Abstract
Cultivated rice, Oryza sativa L., represents the world’s most important staple food crop, feeding more than half of the human population. Despite this essential role in world agriculture, the history of cultivated rice’s domestication from its wild ancestor, Oryza rufipogon, remains unclear. In this study, DNA sequence variation in three gene regions is examined in a phylogeographic approach to investigate the domestication of cultivated rice. Results indicate that India and Indochina may represent the ancestral center of diversity for O. rufipogon. Additionally, the data suggest that cultivated rice was domesticated at least twice from different O. rufipogon populations and that the products of these two independent domestication events are the two major rice varieties, Oryza sativa indica and Oryza sativa japonica. Based on this geographical analysis, O. sativa indica was domesticated within a region south of the Himalaya mountain range, likely eastern India, Myanmar, and Thailand, whereas O. sativa japonica was domesticated from wild rice in southern China.
The domestication of cultivated plant species from their wild ancestors represents one of the most important events of human history, allowing early human populations access to greater food resources than were previously available (1). The origins of most cultivated species of plants are associated with different geographic centers of domestication (the Fertile Crescent, sub-Saharan Africa, Mesoamerica, the Andes of South America, and east Asia, among others) (1, 2). In each center, at least one grass and one legume species were domesticated, supplying a starch source and a protein source for early agricultural societies (3). For example, in the Fertile Crescent, wheat, barley, peas, and lentils were domesticated, and, in eastern Asia, rice and soybeans were developed (4). In each of the major centers of domestication a different grass species was domesticated. Today the descendents of these early domesticates, wheat, barley, maize, sorghum, and rice, feed the majority of the human population. An inadvertent result of the strong selection and genetic bottlenecks associated with domestication is a concurrent reduction in genetic diversity, as only a subset of wild plants are selectively propagated (5). Past studies of nucleotide diversity have demonstrated that, in general, domesticated grasses contain approximately two-thirds of the genetic diversity found within their wild ancestors (6).
One of the most fundamental and debated questions regarding the evolution of cultivated plants and the domestication of crop species is the geographic origin and the number of times domestication has occurred for a given crop (1). For each of the major grass crops the wild progenitor has been identified through a combination of morphological, biochemical, and genetic studies (6). Coupled with the geographic ranges of ancestral species and phylogenetic data, the origins of these crops are now being uncovered. Wheat and barley appear to have been each domesticated once from their wild ancestors (Triticum dicoccoides and Hordeum spontaneum, respectively) in the Fertile Cresent (7, 8). Similarly, corn arose from a single domestication in Mexico from its wild ancestor, teosinte (Zea mays spp. parviglumis) (9, 10). However, the domestication of sorghum and rice is less clear. Sorghum appears to have been domesticated in northeastern Africa from its progenitor Sorghum bicolor ssp. verticilliflorum, although the hypothesized number of events varies from a single domestication to three (11). Finally, although the domestication of rice from its wild ancestor Oryza rufipogon is documented and occurred somewhere in east Asia, the number of domestications and precise geographical location of domestication are debated.
In the east Asian center, a region of plant domestication dominated by southern and eastern China, some 97 different agricultural plants have been domesticated, including several important crop species such as soybeans, adzuki beans, foxtail millet, Citrus sp., tea, and rice, among others (4). The earliest archaeological evidence for domestication of crops in this region is estimated to ≈7,000–10,000 B.C. and includes early evidence for domesticated melon, cucumber, almond, mango, root crops such as taro, and rice (4). Of all of the crops domesticated in the east Asian center, cultivated Asian rice, Oryza sativa, represents the world’s most important agricultural species, feeding more people since the time of its domestication than any other crop (12).
Study System
The genus Oryza includes 21 wild species and 2 cultivated species, Oryza glaberrima and O. sativa. Whereas O. glaberrima is cultivated in restricted areas of western Africa, O. sativa is cultivated globally. Since the time of its initial domestication, Asian cultivated rice has been moved across the globe with migrating human populations (13); rice cultivation can now be found on all continents except Antarctica and feeds more than half of the world’s population (12). Each year an estimated 408,661 million metric tons of rice is consumed, supplying 20% of the world’s total caloric intake (14).
Previous research has identified O. rufipogon as the wild ancestor of cultivated rice (13). However, because O. rufipogon is a perennial species and cultivated rice is an annual species, it has been proposed that the annually occurring form of O. rufipogon, Oryza nivara, may represent the most recent ancestor of O. sativa (13, 15). Here we treat both O. rufipogon and O. nivara as part of the same ancestral gene pool and do not differentiate between these names when referring to wild rice. Through the process of domestication, cultivated rice has lost many of the traits associated with other wild grass species; in addition to a shift from perennial to annual populations, cultivated rice differs from O. rufipogon in its nonshattering seeds, lack of awns, erect habit, and high grain yield (16).
