition of the protease with EDTA protected PfoTPT (Fig. 5). Lysis of the apicoplasts with Triton X-100 detergent allowed the protease access to the stromal and innermost membrane proteins (Fig. 5). In concert with the immunocytochemical localizations, the proteolytic analyses provide strong evidence for PfoTPT being localized in the outermost apicoplast membrane. The related cattle parasite Theileria parva also has a TPT with a bipartite leader plus two additional TPTs with no leaders (29), suggesting a similar arrangement in the Theileria apicoplast.
Fig. 5.
Proteolysis of free, intact apicoplasts demonstrating exposure of the N and C-termini of PfoTPT on the outside of apicoplasts. (A) In free, intact apicoplasts from wild-type parasites (3D7), thermolysin degrades PfoTPT but not the innermost membrane protein PfiTPT or the apicoplast stromal protein ACP. (B) Thermolysin digestion of free, intact apicoplasts from parasites transfected with a C-terminally tagged copy of PfiTPT (iTPT-HA) shows that PfiTPT is protected whereas PfoTPT is exposed. (C) Thermolysin digestion of free, intact apicoplasts from parasites transfected with a C-terminally tagged copy of PfoTPT (oTPT-HA) shows that the innermost membrane protein PfiTPT and the apicoplast stromal protein ACP are protected, whereas oTPT-HA is exposed on the apicoplast surface. (D) Thermolysin digestion of free, intact apicoplasts from parasites transfected with an N-terminally tagged copy of PfoTPT (HA-oTPT) shows that the innermost membrane protein PfiTPT and the apicoplast stromal protein ACP are protected whereas HA-oTPT is exposed on the apicoplast surface.
Fuelling the Apicoplast.
PfiTPT and PfoTPT have the correct substrate recognition signatures and are appropriately situated to act in tandem to transport phosphorylated sugars across the apicoplast membranes. We predict that PfiTPT and PfoTPT will import, but not export, sugars because there is no apparent internal source of reduced carbon compounds to export (10). We also predict that the apicoplast transporters will import phosphorylated C3, but not C5 or C6, compounds because there are no candidate apicoplast enzymes to handle such sugars; the apicoplast apparently lacks chloroplast type pentose phosphate and glycolytic pathways (10). Thus, PfoTPT could transport trioses across the outermost apicoplast membrane and PfiTPT could mediate transport across the innermost membrane. How these trioses, or any other molecules, cross the two middle membranes is not clear, but we note that no specific transporter seems to be required for trioses to cross the outermost of the two membranes of plant plastids (11, 30, 31), which is believed to be homologous to the apicoplast third membrane counting from the outside (32–34). Two additional TPT-like proteins are also encoded by the Plasmodium falciparum genome (PFL0890c and PFE0785c), and they might also be located to the apicoplast and involved in triose transport.
The PfiTPT and PfoTPT transporters could thus fuel the apicoplast pathways by providing carbon, reducing power, and ATP. For instance, imported PEP would be converted to pyruvate by pyruvate kinase, which we have shown to be apicoplast targeted (L.L., K.A.M., and G.I.M., unpublished data), which can in turn be converted to acetyl CoA by apicoplast pyruvate dehydrogenase (21), producing carbon input, ATP, and NADH to power apicoplast type II fatty acid synthesis (6, 8, 10, 19, 24, 35, 36). Dihydroxyacetonephosphate could also be imported and converted by apicoplast triose phosphate isomerase to glyceraldehyde-3-phosphate to be combined with pyruvate by deoxyxylulose phosphate synthase in the first step of apicoplast-based isoprene subunit synthesis (9, 10). A further role for imported dihydroxyacetonephosphate is to be converted to glycerol-3-phosphate (G3P) by G3P dehydrogenase (GpdA) to generate backbones for apicoplast phospholipid biosynthesis (10). The linking of cytosolic glycolysis and apicoplast anabolism by the PfiTPT and PfoTPT plant-like transporters makes them potentially excellent drug targets.