Despite rice’s central role in world agriculture, the evolutionary history of O. sativa is not well understood. Within O. sativa, extensive morphological, ecological, and physiological variation exists, the result of selection for adaptations to different habitats and growing conditions across the globe. O. sativa includes an estimated 120,000 different, named cultivars (13) ranging from traditional rice varieties preserved by local indigenous farmers to the commercially bred “elite” cultivars developed during the green revolution. The majority of rice cultivars can be placed within two subspecies or races of rice, Oryza sativa japonica and Oryza sativa indica, based on a number of physiological and morphological traits such as drought tolerance, potassium chlorate resistance, phenol reaction, plant height, and leaf color (17, 18). These two subspecies are commonly associated with differences in growth habitat as well, with indica rice usually found in the lowlands of tropical Asia and japonica rice typically found in the upland hills of southern China, southeast Asia, and Indonesia, as well as rice outside of Asia (Africa, North America, and South America) (13). O. sativa japonica can be further differentiated into tropical (javanica) and temperate (japonica) forms, with the temperate form appearing to be a derivative of tropical japonica (19). In addition to the two major races, several other minor rice types have been identified with genetic markers (19–21) and include the upland drought-tolerant Aus cultivars of India and Bangladesh, the deep-water Ashinas varieties of Bangladesh, and the aromatic basmati rice of India as well as many others. Whether all these different rice types have arisen from a single domestication event or are the result of multiple domestications remains unknown.
Rice domestication is thought to have begun ≈9,000 years ago within a broad geographic range spanning eastern India, Indochina, and portions of southern China (13). The wild ancestor of cultivated rice, O. rufipogon, grows across this entire range. Two major hypotheses, suggesting either a single origin of cultivated rice or multiple, geographically independent domestications from its wild ancestor, have been proposed. Both archaeological evidence, based on ancient rice grains (22), and some genetic markers (23) support a single domestication of O. sativa indica from O. rufipogon populations growing in the lowland regions of southern China. In this scenario, O. sativa japonica varieties were later developed in upland growing regions, selected from the indica rice (24, 25). The alternative hypothesis of at least two separate domestication events leading to indica and japonica rice has been suggested by genetic distance studies (15, 20, 21, 26, 27). The genetic distance data reveal that japonica and indica are genetically distinct from each other, raising the possibility that the two major rice types may have arisen from different ancestral gene pools; however, these studies lack the resolution to identify unambiguously the geographic region(s) associated with the domestication of O. sativa. The domestication history of cultivated rice has been complicated by large-scale movement associated with the trading of cultivated rice varieties and movement of people across Asia and the world. However, by examining the spatial variation of wild rice haplotypes we are able to identify the geographic regions from which cultivated rice arose (28). Such an approach has been successfully used to understand the domestication of both plants (29–31) and animals (32–34).
In this study we employ a phylogeographic approach using DNA sequence variation from one chloroplast and two nuclear gene regions to elucidate the evolutionary history of cultivated rice. Our objectives are to (i) examine the DNA sequence variation in cultivated and wild rice, (ii) examine the geographic pattern of cultivated and wild haplotypes, and (iii) determine the number of potential domestication events and regions within south and southeast Asia.
Results and Discussion
DNA Sequence Variation.
Three gene regions, one chloroplast and two nuclear, were chosen to examine the relationship between cultivated and wild rice. The chloroplast region (atpB-rbcL) was chosen as a maternally inherited (35) marker that contains a good representation of genetic variation in cultivated and wild rice. Nuclear gene regions offer a biparental view of genetic change and provide higher resolution of the relationships between rice samples. We chose a region of a nuclear pseudogene, V-ATPase B-subunit (p-VATPase), that is presumed to be under little selective constraint (36). Examining this neutral region allows for historical inference of the differences between the O. sativa and O. rufipogon, where evolutionary forces such as gene flow and genetic drift predominate. The other nuclear gene, S-adenosyl methionine synthetase (SAM), is functional (37) and was chosen as a representative of gene regions that have been influenced by the forces of selection. All three gene regions were variable within and between O. sativa and O. rufipogon samples, although the level of variation differed. The atpB-rbcL chloroplast spacer region in O. rufipogon and O. sativa included 801 aligned base pairs of noncoding DNA (797–801 bp before alignment) of which 50 sites were polymorphic (6.24%). For the nuclear gene regions p-VATPase comprised 1,300 aligned base pairs of noncoding DNA (1,250–1,280 bp before alignment) and included 66 polymorphic sites (5.07%). The second nuclear region, SAM, comprised 1,166 base pairs of coding and noncoding DNA and included 53 polymorphic sites (4.55%), excluding Oryza officinalis.