Transporters and Multimembraned Plastids.
PfoTPT is a clear homologue of plant plastid transporters that evolved in the eukaryotic host genome subsequent to the establishment of the primary plastid endosymbiont (11, 13–15, 31). The transporters integrate host and symbiont metabolisms acting as gatekeepers for the transfer of reduced carbon compounds; sugars come out of the endosymbiont during times of production (photosynthesis), and sugars go in during nonproductive times (i.e., darkness) (11, 31). The ability to “feed” nonphotosynthesizing plastids with cytosolic sugars also permitted plants to develop underground tissues that would never see plastid-sustaining light (31). We propose that a similar ability to feed nonphotosynthetic plastids has been part of the evolution of the apicoplast.
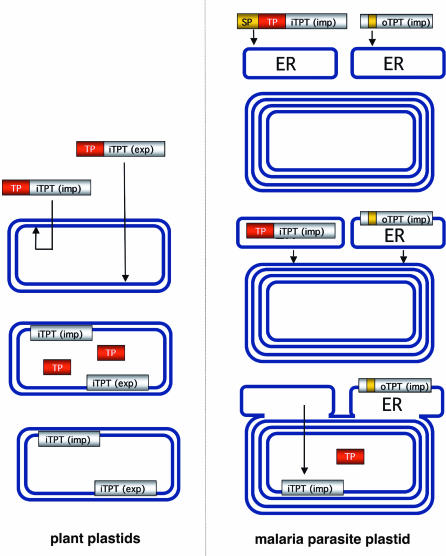
Acquisition of plastids by secondary endosymbiosis results in multiple envelope membranes, typically four, and the problems of transporting substrates across these membranes are inherently more complex. Here, we show that a malaria parasite homologue (PfoTPT) of a plant plastid innermost membrane transporter is located in the outermost membrane of a secondary plastid. PfoTPT apparently inserts into the endomembrane and lodges in the outermost apicoplast membrane. We hypothesize that the first TMD acts as a recessed, noncleavable signal peptide that initiates import into the endomembrane by the Sec machinery (Fig. 6). Little is known about the mechanism of insertion of multispan membrane proteins, but it is known that the outermost apicoplast membrane is part of the endomembrane pathway (18, 26, 27), so PfoTPT could travel to the apicoplast by vesicular transfer (Fig. 6). Mutagenesis of epitope-tagged PfoTPT should allow us to test models of insertion and apicoplast retention.
Fig. 6.
Model for the evolution of plastid transporters. Both importers and exporters are targeted to the innermost membrane of photosynthetic primary plastids (such as those of plants) courtesy of removable transit peptides (TP) (31). During establishment of a secondary plastid, such as that of the malaria parasite, the transporter genes (complete with transit peptide) are relocated from the endosymbiont nucleus to the host nucleus and must acquire a signal peptide (SP) in order for the protein to traffic through the outermost membrane of the secondary plastid and into its innermost membrane (e.g., PfiTPT) (39). However, loss of the transit peptide and no acquisition of an N-terminal signal peptide for a copy of iTPT (exp) results in targeting into the endomembrane (ER) courtesy of an internal signal peptide that also does duty as the first transmembrane domain. ER-derived vesicles then fuse with the outermost apicoplast membrane delivering oTPT to the outermost secondary plastid membrane. Because PfoTPT now enters the membrane in reverse orientation, it has been converted from an exporter to an importer.