Tests of selection and neutrality (Tajima’s D and Fu and Li’s D*) were performed to assess the evolutionary processes influencing each gene region. These tests indicate significant values for O. rufipogon for atpB-rbcL (D = −2.3), p-VATPase (D* = −3.02), and SAM (D = −1.94), but not for O. sativa. Significant values can be indicative of purifying selection or may be the result of population expansion in the wild species, a likely scenario given geographical range expansion of Asian plants after the last glaciation (38). However, the genome level measures of these neutrality tests have not been examined, and further work in this area is needed to test this hypothesis. Additionally, in cultivated rice, Tajima’s D values (D = −1.8) for nonsynonymous mutations in the functional gene, SAM, suggest purifying selection against mutations (Table 1).
Table 1.
Tests of neutrality
| Gene | n | Haplotype no. | Tajima’s D value | Fu and Li’s D* value |
|---|---|---|---|---|
| atpB-rbcL | ||||
| O. rufipogon | 677 | 38 | −2.2889 | −2.41145 |
| O. sativa | 211 | 11 | −1.3107 | −1.10128 |
| p-VATPase | ||||
| O. rufipogon | 94 | 32 | −1.75119 | −3.01946 |
| O. sativa | 194 | 12 | 0.66369 | −0.01036 |
| SAM | ||||
| O. rufipogon | 232 | 55 | −1.93608 | −2.20856 |
| Synonymous | −1.86135 | −2.34464 | ||
| Nonsynonymous | −1.0890 | −0.81395 | ||
| O. sativa | 240 | 19 | −0.53775 | −1.41569 |
| Synonymous | 0.76085 | −0.29329 | ||
| Nonsynonymous | −1.79207 | −2.32646 |
Tajima’s D and Fu and Li’s D* measures of neutrality were performed by using the computer program dnasp. Significant values are displayed in bold. Significant values for O. rufipogon suggest either purifying selection or range expansion. Significant values were found only for O. sativa for nonsynonymous mutations in the functional gene SAM. This result suggests purifying selection against mutations, which may cause differences in amino acid composition. Values in bold indicate P < 0.05.
Distribution of Genetic Diversity.
Comparison of levels of diversity between wild and cultivated rice indicate that cultivated rice has a reduced subset of the total genetic variation of O. rufipogon; these results are consistent with other genetic studies on rice diversity (19). O. sativa cultivars, whether elite or landrace, share only a subset of haplotypes with O. rufipogon and are most often associated with tip positions in the gene networks (Figs. 1 and 2). As in many domesticated species, the domestication history of O. sativa appears to involve an intense genetic bottleneck, which is detected in all three gene regions. Using haplotype number as a proxy for diversity, cultivated rice contains 22–28% of the total haplotype diversity and wild rice comprises 82–88% of the total haplotype diversity in these three genes (Table 2). This level of diversity is much lower than the approximately two-thirds level of diversity preserved in wheat and maize when compared with their wild relatives (6). However, these species are outcrossing species, whereas rice is primarily a selfing species (39), potentially creating a stronger genetic bottleneck during the domestication of rice. Moreover, landrace varieties contain greater haplotype diversity than do elite varieties, suggesting that landrace varieties represent an intermediate stage in domestication between wild and elite cultivars (Table 2). The higher variation in landrace varieties may indicate that they hold variation in important agronomic traits and suggest that landrace varieties are an important reservoir of genetic diversity as well as a potential source of beneficial alleles for crop breeding and improvement.
Fig. 1.
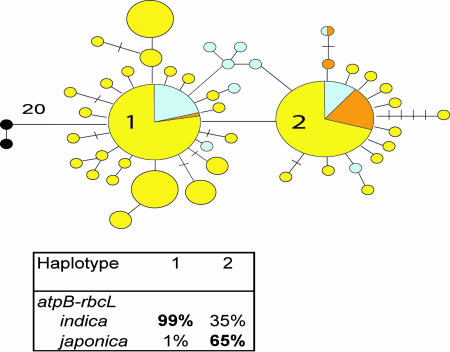
Haplotype network of the chloroplast atpB-rbcL region representing two common cultivated haplotypes. Circle size is proportional to the number of samples within a given haplotype, and dashes between haplotypes represent unobserved, inferred haplotypes. Lines between haplotypes represent mutational steps between alleles. Colors denote rice designation: yellow, O. rufipogon; orange, O. sativa japonica; blue, O. sativa indica; black, O. granulata. Branch length between haplotype 1 and the outgroup O. officinalis is 20 mutational steps. Haplotype composition is shown as a percentage of cultivated sample.
Fig. 2.