In plants, TPTs are triose phosphate exporters/importers that are inserted into the innermost plastid membrane courtesy of cleavable transit peptides (14). The apicoplast innermost membrane protein PfiTPT likely inserts in a similar manner after first gaining entry to the endomembrane lumen courtesy of a signal peptide (Fig. 6). However, PfoTPT uses no transit peptide and instead inserts into the outermost membrane. We hypothesize that relocation of a TPT exporter into the outermost secondary plastid membrane came about through loss of the transit peptide (Fig. 6). This relocation event could also have reversed iTPT(exporter) polarity effectively converting an exporter to an importer (Fig. 6). It will now be interesting to characterize TPT-like transporters in a photosynthetic secondary plastid such as that of the recently sequenced diatom Thallasiosira pseudonana (37), because it should have both importers and exporters in the outermost membrane as well as the innermost membrane.
Materials and Methods
Predicted proteins PFE1510c and PFE0410w, annotated as phosphoenolpyruvate/phosphate transporter and TPT, were retrieved from PlasmoDB (www.plasmodb.org) and compared to GenBank. In this paper, we refer to PFE1510c as PfiTPT (Plasmodium falciparum innermost triose phosphate transporter) and PFE0410w as PfoTPT (Plasmodium falciparum outermost triose phosphate transporter), reflecting their membrane location and presumed substrate specificity.
Hemagglutinin Tagging.
To determine the localization of the multitransmembrane domain transporters, a hemagglutinin (HA) tagging approach was taken (Table 1, which is published as supporting information on the PNAS web site). This triple tag is only 3 kDa compared to the 25-kDa GFP, and therefore would be less likely to disrupt protein folding and correct localization. Destination vectors containing HA-tagged transporters were created by using Invitrogen’s Multisite Gateway technology as described (22). The CRT promoter was used for expression of transporters as it drives lower levels of expression than the HSP86 promoter (22). These plasmids were transfected into P. falciparum (3D7) as described (20). After 20–30 days, drug-resistant parasites selected on 5 nM WR99210 expressing the tagged transporters were detected in cultures.
Production of Antibodies.
PfiTPT (RNQPELFYDEQELKRINS) and PfoTPT (MKDNEKKNEYGTFPIS) peptides were synthesized and conjugated to keyhole limpet hemocyanin and Diphtheria toxin, respectively (Mimotopes). Rabbits were immunized with the conjugates in Freund’s complete adjuvant. Antibodies were affinity-purified from serum by using peptide bound to Sepharose columns as described (38).
Immunoblotting and Immunofluorescence.
Parasite membrane proteins were separated from water-soluble proteins by heat-induced phase partitioning followed by centrifugation through a sucrose cushion (17), separated by SDS/PAGE and transferred to nitrocellulose. Membranes were probed with rat anti-HA (1:1,000; Roche), rabbit anti-PfiTPT (1:100), rabbit anti-PfoTPT (1:100), rabbit anti-ACP (1:500) (18), or mouse anti-YFP (1:1,000; Clontech Living Colors monoclonal) antibodies, followed by goat anti-rat (1:1,000; DakoCytomation), goat anti-rabbit (1:5,000; Pierce), or goat anti-mouse (1:5,000; Pierce) antibodies before detection by chemiluminescence (Pierce). Antibodies were diluted in 5% skim milk powder in Tris-buffered saline containing 0.05% Tween 20. Immunofluorescence assays (IFAs) were performed as described (20) using rat anti-HA (1:200; Roche), rabbit anti-iTPT (1:50), rabbit anti-oTPT (1:25), and rabbit anti-ACP (1:1,000). Proteins were detected by Alexa Fluor 488-conjugated goat anti-rat and Alexa Fluor 546-conjugated goat anti-rabbit antibodies (1:1,000; Molecular Probes).
Pulse–Chase and N-Terminal Sequencing.
P. falciparum 3D7 cultures (5–8% parasitemia) were metabolically labeled and PfiTPT and PfoTPT were immunoprecipitated and detected by fluorography as described (23). Processed PfiTPT (43 kDa) was isolated for N-terminal sequencing at the Australian Proteomic Analysis Facility at Macquarie University (Sydney, New South Wales, Australia) as described (23)
Immunomicroscopy of Free, Intact Apicoplasts.