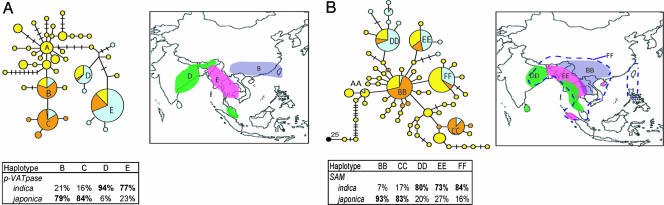
Haplotype network of the nuclear gene regions and geographic ranges of wild rice found within common cultivated haplotypes. Colors denote rice designation: yellow, O. rufipogon; orange, O. sativa japonica; blue, O. sativa indica; black, O. officinalis. Circle size is proportional to number of samples within a given haplotype, and dashes between haplotypes represent unobserved, inferred haplotypes. Lines between haplotypes represent mutational steps between alleles. (A) Haplotype network of the neutral, nuclear p-VATPase region and geographic range of wild rice found within cultivated haplotypes. Three separate geographic ranges represent independent domestication centers for cultivated rice: purple, japonica; green, Aus indica; pink, indica. (B) Haplotype network of the functional, nuclear SAM region and geographic ranges of wild rice found within major cultivated haplotypes. Colors of ranges correspond to those in A except for the addition of a pan-Asia gene pool (FF) for a novel haplotype of indica rice (blue dash). Major domestication regions are still identified despite the different (selection) evolutionary history of the SAM region. Haplotype letter designations describe congruence between both geography and sample identification in both haplotype networks (e.g., B corresponds to BB).
Table 2.
Genetic diversity of cultivated and wild rice
| Gene | Haplotype variation |
|||||
|---|---|---|---|---|---|---|
| Haplotype no. | rbcL-atpB, % | Haplotype no. | p-VATPase, % | Haplotype no. | SAM, % | |
| O. sativa | 10 | 0.23 | 11 | 0.28 | 15 | 0.22 |
| Elite | 7 | 0.16 | 5 | 0.13 | 6 | 0.09 |
| Landrace | 6 | 0.14 | 10 | 0.25 | 15 | 0.22 |
| O. rufipogon | 26 | 0.82 | 33 | 0.83 | 59 | 0.88 |
| Total | 44 | 40 | 67 | |||
Total haplotype number is used as a proxy for genetic diversity in both the cultivated and wild species of rice. O. sativa represents a subset of the total genetic diversity present in O. rufipogon. Landrace varieties of rice contain greater diversity in haplotypes for both nuclear genes, suggesting that landraces may serve as a reservoir of genetic diversity in cultivated rice. Values in bold indicate P < 0.05.
Chloroplast atpB-rbcL Haplotype Network.
The atpB-rbcL gene region is used as a marker for the maternally inherited chloroplast in rice. This gene region includes the noncoding spacer DNA between the atpB and rbcL genes and in this study functions as a maternal marker. Forty-four haplotypes were identified for this gene region in the O. rufipogon and O. sativa samples (Fig. 1). The haplotype network is dominated by two common haplotypes that were detected in the majority of the cultivated and wild samples. Haplotype 1 includes primarily the indica type cultivated rices, whereas haplotype 2 was detected primarily in the japonica type cultivated rices, although a few indica type rices carried this haplotype as well. This network supports previously reported results suggesting japonica rice is less diverse than indica rice and also demonstrates the origin of O. sativa from O. rufipogon (13, 21). However, this gene region lacks the variation and historical resolution necessary to infer separate geographic gene pools for the domestication of O. sativa indica and O. sativa japonica.
Nuclear p-VATPase Haplotype Network.
The nuclear p-VATPase region represents a neutrally evolving pseudogene of the V-ATPase gene. This gene region appears to have evolved into a pseudogene through both DNA transfer from the mitochondria and exon shuffling. Because of these events, the gene region represents a functionally neutral gene region (36). Because this region is not under selection, we expect the evolutionary forces of mutation, drift, and gene flow to be reflected in the haplotype network topography.
Forty unique haplotypes were identified with the p-VATPase gene region. The haplotype network can be split into three clusters, one associated with wild rice and two associated with cultivated rice (Fig. 2A). The first cluster, surrounding haplotype A, represents the center of the unrooted p-VATPase network and includes only samples of O. rufipogon. The wild rice accessions carrying haplotype A were found primarily in India and Thailand. The second cluster centers on haplotypes B and C, which were found primarily in O. sativa japonica cultivars (79% and 84%, respectively). The third cluster includes haplotypes D and E and surrounding haplotypes, which were carried primarily by O. sativa indica cultivars (94% and 77%, respectively). Haplotypes recovered in O. sativa japonica and O. sativa indica occur in different parts of the p-VATPase haplotype network and are separated by a long chain of mutational events (Fig. 2A). The two major haplotype clusters represent not only the two major races of cultivated rice but also two geographically distinct regions, supporting the hypothesis of multiple domestications of cultivated rice.