To demonstrate that PfoTPT is located in the outermost apicoplast membrane, and to determine the orientation of the N and C termini, free organelles were incubated with antibodies before fixation and then analyzed by IFA or EM. Saponin-released P. falciparum parasites expressing either ACP(l)-GFP (18), PfiTPT-HA, PfoTPT-HA, or HA-PfoTPT were washed three times with PBS followed by one wash in hypotonic buffer (1 mM Hepes-NaOH, pH 7.4/2 mM EGTA and protease inhibitors; Roche). Parasites were permeabilized by suspension in hypotonic buffer and lysed by expulsion through a 27-gauge needle (20×). The lysate was made isotonic with the addition of 1/4 volume of 4× assay buffer (200 mM Hepes-NaOH, pH 7.4/200 mM NaCl/1 M sucrose). Unlysed parasites and nuclei were removed by centrifugation (1,500 × g, 10 min, 4°C). Organelles, including the apicoplasts, in the postnuclear supernatant were pelleted by centrifugation (13,000 × g, 10 min, 4°C). The organelle pellet was suspended in 1× assay buffer containing 1% BSA and mixed at 4°C for 30 min. Primary antibodies were added to the organelle preparation (1:200 rat anti-HA, 1:25 rabbit anti-oTPT) and mixed for at least 1 h at 4°C. After one wash with 1× assay buffer containing 1% BSA, organelles were suspended in the above buffer containing secondary antibodies conjugated to fluorophores as above for IFA or colloidal gold conjugates for EM (10 nm goat anti-rat, 10 nm goat anti-rabbit, 1:20; Sigma) and mixed at 4°C for at least 1 h. Organelles were then washed with 1× assay buffer (without 1% BSA) before fixation.
Samples for IFA analysis were fixed in 1× assay buffer containing 4% paraformaldehyde and 0.0075% glutaraldehyde for 30 min on ice. To detect proteins within the apicoplast, organelles were permeabilized with 0.1% Triton X-100 in 1× assay buffer for 10 min on ice before blocking and incubation with primary antibodies (1:1,000 rabbit anti-iTPT or rabbit anti-ACP, 1:200 rat anti-HA) then fluorophore-conjugated secondary antibodies as above. Samples for electron microscopy analysis were fixed in 1× assay buffer containing 1% glutaraldehyde and 1% OsO4. Samples were dehydrated in ethanol and embedded in LR white resin (London Resin), and ultrathin sections were viewed on a Philips BioTwin electron microscope.
Thermolysin Treatment.
Organelle preparations from 3D7 parasites and parasites expressing PfiTPT-HA, PfoTPT-HA, and HA-PfoTPT were generated as described above, except that the hypotonic buffer did not contain EGTA or protease inhibitors and the 4× assay buffer was 200 mM Hepes-NaOH, (pH 7.4), 1.2 M sorbitol, and 2 mM CaCl2. The organelle preparation was split into five tubes. One sample was used to determine protein concentration by Bradford assay. The remaining four samples were treated as follows: (i) no protease (− thermolysin), (ii) 25 μg of thermolysin per mg of protein (+ thermolysin), (iii) 10 mM EDTA and 25 μg of thermolysin per mg of protein (inhibited thermolysin), and (iv) 0.1% Triton X-100 and 25 μg of thermolysin per mg of protein (permeabilized organelles + thermolysin). After 20-min incubation on ice, proteolysis was stopped by addition of EDTA to a final concentration of 10 mM, and proteins were precipitated by using chloroform/methanol/water and analyzed by immunoblotting.
Supplementary Material
Acknowledgments
We thank Kiaran Kirk and Megan Downie for help with oocyte expression and support from the Australian Research Council/National Health and Medical Research Council Research Network for Parasitology. This work was supported by a National Health and Medical Research Council Program Grant. S.A.R. was supported by a Melbourne Research Scholarship. G.I.M. is a Howard Hughes Medical Institute International Research Scholar and an Australian Research Council Professorial Fellow. The Australian Red Cross generously supplied red blood cells.
Abbreviations
- TPT
triosephosphate/phosphate transporter
- YFP
yellow fluorescent protein
- IFA
immunofluorescence assay.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wilson R. J. Biol. Rev. Camb. Philos. Soc. 2005;80:129–153. doi: 10.1017/s1464793104006591. [DOI] [PubMed] [Google Scholar]
- 2.Fichera M. E., Roos D. S. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 3.He C. Y., Shaw M. K., Pletcher C. H., Striepen B., Tilney L. G., Roos D. S. EMBO J. 2001;20:330–339. doi: 10.1093/emboj/20.3.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ralph S. A., D’Ombrain M. C., McFadden G. I. Drug Resistance Updates. 2001;4:145–151. doi: 10.1054/drup.2001.0205. [DOI] [PubMed] [Google Scholar]
- 5.Foth B. J., McFadden G. I. Int. Rev. Cytol. 2003;224:57–110. doi: 10.1016/s0074-7696(05)24003-2. [DOI] [PubMed] [Google Scholar]
- 6.Lu J. Z., Lee P. J., Waters N. C., Prigge S. T. Comb. Chem. High-Throughput Screen. 2005;8:15–26. doi: 10.2174/1386207053328192. [DOI] [PubMed] [Google Scholar]
- 7.Ramya T. N., Surolia N., Surolia A. IUBMB Life. 2005;57:371–373. doi: 10.1080/15216540500091460. [DOI] [PubMed] [Google Scholar]
- 8.Waller R. F., Ralph S. A., Reed M. B., Su V., Douglas J. D., Minnikin D. E., Cowman A. F., Besra G. S., McFadden G. I. Antimicrob. Agents Chemother. 2003;47:297–301. doi: 10.1128/AAC.47.1.297-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jomaa H., Wiesner J., Sanderbrand S., Altincicek B., Weidemeyer C., Hintz M., Turbachova I., Eberl M., Zeidler J., Lichtenthaler H. K., et al. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- 10.Ralph S. A., van Dooren G. G., Waller R. F., Crawford M. J., Fraunholz M., Foth B. F., Tonkin C. J., Roos D. S., McFadden G. I. Nat. Rev. Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- 11.Weber A. P., Schwacke R., Flugge U. I. Annu. Rev. Plant Biol. 2005;56:133–164. doi: 10.1146/annurev.arplant.56.032604.144228. [DOI] [PubMed] [Google Scholar]
- 12.Gardner M. J., Hall N., Fung E., White O., Berriman M., Hyman R. W., Carlton J. M., Pain A., Nelson K. E., Bowman S., et al. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weber A. P., Linka M., Bhattacharya D. Eukaryot. Cell. 2006;5:609–612. doi: 10.1128/EC.5.3.609-612.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knappe S., Flugge U. I., Fischer K. Plant Physiol. 2003;131:1178–1190. doi: 10.1104/pp.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin R. E., Kirk K. Mol. Biol. Evol. 2004;21:1938–1949. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 16.Kirk K. Acta Trop. 2004;89:285–298. doi: 10.1016/j.actatropica.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Bordier C. J. Biol. Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 18.Waller R. F., Cowman A. F., Reed M. B., McFadden G. I. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waller R. F., Keeling P. J., Donald R. G. K., Striepen B., Handman E., Lang-Unnasch N., Cowman A. F., Besra G. S., Roos D. S., McFadden G. I. Proc. Natl. Acad. Sci. USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonkin C. J., Van Dooren G. G., Spurck T. P., Struck N. S., Good R. T., Handman E., Cowman A. F., McFadden G. I. Mol. Biochem. Parasitol. 2004;137:13–21. doi: 10.1016/j.molbiopara.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Foth B. J., Stimmler L. M., Handman E., Hodder A. N., Crabb B. S., McFadden G. I. Mol. Microbiol. 2004;55:39–53. doi: 10.1111/j.1365-2958.2004.04407.x. [DOI] [PubMed] [Google Scholar]