Our results support the hypothesis that japonica rice was domesticated from O. rufipogon in south China. Haplotype B occurs in O. rufipogon from south China and Taiwan and is not found in any other region (Fig. 2A). Haplotype B occurs predominantly in tropical japonica cultivars often referred to as javanicas. These cultivated varieties are found in southeast Asia and also include much of the rice grown in the United States, Latin America, and Africa. Haplotype C occurs in the temperate form of japonica rice, which is commonly grown in the temperate/subtropical climates of China and upland regions of southeast Asia (13, 16). With only a single sample of wild rice possessing haplotype C, the geographical origin of haplotype C is unclear. However, temperate japonica rice has been shown previously to be a subset of total japonica diversity and likely shares a common domestication event with the javanica rice sampled in haplotype B (19).
This haplotype network also supports the hypothesis of indica rice being domesticated in a separate region from japonica rice, south and southwest of the Himalayan mountain range. Haplotypes D and E are found in indica rice, and the wild rice that contains these two haplotypes is found within India and Indochina. Haplotype D occurs in a group of indica rice known as Aus cultivars grown primarily in Bangladesh and India under short-season and upland conditions (13). Wild rice populations that contain haplotype D are found primarily in India and Malaysia, suggesting that Aus cultivars were domesticated in this region. Considering that Aus cultivars are cultivated exclusively in India during the short growing season, they were likely domesticated from the Indian samples within haplotype D and not wild rice sampled from Malaysia. Haplotype E occurs primarily in samples of O. sativa indica and also in wild rice found within southeast Asia, primarily Thailand and Myanmar (Fig. 2A). The close association of haplotypes D and E in the network and the long mutational separation from haplotype B support the hypothesis that O. sativa indica rice was domesticated in a separate area (Thailand, Myanmar, and India) from that of japonica rice (south China). If Aus rice is considered distinct from other indica rice, three possible geographic regions are supported as domestication centers for cultivated rice.
Finally, a contingency test was used to test for a random association between major haplotype class (B, D, and E) and major geographic region (India, Indochina, and China) (see supporting information, which is published on the PNAS web site). The geographic distribution of wild rice variation supports the hypothesis of separate and distinct geographic gene pools as centers for indica and japonica domestication (P < 0.01).
Nuclear SAM Haplotype Network.
The second nuclear gene, SAM, is part of an essential plant biochemical pathway and is involved in the biosynthesis of polyamines and the phytohormone ethylene (37). We expect that this gene could be under selection, which could distort the topography of the haplotype network from that of the neutral gene network, yet it also carries the signature of multiple gene pools associated with separate domestications. The haplotype network of the SAM region contains six major clades, AA–FF (Fig. 2B). The presence of an outgroup in this haplotype network, O. officinalis, allows us to root the network and infer ancestral haplotypes. Haplotype AA is restricted to wild rice found in India and southeast Asia, inferred as one of the oldest haplotypes in the network, and is consistent with the interior haplotype A in the p-VATPase data set, also inferred as one of the oldest haplotypes in the network based on coalescent theory (40). These results suggest that India and Indochina may represent the ancestral geographic center of O. rufipogon (Fig. 2).
Among the cultivated subspecies, haplotypes BB and CC occur primarily in O. sativa japonica rice (93% and 83%, respectively). O. sativa indica rice possess three distinct haplotypes, DD, EE, and FF (80%, 73%, and 84%, respectively), each of which also occurs in wild rice. Whereas the indica lineage arises once in the p-VATPase data set, it is associated with three separate regions in the SAM network (Fig. 2B). This difference in network topography is likely due to the differences in evolutionary history between our neutral and functional gene, although this pattern could represent lineage sorting or multiple origins of the O. sativa indica lineage.
O. rufipogon accessions that carried haplotype BB (recovered primarily in O. sativa japonica) originated in China, Indochina, and India (Fig. 2B). Whereas japonica-like wild rice was restricted to south China in the p-VATPase network, the SAM data set does not convey geographic specificity for ancestral japonica rice. However, haplotype BB contains the highest percentage of samples originating from China, compared with other haplotypes, and is consistent with the hypothesis of south China as the domestication region of japonica rice. Haplotype CC was not recovered in any accessions of O. rufipogon, excluding the possible inference of a separate domestication for this haplotype.
SAM haplotypes DD and EE have distributions largely similar to haplotypes D and E in the p-VATPase data set. The Aus rice SAM haplotype, DD, occurs in wild rice found in India, Sri Lanka, Burma, and Thailand. The wild rice accessions carrying haplotype EE (recovered primarily in the indica rice accessions) are found in Thailand, Myanmar, Bangladesh, and Indonesia. Finally, haplotype FF, common in O. sativa indica accessions, is primarily concentrated in Indochina but occurs in some samples of wild rice throughout from the range of O. rufipogon (Fig. 2B). The occurrence of haplotype FF in wild rice samples from a broad geographic range suggests that, during domestication, selection on the SAM loci may have preserved an altered signature from that of the p-VATPase region.
Conclusions
Phylogeographic analysis of three distinct gene regions suggests that cultivated rice, O. sativa, was domesticated from its wild progenitor, O. rufipogon, at least twice in at least two different geographic regions in eastern Asia. Although at first these three gene regions appear to yield some inferences that potentially conflict, they in fact provide a consistent signal of domestication. The genome regions are under different evolutionary histories (maternal, neutral, and functional), creating the different network topographies. In addition, differences in the pattern between major haplotype clusters (japonica vs. indica) are expected because of differences in evolutionary history; genes that affect important phenotypes are affected by human-mediated and natural selection. The variation in the chloroplast gene region allows for an examination of the genetic bottleneck experienced by the cultivated rice gene pool during domestication. The paucity of haplotypes in cultivated rice as well as the nesting of these haplotypes within wild rice haplotypes support O. sativa domestication from O. rufipogon, and this signature can be observed in all three haplotype networks. Additionally, it is clear that the major cultivated haplotypes identified in each haplotype network are shared with different wild rice gene pools. The two nuclear gene regions have a more resolved network structure than the chloroplast network, allowing for inferences into the domestication history of cultivated rice. The variation and structure of the haplotype network for the pseudogene region show two major cultivated haplotype clusters separated by a long chain of mutations. The p-VATPase haplotype network suggests that the two major subspecies of rice were domesticated from geographically different wild rice gene pools. The variation and structure of the SAM network demonstrate a closer link between domestication regions and indica/japonica differentiation. It is likely that the evolutionary dynamics affecting functional genes have played a role in suppressing the domestication signal in the SAM data, whereas the p-VATPase region, under more neutral dynamics, provides high resolution of historical events.
The combined genetic and geographic data provide strong evidence for multiple domestications of cultivated rice. O. sativa japonica and O. sativa indica rice each appear to have arisen separately from ancestral wild rice gene pools: japonica from southern China and indica from India/Indochina. An additional domestication event may have occurred for Aus rice in India. Further studies that examine the finer-scale diversification of the minor races of rice (Aus, Ashinas, and aromatics) are needed to resolve the evolutionary history of these rice types as well as to identify the important genetic resource they may represent for the future of crop improvement.
Materials and Methods
Sample Collections.
Rice seeds were obtained from the International Rice Research Institute (Manila, Philippines; U.S. Department of Agriculture Permit 58359) and from the U.S. Small Grain Center (National Plant Germplasm System, United States Department of Agriculture, Agricultural Research Service, Aberdeen, ID) and included 203 cultivars of O. sativa and 129 populations of O. rufipogon, which span the entire geographic range of O. rufipogon. Thirty-two additional populations of O. rufipogon (leaf tissue) and two outgroup species, Oryza granulata and O. officinalis, were included. All collection information can be found in the supporting information.
The accessions of cultivated rice included 114 samples designated as landrace varieties by the International Rice Research Institute. Landraces are cultivated rice varieties that are not in widespread commercial production but are grown and preserved by traditional farmers across south and east Asia and potentially represent a step in domestication between wild rice and elite rice cultivation (41). The remaining accessions of cultivated rice were chosen to represent the major subspecies of rice as well as to cover the geographic and environmental range of rice cultivation. Identification of indica and japonica designation for cultivated rice samples was assigned based on published material obtained from the International Rice Research Institute as well as phenotypical seed hull reaction to phenol staining (42, 43).
DNA Extraction and DNA Sequence Analysis.
Dried leaf material was frozen with liquid nitrogen and crushed by using a mortar and pestle. DNA was extracted by using a modified cetyltrimethyl-ammonium bromide extraction method (44).
Multiple individuals (1–5) of each accession were sequenced for the chloroplast atpB-rbcL gene region and for the nuclear SAM gene region. The total number of sequences for the atpB-rbcL region included 677 O. rufipogon, 211 O. sativa, and 2 O. officinalis. The total number of sequences for the SAM region included 230 O. rufipogon, 240 O. sativa, and 2 O. officinalis. A single individual per cultivar/population was sequenced for the pseudogene p-VATPase B-subunit. The total number of sequences for the p-VATPase region included 94 O. rufipogon and 194 O. sativa. Samples of O. officinalis and O. granulata are not included because of lack of the p-VATPase gene region amplicon in these species.