- 22.van Dooren G. G., Marti M., Tonkin C. J., Stimmler L. M., Cowman A. F., McFadden G. I. Mol. Microbiol. 2005;57:405–419. doi: 10.1111/j.1365-2958.2005.04699.x. [DOI] [PubMed] [Google Scholar]
- 23.van Dooren G. G., Su V., D’Ombrain M. C., McFadden G. I. J. Biol. Chem. 2002;277:23612–23619. doi: 10.1074/jbc.M201748200. [DOI] [PubMed] [Google Scholar]
- 24.Surolia N., Surolia A. Nat. Med. 2001;7:167–173. doi: 10.1038/84612. [DOI] [PubMed] [Google Scholar]
- 25.Sato S., Wilson R. J. Curr. Genet. 2002;40:391–398. doi: 10.1007/s00294-002-0273-3. [DOI] [PubMed] [Google Scholar]
- 26.DeRocher A., Gilbert B., Feagin J. E., Parsons M. J. Cell Sci. 2005;118:565–574. doi: 10.1242/jcs.01627. [DOI] [PubMed] [Google Scholar]
- 27.DeRocher A., Hagen C. B., Froehlich J. E., Feagin J. E., Parsons M. J. Cell Sci. 2000;113:3969–3977. doi: 10.1242/jcs.113.22.3969. [DOI] [PubMed] [Google Scholar]
- 28.Cline K., Werner-Washburne M., Andrews J., Keegstra K. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardner M. J., Bishop R., Shah T., de Villiers E. P., Carlton J. M., Hall N., Ren Q., Paulsen I. T., Pain A., Berriman M., et al. Science. 2005;309:134–137. doi: 10.1126/science.1110439. [DOI] [PubMed] [Google Scholar]
- 30.Clausen C., Ilkavets I., Thomson R., Philippar K., Vojta A., Mohlmann T., Neuhaus E., Fulgosi H., Soll J. Planta. 2004;220:30–37. doi: 10.1007/s00425-004-1325-3. [DOI] [PubMed] [Google Scholar]
- 31.Flugge U. I. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:27–45. doi: 10.1146/annurev.arplant.50.1.27. [DOI] [PubMed] [Google Scholar]
- 32.Roos D. S., Crawford M. J., Donald R. G., Kissinger J. C., Klimczak L. J., Striepen B. Curr. Opin. Microbiol. 1999;2:426–432. doi: 10.1016/S1369-5274(99)80075-7. [DOI] [PubMed] [Google Scholar]
- 33.Cavalier-Smith T. Philos. Trans. R. Soc. London B. 2002;358:109–134. doi: 10.1098/rstb.2002.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavalier-Smith T. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- 35.Waters N. C., Kopydlowski K. M., Guszczynski T., Wei L., Sellers P., Ferlan J. T., Lee P. J., Li Z., Woodard C. L., Shallom S., et al. Mol. Biochem. Parasitol. 2002;123:85–94. doi: 10.1016/s0166-6851(02)00140-8. [DOI] [PubMed] [Google Scholar]
- 36.Prigge S. T., He X., Gerena L., Waters N. C., Reynolds K. A. Biochemistry. 2003;42:1160–1169. doi: 10.1021/bi026847k. [DOI] [PubMed] [Google Scholar]
- 37.Armbrust E. V., Berges J. A., Bowler C., Green B. A., Martinez D., Putnam N. H., Zhou S., Allen A. E., Apt K. E., Bechner M., et al. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 38.Goding J. Monoclonal Antibodies: Principles and Practice. London: Academic; 1996. [Google Scholar]
- 39.Nassoury N., Morse D. Biochim. Biophys. Acta. 2005;1743:5–19. doi: 10.1016/j.bbamcr.2004.09.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.