DNA amplification was performed in PTC-1000 (MJ Research, Cambridge, MA) and ABI 9700 (Applied Biosystems) thermocyclers. Cycling conditions were 94°C for 5 min followed by 35 cycles of 94°C (1 min), 55°C (35 s), 72°C (1 min), and concluding extension at 72°C (5 min). Primers used for amplification of the DNA regions are as follows: atpB-rbcL, 5′-AGTGACATCCAGCACGGGTC-3′ and 5′-CCTTAACACCAGCTTTAAATCC; p-VATPase, 5′-ATTCGGTGACTTTCGCGG-3′ and 5′-CAATTCAGAAATCATGGTTTGC; SAM, 5′-AGAAGATGGCCGCACTTGAT-3′ and 5′-GAAAGGGAGCTTTTAGGCAGA-3′. Additional internal primers were used for sequencing for both p-VATPase and SAM gene regions and are as follows: p-VATPase, 5′-GGCAATACAATATATCATGC-3′ and 5′-TAGCAGCATCAATGAAGAC-3′; SAM, 5′-TCAGCCATGTCCTTGCTAC-3′ and 5′-TCATTGCGGTACTCAACAG-3′. PCRs were cleaned for direct sequencing by using the Viogene Gel Extraction kit (Viogene, Illkirch, France). Samples were then sequenced in both directions by using the dideoxy chain termination method using fluorescent labeling with Big Dye Terminator (version 1.1). Sequences were analyzed with a Base Station DNA Fragment Analyzer (MJ Research) withcartographer software and an ABI 3130 Capillary Sequencer (Applied Biosystems). All sequence information has been deposited in the GenBank database (accession nos. AM177181–AM177311 and AM179944–AM179987).
Tests of neutrality using Tajima’s D and Fu and Li’s D* statistics were conducted by using the computer program dnasp 4.0 (45). Phylogeographic analysis was used to test for a spatial pattern associated with DNA sequence variation. A contingency test (χ2) was used to test for a random association between major allele classes (haplotypes) and major geographic regions of south and southeast Asia. Minimum spanning (haplotype) networks, representing unique DNA sequences or alleles separated by mutational steps, were constructed by using the computer programs minspnet and tcs (46) using genetic distance and statistical parsimony methods.
Supplementary Material
Acknowledgments
We thank the members of the B.A.S. laboratory, Dr. Allison Miller, and the Washington University greenhouse staff. We also thank the International Rice Research Institute and the U.S. National Plant Germplasm System for providing cultivated and wild rice samples, and the McKnight Foundation and the U.S. Agency for International Development for partial financial support of this project.
Footnotes
References
- 1.Diamond J. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 2.Vavilov N. I. Origin and Geography of Cultivated Plants. Cambridge, U.K.: Cambridge Univ. Press; 1987. pp. 22–136. [Google Scholar]
- 3.Simpson B. B., Conner-Ogorzaly M. Economic Botany: Plants in our World. New York: McGraw-Hill; 1986. pp. 182–183. [Google Scholar]
- 4.MacNeish R. S. The Origins of Agriculture and Settled Life. Norman: Univ. of Oklahoma Press; 1992. pp. 150–156. [Google Scholar]
- 5.Harlan J. Crops and Man. Madison, WI: Am. Soc. of Agronomy, Crop Science Soc. of America; 1992. pp. 117–130. [Google Scholar]
- 6.Buckler E. D., IV, Thornsberry J. M., Kresovich S. Genet. Res. 2001;77:213–218. doi: 10.1017/s0016672301005158. [DOI] [PubMed] [Google Scholar]
- 7.Ozkan H., Brandolini A., Schafer-Pregl R., Salamini F. Mol. Biol. Evol. 2002;19:1797–1801. doi: 10.1093/oxfordjournals.molbev.a004002. [DOI] [PubMed] [Google Scholar]
- 8.Badr A., Muller K., Schafer-Pregl R., El Rabey H., Effgen S., Ibrahim H. H., Pozzi C., Rohde W., Salamini F. Mol. Biol. Evol. 2000;17:499–500. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- 9.Doebley J. Annu. Rev. Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 10.Matsuoko Y., Vigouroux Y., Goodman M. M., Sanchez J., Buckler E., Doebley J. Proc. Natl. Acad. Sci. USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahlberg J. Afr. Crop Sci. J. 1995;3:143–151. [Google Scholar]
- 12.Sasaki T. Photosynth. Res. 2001;70:119–127. doi: 10.1023/A:1013844110497. [DOI] [PubMed] [Google Scholar]
- 13.Khush G. Plant Mol. Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- 14.Clay J. W. World Agriculture and the Environment: A Commodity-By-Commodity Guide to Impacts and Practices. Washington, DC: Island; 2004. pp. 388–390. [Google Scholar]
- 15.Yamanaka S., Nakamura I., Nakai H., Sato Y. Genet. Resour. Crop Evol. 2003;50:529–538. [Google Scholar]
- 16.Hoshikawa K. The Growing Rice Plant: An Anatomical Monograph. Nobunkyo, Tokyo: 1989. pp. 3–7. [Google Scholar]
- 17.Oka H. I. Ind. J. Genet. Plant Breed. 1958;18:79–89. [Google Scholar]
- 18.Oka H. I. Origin of Cultivated Rice. Tokyo: JSSP/Elsevier; 1988. [Google Scholar]
- 19.Garris A. J., Tai T. H., Coburn J., Kresovich S., McCouch S. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bautista N. S., Solis R., Kamijima O., Ishii T. Genes Genet. Syst. 2001;76:71–79. doi: 10.1266/ggs.76.71. [DOI] [PubMed] [Google Scholar]
- 21.Second G. Jpn. J. Genet. 1982;57:25–57. [Google Scholar]
- 22.Chen B. Agric. Archaeol. 1999;1:55–57. [Google Scholar]
- 23.Joshi S. P., Gupta V. S., Aggarwal R. K., Ranjekar P. K., Brar D. S. Theor. Appl. Genet. 2000;100:1311–1320. [Google Scholar]
- 24.Ting Y. China Acta Agron. Sinica. 1957;8:243–260. [Google Scholar]
- 25.Oka H. I., Morishima H. Euphytica. 1982;31:41–50. [Google Scholar]
- 26.Cheng C., Motohashi R., Tsuchimoto S., Fukuta Y., Ohtsubo H., Ohtsubo E. Mol. Biol. Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- 27.Vitte C., Ishii T., Lamy F., Brar D., Panaud O. Mol. Genet. Genom. 2004;272:504–511. doi: 10.1007/s00438-004-1069-6. [DOI] [PubMed] [Google Scholar]
- 28.Schaal B. A., Olsen K. Proc. Natl. Acad. Sci. USA. 2000;97:7024–7029. doi: 10.1073/pnas.97.13.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen K., Schaal B. A. Am. J. Bot. 2001;88:131–142. [PubMed] [Google Scholar]
- 30.Miller A., Schaal B. A. Proc. Natl. Acad. Sci. USA. 2005;102:12801–12806. doi: 10.1073/pnas.0505447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chacon M. I., Pickersgill S. B., Debouck D. G. Theor. Appl. Genet. 2005;110:432–444. doi: 10.1007/s00122-004-1842-2. [DOI] [PubMed] [Google Scholar]
- 32.Larson G., Dobney K., Albarella U., Fang M., Matisoo-Smith E., Robins J., Lowden S., Finlayson H., Brand T., Willerslev E., et al. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 33.Bradley D. G., MacHugh D. E., Cunningham P., Loftus R. T. Proc. Natl. Acad. Sci. USA. 1996;93:5131–5135. doi: 10.1073/pnas.93.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joshi M. B., Rout P. K., Mandal A. K., Tyler-Smith C., Singh L., Thangaraj K. Mol. Biol. Evol. 2004;21:454–462. doi: 10.1093/molbev/msh038. [DOI] [PubMed] [Google Scholar]
- 35.Ennos R. A., Sinclair W. T., Hu X. S., Langdon A. In: Molecular Systematics and Plant Evolution. Hollingsworth P. M., Bateman R. M., Gornall R. J., editors. London and New York: Taylor and Francis; 1999. pp. 1–19. [Google Scholar]
- 36.Kubo N., Takano M., Nishiguchi M., Kadowaki K. Gene. 2001;271:193–201. doi: 10.1016/s0378-1119(01)00537-6. [DOI] [PubMed] [Google Scholar]
- 37.Walden R., Cordeiro A., Tiburcio A. Plant Physiol. 1997;113:1009–1013. doi: 10.1104/pp.113.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu G., Chen X., Ni J., Cheddadi R., Guiot J., Han H., Harrison S. P., Huang C., Ke M., Zang Z., et al. J. Biogeogr. 2000;27:635–664. [Google Scholar]
- 39.Beachell H. M., Adair C. R., Jodon N. E., Davis L. L., Jones J. J. Am. Soc. Agron. 1938;30:743–753. [Google Scholar]
- 40.Castelloe J., Templeton A. Mol. Phylogenet. Evol. 1994;3:102–113. doi: 10.1006/mpev.1994.1013. [DOI] [PubMed] [Google Scholar]
- 41.Zeven A. C. Euphytica. 1998;104:127–139. [Google Scholar]
- 42.Morishima H., Oka H. I. Jpn. J. Breed. 1981;31:402–413. [Google Scholar]
- 43.Oka H. I. Jpn. J. Breed. 1953;3:33–43. [Google Scholar]
- 44.Doyle J. J., Doyle J. L. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 45.Rozas J., Sánchez-DelBarrio J. C., Messeguer X., Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 46.Clement M., Posada D., Crandall K. Mol. Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.